Abstract
NG-monomethyl-l-arginine (L-NMMA) has been reported to be elevated in uremic patients. Based on the hypothesis that the pathogenesis of the anemia of renal disease might be due to the perturbation of transcription factors of the erythropoietin (Epo) gene by L-NMMA, the present study was designed to investigate the effect of L-NMMA on Epo gene expression through the GATA transcription factor. L-NMMA caused decreased levels of NO, cyclic guanosine monophosphate (cGMP), and Epo protein in Hep3B cells. L-NAME (analogue of L-NMMA) also inhibited Epo production in anemic mice. Transfection of the Epo promoter-luciferase gene into Hep3B cells revealed that L-NMMA inhibited the Epo promoter activity. However, L-NMMA did not inhibit the Epo promoter activity when mutated Epo promoter (GATA to TATA) was transfected, and L-NMMA did not affect the enhancer activity. Electrophoretic mobility shift assays demonstrated the stimulation of GATA binding activity by L-NMMA. However, L-NMMA had no effect on the binding activity of hepatic nuclear factor-4, COUP-TF1, hypoxia-inducing factor-1, or NF-κB. Furthermore, cGMP inhibited the L-NMMA–induced GATA binding activity. L-NMMA also increased GATA-2 messenger RNA expression. These results demonstrate that L-NMMA suppresses Epo gene expression by up-regulation of the GATA transcription factor and support the hypothesis that L-NMMA is one of the candidate substances that underlie the pathogenesis of renal anemia.
Introduction
In humans and mammals, erythropoiesis is regulated by the 30.4-kd glycoprotein hormone erythropoietin (Epo).1Epo gene expression is regulated by hypoxia through an oxygen sensor.1 The major sites of Epo production are the liver in the fetus2 and the kidney in the adult.3 Peritubular capillary interstitial cells are thought to be the major site of production of Epo in the kidney.4 The cause of the anemia of renal disease is believed to be damage to this site of the Epo production by renal failure.5 In this regard, however, it is interesting to note that some patients with the anemia of renal disease still have the ability to produce Epo in response to acute blood loss and hypoxia.6 On the other hand, other patients with renal failure do not have anemia.7 These observations suggest that chronic perturbation of oxygen sensing or signal transduction or both underlie the pathogenesis of the anemia of renal disease rather than damage at the site of Epo production. Recently, NG-monomethyl-l-arginine (L-NMMA) was reported to be undetectable in nonuremic subjects, whereas the concentration of L-NMMA was markedly elevated in uremic patients.8 Based on this observation, we hypothesized that this substance may be a candidate uremic toxin responsible for renal anemia. However, the precise function of L-NMMA in mediating expression of theEpo gene remains to be elucidated. Because L-NMMA functions as an inhibitor of nitric oxide synthase (NOS),9 it is expected to suppress the production of nitric oxide (NO) and cyclic guanosine 3′, 5′-monophosphate (cGMP). We have found that GATA transcription factors bind to a GATA site in the Epo gene promoter and negatively regulate the gene expression in Hep3B cells (Figure 1A).10 In this study, we demonstrate that L-NMMA suppresses Epo gene expression by up-regulation of the GATA transcription factor.
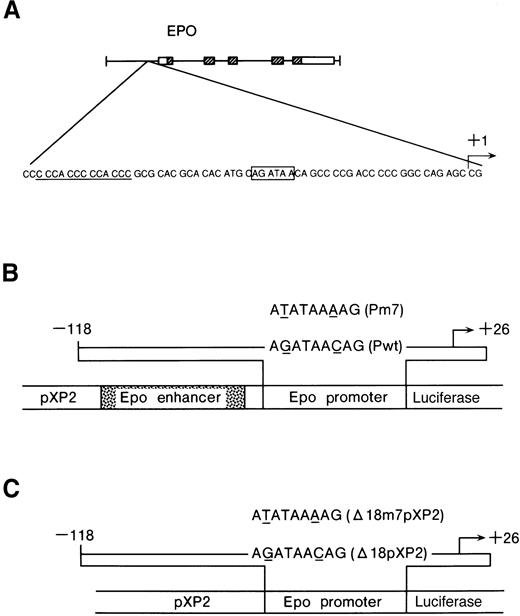
Constructs used in this study.
(A) The GATA-binding site in the Epo 5′ promoter region (boxed) and repeated CACCC elements (underlined). (B) Diagrams of the reporter construct, the wild-type (Pwt) and the mutated GATA (Pm7) used in this study. The pEPLuc reporter construct is shown in the center. Shown above is the 144-bp insert from the Epo promoter. The mutation is indicated by underlining. The Epo enhancer contains an HIF-1 binding site. (C) Diagrams of the reporter constructs, Δ18pXP2, Δ18m7pXP2, used in this study. The mutation is indicated by underlining.
Constructs used in this study.
(A) The GATA-binding site in the Epo 5′ promoter region (boxed) and repeated CACCC elements (underlined). (B) Diagrams of the reporter construct, the wild-type (Pwt) and the mutated GATA (Pm7) used in this study. The pEPLuc reporter construct is shown in the center. Shown above is the 144-bp insert from the Epo promoter. The mutation is indicated by underlining. The Epo enhancer contains an HIF-1 binding site. (C) Diagrams of the reporter constructs, Δ18pXP2, Δ18m7pXP2, used in this study. The mutation is indicated by underlining.
Materials and methods
Cell culture and RNA preparation
The Hep3B cell line was obtained from the American Type Tissue Culture Collection (Rockville, MD). This cells were cultured in Dulbecco modified Eagle medium (Life Technologies, Gaithersburg, MD), supplemented with penicillin (100 U/mL), streptomycin (100 μg/mL), and 10% heat-inactivated fetal bovine serum (FBS; Hyclone, Logan, UT) in 10-cm dishes. Cells were maintained in a humidified 5% CO2/95% air incubator at 37°C. The cells were grown under conditions of hypoxia (1% oxygen) or normoxia as previously described.11 These cells were stimulated where appropriate by the addition of L-NMMA to the culture medium. After incubation under hypoxic/normoxic or L-NMMA–stimulated/unstimulated conditions or both, the cells were harvested and cellular extracts were prepared. Total cellular RNA was also prepared by conventional methods.12
Transfection
Electroporation of the plasmids into Hep3B cells was performed as previously described.10 A total of 3 to 10 × 106 cells were resuspended in 1 mL of 20 mmol/L HEPES buffer (pH 7.05) with 137 mmol/L NaCl, 5 mmol/L KCl, 0.7 mmol/L Na2HPO4, 6 mmol/L dextrose, 20 μg of vector DNA (circular DNA), and 500 μg carrier salmon sperm DNA, and then electroporated at a voltage of 250 V and a capacitance of 960 μF (Bio-Rad, Hercules, CA). The RSVCAT (Chloramphenicolacetyltransferase; 10 μg) plasmid or β-galactosidase was co-transfected as an internal standard for all transfection reactions.10 The time constant of the shock was approximately 12 to 14 ms.
DNA binding assay
Nuclear extracts were prepared as previously described.13 Protein concentrations were determined by assay (Bio-Rad) using bovine serum albumin (BSA) as a standard. Sense-strand oligonucleotide (wild-type: CATGCAGATAA CAGCCCCGAC) was end-labeled with T4 polynucleotide kinase (Toyobo, Tokyo, Japan) and annealed to a 4-fold excess of the unlabeled antisense oligonucleotide. Two nanograms of labeled probe was used in each binding reaction. The binding buffer consisted of 10 mmol/L Tris HCl (pH 7.5), 1 mmol/L EDTA, 4% Ficoll, 1 mmol/L dithiothreitol, and 75 mmol/L KCl. An equimolar mixture of poly[d(I-C)] and poly[d(A-T)] (25 ng; Sigma, St Louis, MO) was used as a nonspecific competitor. The reaction mixtures (25 μL) were incubated for 15 minutes at 4°C and then electrophoresed on 5% nondenaturing polyacrylamide gels in 0.25 × TBE buffer (22 mmol/L Tris borate, 22 mmol/L boric acid, 0.5 mmol/L EDTA) at room temperature at 150 V for 1.5 hours as previously described.13 Gels were vacuum dried and then autoradiography was performed using intensifying screens at −80°C for 24 hours. Monoclonal antibodies to hGATA-1, -2, and -3 were prepared as previously described.14
Plasmid vectors
We used the reporter plasmid pEPLuc described by Blanchard and coworkers15 as a basic plasmid construct, in which both the 126-bp 3′ Epo enhancer (120 to 245-bp 3′ of the poly(A) addition site) and the 144-bp minimal Epo promoter (from −118 to +26 relative to the transcription initiation site) were placed upstream of the firefly luciferase (Luc) gene in pXP2,16 resulting in Pwt17 or V2-Ewt-Pwt-pXP217 (Figure 1B). This enhancer contained hypoxia-inducible factor 1 (HIF-1) binding site and steroid receptor response element (SRRE). In the mutant construct, the GATA sequence in the Epo promoter was mutated to TATA (AGATAACAG to ATATAAAAG). This mutant construct is called Pm717 or V3-Ewt-Pm7-pXP217 (Figure 1B). The 144-bp minimal Epo promoter (from −118 to +26 relative to the transcription initiation site) was placed upstream of theLuc gene in pXP2, resulting in Δ18pXP2 (Figure 1C). In the mutant construct, the GATA sequence in the Epo promoter was mutated to TATA (AGATAACAG to ATATAAAAG). This mutant construct is called Δ18m7pXP2 (Figure 1C). Construction of the hGATA-2 expression plasmid has been previously described.14
Northern blot analysis
Probes were labeled with [α-32P]deoxycytidine triphosphate by random priming and used in RNA blot hybridization.18 Formaldehyde gels for RNA electrophoresis were prepared as described.18 RNA blot hybridization was performed using 25 μg of total RNA from Hep3B cells. The filter was hybridized to probe of hGATA-2 complementary DNA (cDNA). The same filter was stripped and rehybridized to probe consisting of the Epo cDNA, and a probe for ribosomal RNA to determine the level of RNA in each lane. Autoradiography was performed at −80°C and quantitated by densitometric scanning.
Anemic mice
The BDF1 mice were injected intraperitoneally with 0.2 mL of 10 mg NG-nitro-l-arginine methyl ester (L-NAME)/mL phosphate-buffered saline (PBS) or 0.2 mL of PBS. Blood samples (0.3 mL) were obtained from the orbital vein at 0, 12, and 24 hours after injection of L-NAME. Epo levels in the serum were determined by radioimmunosorbent assay (RIA).
Other assays
Transfected Hep3B cells were washed with PBS and lysed in 10-cm dishes with 800 μL of cell lysis buffer (PicaGene, Toyo Ink, Tokyo). Luc activity in 20 μL of the cell extract was determined by Autolumat luminometer (Berthorude, Tokyo, Japan) for 10 seconds. Each measurement of relative light units was corrected by subtraction of the background and standardized to the RSVCAT or β-galactosidase internal transfection control activity. Hypoxic inducibility was defined as the ratio of the corrected relative light units of the hypoxic (1% O2) dish to those of the normoxic (21% O2) dish. CAT activity was determined as described by Neumann and colleagues.19 NO was detected by the 2,3-diaminonaphthalene method,20 cGMP was measured by enzyme immunosorbent assay (EIA),21 and α-fetoprotein (AFP) was measured by RIA.
Results
Inhibition of Epo protein by L-NMMA
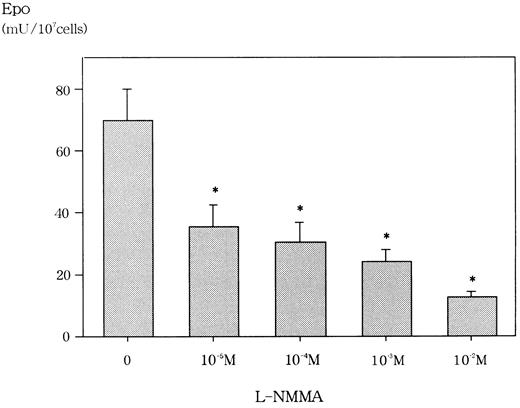
We first confirmed that L-NMMA was not cytotoxic at concentrations of up to 10−2 mol/L for Hep3B cells by the trypan blue dye exclusion method and the methyl-thiazol-diphenyl-tetrazolium (MTT) method (data not shown). Similarly, Fisher and coworkers reported that L-NMMA concentrations of up to 10−3 mol/L were not cytotoxic for Hep3B cells.22 We then examined the effect of L-NMMA on the production of Epo protein in Hep3B cells. Incubation for 24 hours with 10−2 mol/L L-NMMA under hypoxic conditions showed an 80% inhibition of Epo, whereas 10−5 mol/L,10−4 mol/L, and 10−3mol/L L-NMMA each showed a 60% inhibition of Epo (Figure2). To make sure that this inhibition of Epo protein by L-NMMA was specific, AFP was measured by RIA. Up to 10−2 mol/L L-NMMA did not inhibit the level of AFP (data not shown). These results suggest that L-NMMA specifically inhibited the production of Epo protein in Hep3B cells.
Effect of L-NMMA on Epo protein from Hep3B cells stimulated by hypoxia.
Hep3B cells were incubated with different concentrations of L-NMMA under hypoxic conditions (1% O2) for 24 h. Epo protein was measured by RIA. Four separate experiments were performed (n = 4). Error bars represent 1 SD. *P < .01.
Effect of L-NMMA on Epo protein from Hep3B cells stimulated by hypoxia.
Hep3B cells were incubated with different concentrations of L-NMMA under hypoxic conditions (1% O2) for 24 h. Epo protein was measured by RIA. Four separate experiments were performed (n = 4). Error bars represent 1 SD. *P < .01.
Inhibition of NO and cGMP by L-NMMA
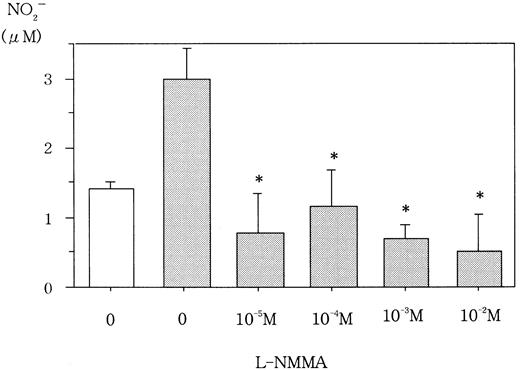
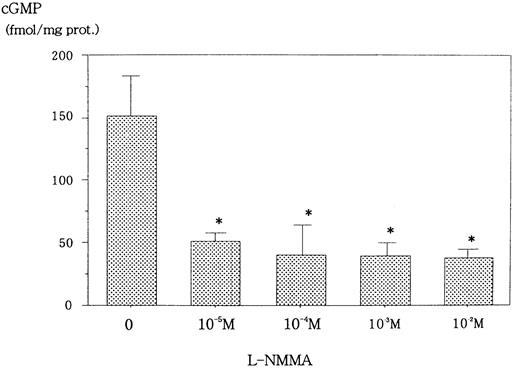
L-NMMA is known to be an NOS inhibitor, and, therefore, a decrease in NO from cells incubated with L-NMMA was expected. To this end, Hep3B cells were incubated with different concentrations of L-NMMA. Hypoxia induced the secretion of NO, but the addition of L-NMMA inhibited this induction (Figure 3). Because NO stimulates guanylate cyclase (GC) to produce cGMP,23 a decrease in cGMP from cells incubated with L-NMMA was also expected. Hypoxic Hep3B cells were incubated with different concentrations of L-NMMA. As shown in Figure 4, L-NMMA inhibited the secretion of cGMP from the cells.
Effect of L-NMMA on NO from Hep3B cells.
Hep3B cells were incubated under normoxic (21% O2, ■) or hypoxic (1% O2, ▪) conditions for 4 hours in the presence or absence of L-NMMA. NO was measured by the 2,3-diaminonaphthalene method. Three separate experiments were performed (n = 3). Error bars represent 1 SD. *P < .01.
Effect of L-NMMA on NO from Hep3B cells.
Hep3B cells were incubated under normoxic (21% O2, ■) or hypoxic (1% O2, ▪) conditions for 4 hours in the presence or absence of L-NMMA. NO was measured by the 2,3-diaminonaphthalene method. Three separate experiments were performed (n = 3). Error bars represent 1 SD. *P < .01.
Effect of L-NMMA on cGMP from Hep3B cells stimulated by hypoxia.
Hep3B cells were incubated with different concentrations of L-NMMA under hypoxia (1% O2) for 2 hours. cGMP was measured by EIA. Two separate experiments were performed (n = 2). Error bars represent 1 SD. *P < .01.
Effect of L-NMMA on cGMP from Hep3B cells stimulated by hypoxia.
Hep3B cells were incubated with different concentrations of L-NMMA under hypoxia (1% O2) for 2 hours. cGMP was measured by EIA. Two separate experiments were performed (n = 2). Error bars represent 1 SD. *P < .01.
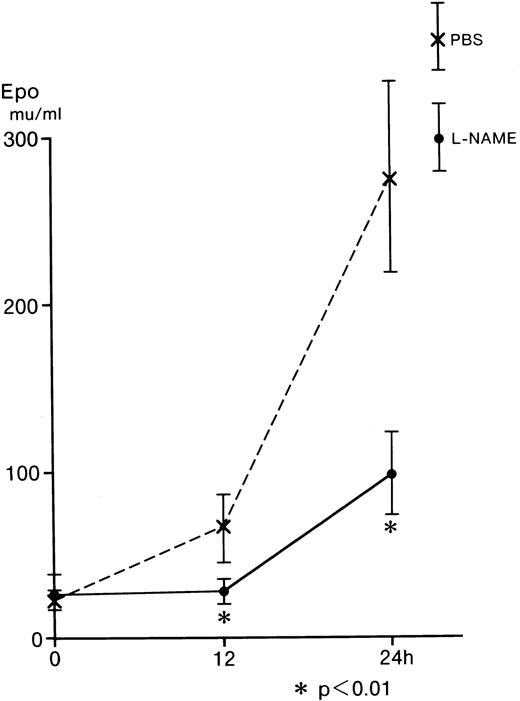
Inhibition of serum Epo by L-NAME from L-NAME–injected mice
L-NAME was examined using an in vivo mouse assay, because L-NMMA is reported to be catabolyzed by NG-dimethylarginine dimethylaminohydrolase (DDHA) in the intact kidney24,25; however, L-NAME is not catabolyzed by this enzyme.26Hecker and associates observed that L-NMMA was rapidly hydrolyzed tol-citrulline and lost the inhibitory effect in endothelial cells.27 Furthermore, L-NMMA is continuously released into body fluids during the in vivo breakdown of the proteins and is assumed to be readily excreted in urine without reincorporation into proteins or further degradation in intact animals.24 To identify the effect of L-NAME on Epo production in vivo, BDF1 mice were injected intraperitoneally with 0.2 mL of 10 mg L-NAME/mL PBS or 0.2 mL of PBS as a control. Blood samples (0.3 mL) were obtained from the orbital vein immediately after (0 hours) and at 12 and 24 hours after the injection of L-NAME (Figure 5). The serum Epo from the control (dashed line) increased to 66.4 mU/mL at 12 hours and to 276.8 mU/mL at 24 hours after the injection. The serum Epo from the mice injected with L-NAME (straight line) was 28.5 mU/mL at 12 hours and 98.8 mU/mL at 24 hours after the injection. These values were significantly lower than those of the control (Figure 5). These in vivo results are comparable with those obtained from the in vitro incubation of Hep3B cells (Figure 2).
Effect of L-NAME on serum Epo from anemic mice.
Five BDF1 mice were injected intraperitoneally with 0.2 mL of 10 mg L-NAME/mL PBS or 0.2 mL of PBS. Blood samples (0.3 mL) were taken from the orbital vein at 0, 12, and 24 hours after injection of L-NAME. Serum Epo levels were determined by RIA. Error bars represent 1 SD. *P < .01.
Effect of L-NAME on serum Epo from anemic mice.
Five BDF1 mice were injected intraperitoneally with 0.2 mL of 10 mg L-NAME/mL PBS or 0.2 mL of PBS. Blood samples (0.3 mL) were taken from the orbital vein at 0, 12, and 24 hours after injection of L-NAME. Serum Epo levels were determined by RIA. Error bars represent 1 SD. *P < .01.
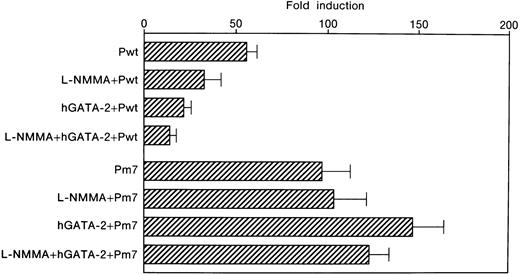
Inhibition of Epo promoter activity by L-NMMA
Expression of the Epo gene has been shown to be induced by hypoxia.1 To elucidate the molecular mechanisms underlying the hypoxic induction of the Epo gene, plasmids containing both the promoter and enhancer of the Epo gene were used in a transient transfection assay into Hep3B cells.10 One GATA and 2 CACCC motifs are in the promoter region, and one HIF-1 and one hepatic nuclear factor 4 (HNF-4) binding site are in the enhancer (Figure 1A). Both the promoter and enhancer were inserted upstream of the Luc gene to give rise to Pwt and Pm7 plasmids (Figure 1B). The latter contains a mutation in the promoter GATA element. We transfected Pwt and Pm7 into Hep3B cells and incubated the cells in the presence or absence of L-NMMA under 21% (normoxia) or 1% (hypoxia) oxygen for 24 hours. The hypoxic induction of Luc gene expression is represented as a hypoxia/normoxia ratio, as previously described.10 Hypoxic induction from Pwt was 55.8 ± 5.5 fold higher than that from normoxic Pwt (mean ± 1 SD, n = 3) (Table 1 and Figure 6). Interestingly, the addition of L-NMMA inhibited the hypoxic induction of theLuc reporter gene expression from Pwt with hypoxia/normoxia ratio of only 33.0 ± 9.2 fold, 59.1% of that from Pwt incubated without L-NMMA (Table 1 and Figure 6). These results indicate that the hypoxic induction of the Epo gene expression is suppressed by L-NMMA through the Epo gene regulatory regions.
Effect of L-NMMA on the induction of the wild-type and mutated GATA motifs of the Epo promoter/enhancer with Lucreporter constructs in Hep3B cells
| . | % O2 . | Experiment 1 . | Experiment 2 . | Experiment 3 . | Fold induction (mean ± 1 SD) . | |||
|---|---|---|---|---|---|---|---|---|
| RLU/CAT . | Fold induction . | RLU/CAT . | Fold induction . | RLU/CAT . | Fold induction . | |||
| Pwt | 21 | 56.6 | ×50.6 | 75.8 | ×63.4 | 54.3 | ×53.5 | ×55.8 ± 5.5 |
| Pwt | 1 | 2866.4 | 4807.9 | 2902.8 | ||||
| Pwt + L-NMMA | 21 | 42.6 | ×38.0 | 72.9 | ×41.0 | 46.1 | ×20.1 | ×33.0 ± 9.2 |
| Pwt + L-NMMA | 1 | 1619.8 | 2985.7 | 927.0 | ||||
| Pwt + hGATA-2 | 21 | 50.7 | ×24.8 | 80.4 | ×23.4 | 49.4 | ×16.2 | ×21.5 ± 3.8 |
| Pwt + hGATA-2 | 1 | 1257.1 | 1877.8 | 799.8 | ||||
| Pwt + hGATA-2 + L-NMMA | 21 | 57.7 | ×19.1 | 96.3 | ×11.7 | 45.6 | ×11.9 | ×14.2 ± 3.4 |
| Pwt + hGATA-2 + L-NMMA | 1 | 1101.6 | 1130.2 | 544.2 | ||||
| Pm7 | 21 | 125.1 | ×75.9 | 61.0 | ×114.5 | 99.7 | ×100.2 | ×96.9 ± 15.9 |
| Pm7 | 1 | 9498.1 | 6986.5 | 9988.6 | ||||
| Pm7 + L-NMMA | 21 | 113.6 | ×74.9 | 73.7 | ×114.2 | 62.8 | ×122.0 | ×103.7 ± 20.6 |
| Pm7 + L-NMMA | 1 | 8504.7 | 8413.8 | 7660.6 | ||||
| Pm7 + hGATA-2 | 21 | 118.1 | ×138.8 | 112.0 | ×171.2 | 61.0 | ×132.0 | ×147.3 ± 17.1 |
| Pm7 + hGATA-2 | 1 | 16386.4 | 19175.9 | 8050.9 | ||||
| Pm7 + hGATA-2 + L-NMMA | 21 | 182.7 | ×124.6 | 111.5 | ×135.8 | 126.1 | ×110.0 | ×123.5 ±10.6 |
| Pm7 + hGATA-2 + L-NMMA | 1 | 22766.1 | 15145.5 | 13870.4 | ||||
| . | % O2 . | Experiment 1 . | Experiment 2 . | Experiment 3 . | Fold induction (mean ± 1 SD) . | |||
|---|---|---|---|---|---|---|---|---|
| RLU/CAT . | Fold induction . | RLU/CAT . | Fold induction . | RLU/CAT . | Fold induction . | |||
| Pwt | 21 | 56.6 | ×50.6 | 75.8 | ×63.4 | 54.3 | ×53.5 | ×55.8 ± 5.5 |
| Pwt | 1 | 2866.4 | 4807.9 | 2902.8 | ||||
| Pwt + L-NMMA | 21 | 42.6 | ×38.0 | 72.9 | ×41.0 | 46.1 | ×20.1 | ×33.0 ± 9.2 |
| Pwt + L-NMMA | 1 | 1619.8 | 2985.7 | 927.0 | ||||
| Pwt + hGATA-2 | 21 | 50.7 | ×24.8 | 80.4 | ×23.4 | 49.4 | ×16.2 | ×21.5 ± 3.8 |
| Pwt + hGATA-2 | 1 | 1257.1 | 1877.8 | 799.8 | ||||
| Pwt + hGATA-2 + L-NMMA | 21 | 57.7 | ×19.1 | 96.3 | ×11.7 | 45.6 | ×11.9 | ×14.2 ± 3.4 |
| Pwt + hGATA-2 + L-NMMA | 1 | 1101.6 | 1130.2 | 544.2 | ||||
| Pm7 | 21 | 125.1 | ×75.9 | 61.0 | ×114.5 | 99.7 | ×100.2 | ×96.9 ± 15.9 |
| Pm7 | 1 | 9498.1 | 6986.5 | 9988.6 | ||||
| Pm7 + L-NMMA | 21 | 113.6 | ×74.9 | 73.7 | ×114.2 | 62.8 | ×122.0 | ×103.7 ± 20.6 |
| Pm7 + L-NMMA | 1 | 8504.7 | 8413.8 | 7660.6 | ||||
| Pm7 + hGATA-2 | 21 | 118.1 | ×138.8 | 112.0 | ×171.2 | 61.0 | ×132.0 | ×147.3 ± 17.1 |
| Pm7 + hGATA-2 | 1 | 16386.4 | 19175.9 | 8050.9 | ||||
| Pm7 + hGATA-2 + L-NMMA | 21 | 182.7 | ×124.6 | 111.5 | ×135.8 | 126.1 | ×110.0 | ×123.5 ±10.6 |
| Pm7 + hGATA-2 + L-NMMA | 1 | 22766.1 | 15145.5 | 13870.4 | ||||
Wild-type (Pwt) and mutated (Pm7) GATA motifs. Hypoxic induction of the Luc gene expression is represented as a hypoxia/normoxia ratio. Fold induction indicates this hypoxia/normoxia ratio. Three separate experiments (duplicate sample) were performed (n = 3).
Effect of L-NMMA on the induction of the wild-type and mutated GATA motifs of the Epo promoter/enhancer with Luc reporter constructs in Hep3B cells.
Wild-type (Pwt) and mutated (Pm7) GATA motifs. Hypoxic induction of Luc gene expression is represented as a hypoxia/normoxia ratio. Fold induction indicates this hypoxia/normoxia ratio. Three separate experiments (duplicate sample) were performed (n = 3). Error bars represent 1 SD.
Effect of L-NMMA on the induction of the wild-type and mutated GATA motifs of the Epo promoter/enhancer with Luc reporter constructs in Hep3B cells.
Wild-type (Pwt) and mutated (Pm7) GATA motifs. Hypoxic induction of Luc gene expression is represented as a hypoxia/normoxia ratio. Fold induction indicates this hypoxia/normoxia ratio. Three separate experiments (duplicate sample) were performed (n = 3). Error bars represent 1 SD.
We previously found that the GATA element in the promoter plays an important role in limiting the hypoxic induction of the Epogene.10 We therefore examined the contribution of the GATA site to the L-NMMA suppression of hypoxic Epo gene induction. To this end, Hep3B cells were co-transfected with Pwt and an hGATA-2 expression vector. The expression of hGATA-2 resulted in the inhibition of the hypoxic induction of the Epo gene. TheLuc reporter gene was induced in the presence of hGATA-2 by 21.5 ± 3.8 fold, 43.2% of that from Pwt incubated without hGATA-2 (Table 1 and Figure 6). Furthermore, the exposure of Hep3B cells co-transfected with Pwt and hGATA-2 expression vector to L-NMMA resulted in further suppression with hypoxic induction of the reporter gene of only 14.2 ± 3.4 fold, 25.4% of that from the cells incubated without L-NMMA and hGATA-2 (Table 1 and Figure 6). These results suggest that hGATA-2 acts as a repressor of the hypoxic induction of the Epo gene and that the GATA sequence in the promoter mediates L-NMMA suppression.
The GATA element in the promoter only contributes to L-NMMA suppression
The contribution of the GATA element was further tested by using a reporter plasmid, Pm7, which contains GATA site mutations. We previously found that alone this GATA mutation affects the basal level expression of Luc reporter activity.10 As was the case for Pwt, Luc expression was also strongly induced following the exposure of the transfected cells to hypoxia, 96.9 ± 15.9 fold (Table 1 and Figure 6). However, L-NMMA failed to affect the GATA mutant Pm7 Luc activity with hypoxic induction of 103.7 ± 20.6 fold, 107.0% of that from Pm7 only (Table 1 and Figure 6). Transfection of Pm7 into Hep3B cells, which express hGATA-2, resulted in hypoxic induction of 147.3 ± 17.1 fold, 152.0% of that from Pm7 only (Table 1 and Figure6). Furthermore, the exposure of the cells co-transfected with Pm7 and hGATA-2 expression vector to L-NMMA showed 123.5 ± 10.6 fold, and 127.5% of that from Pm7 only (Table 1 and Figure 6). These results suggest that L-NMMA inhibits Epo gene expression through the GATA site in the Epo promoter rather than through the enhancer activity. To clearly identify whether this inhibitory effect of L-NMMA on Epo gene expression was due to a GATA-2 or HIF-1 binding site or both, use of the construct that contained the Epo promoter only was advantageous. To this end, we then transfected Δ18pXP2 (wild type) or Δ18m7pXP2 (GATA site mutant) plasmids into Hep3B cells and incubated the cells both with or without additional L-NMMA under 21% or 1% oxygen for 24 hours (Figure 7). Hypoxic induction from Δ18pXP2 was 7.6 ± 1.0 fold higher than that from normoxic Δ18pXP2 (mean ± 1 SD, n = 4) (Figure 7). The addition of L-NMMA significantly inhibited the hypoxic induction of theLuc reporter gene expression from Δ18pXP2 with hypoxia/normoxia ratio of only 4.0 ± 0.6 fold, 52.6% of that from Δ18pXP2 incubated without L-NMMA (Figure 7). Hypoxic induction from Δ18m7pXP2 was 7.3 ± 0.2 fold higher that from normoxic Δ18m7pXP2. L-NMMA failed to affect the Δ18m7pXP2 Luc activity with hypoxic induction of 8.2 ± 0.5 fold, 112.3% of that from Δ18m7pXP2 only (Figure 7). These results clearly indicate that the inhibitory effect of L-NMMA on Epo gene expression was due to GATA-2, not HIF-1.
Effect of L-NMMA on the induction of the wild-type and mutated GATA motifs of the Epo promoter with Luc reporter constructs in Hep3B cells.
Wild-type (Δ18pXP2) and mutated (Δ18m7pXP2) GATA motifs. Hypoxic induction of Luc gene expression is represented as a hypoxia/normoxia ratio. Fold induction indicates this hypoxia/normoxia ratio. Four separate experiments (duplicate sample) were performed (n = 4). Error bars represent 1 SD. *P < .01.
Effect of L-NMMA on the induction of the wild-type and mutated GATA motifs of the Epo promoter with Luc reporter constructs in Hep3B cells.
Wild-type (Δ18pXP2) and mutated (Δ18m7pXP2) GATA motifs. Hypoxic induction of Luc gene expression is represented as a hypoxia/normoxia ratio. Fold induction indicates this hypoxia/normoxia ratio. Four separate experiments (duplicate sample) were performed (n = 4). Error bars represent 1 SD. *P < .01.
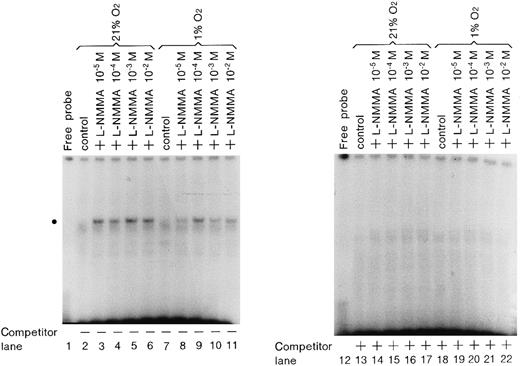
Enhancement of GATA-2 binding activity by L-NMMA
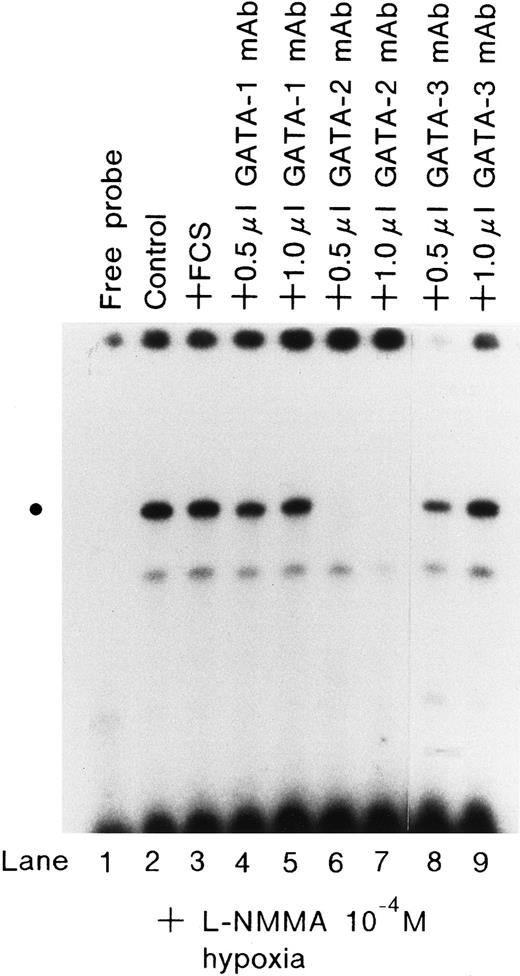
To examine whether L-NMMA treatment affects the binding activity of hGATA-2, nuclear extracts were prepared from the cells stimulated by L-NMMA for 1 hour under normoxic or hypoxic conditions, and electrophoretic mobility shift assays (EMSA) were performed with an oligonucleotide containing the wild-type GATA element (AGATAA) (Figure8). L-NMMA induced the binding activity of GATA-2 (indicated by the circle) under normoxic and hypoxic conditions (lanes 3-6, 9-11). This binding activity was abolished by self-competitor (lanes 14-17, 20-22). To confirm that the band indicated by the circle was GATA-2, nuclear extracts were prepared from Hep3B cells under hypoxia with 10−4 mol/L L-NMMA for 1 hour, and incubated with 0.5 or 1.0 μL monoclonal antibodies of hGATA-1, 2, and 3 and then EMSA was performed under the same conditions as described in Figure 8. The control revealed a band of increased intensity (Figure 9, lane 2), and the addition of FCS further increased the intensity of this band, though the mechanism of this increase is unknown (Figure 9, lane 3). The addition of monoclonal antibodies of hGATA-1 and -3 resulted in bands of similar intensities (lanes 4, 5 and 8, 9); however, the band indicated by the circle disappeared with the addition of monoclonal hGATA-2 antibody (lanes 6 and 7). These results strongly suggest that the band indicated by the circle was a specific GATA-2 band.
Effect of L-NMMA on the expression of GATA-2.
EMSA was performed using 1.5 μg of protein from Hep3B cells under normoxia (lanes 2-6 and 13-17) and hypoxia (lanes 7-11 and 18-22), from Hep3B cells incubated with 10−5 mol/L L-NMMA (lanes 3, 8, 14, and 19), 10−4 mol/L L-NMMA (lanes 4, 9, 15, and 20), 10−3 mol/L L-NMMA (lanes 5, 10, 16, and 21), and 10−2 mol/L L-NMMA (lanes 6, 11, 17, and 22) for 1 hour. The dot at the left indicates the position of the GATA-2 transcription factor. A total of 25 ng (0.5 μL: 12.5-fold molar excess) of competitor DNA was added to each reaction mixture (lanes 13-22).
Effect of L-NMMA on the expression of GATA-2.
EMSA was performed using 1.5 μg of protein from Hep3B cells under normoxia (lanes 2-6 and 13-17) and hypoxia (lanes 7-11 and 18-22), from Hep3B cells incubated with 10−5 mol/L L-NMMA (lanes 3, 8, 14, and 19), 10−4 mol/L L-NMMA (lanes 4, 9, 15, and 20), 10−3 mol/L L-NMMA (lanes 5, 10, 16, and 21), and 10−2 mol/L L-NMMA (lanes 6, 11, 17, and 22) for 1 hour. The dot at the left indicates the position of the GATA-2 transcription factor. A total of 25 ng (0.5 μL: 12.5-fold molar excess) of competitor DNA was added to each reaction mixture (lanes 13-22).
Effect of monoclonal antibodies on the expression of GATA.
EMSA was performed using 1.5 μg of protein from Hep3B cells incubated with 10−4 mol/L L-NMMA under hypoxia (1% O2) for 1 hour; 2 μL of FCS (lane 3) or 0.5 or 1.0 μL monoclonal antibodies of hGATA-1 (lanes 4 and 5), hGATA-2 (lanes 6 and 7), and hGATA-3 (lanes 8 and 9) were incubated with nuclear extracts overnight at 4°C and then EMSA was performed. The dot at the left indicates the position of the respective proteins.
Effect of monoclonal antibodies on the expression of GATA.
EMSA was performed using 1.5 μg of protein from Hep3B cells incubated with 10−4 mol/L L-NMMA under hypoxia (1% O2) for 1 hour; 2 μL of FCS (lane 3) or 0.5 or 1.0 μL monoclonal antibodies of hGATA-1 (lanes 4 and 5), hGATA-2 (lanes 6 and 7), and hGATA-3 (lanes 8 and 9) were incubated with nuclear extracts overnight at 4°C and then EMSA was performed. The dot at the left indicates the position of the respective proteins.
To clarify the effect of cGMP on GATA-2 binding activity, nuclear extracts were prepared from Hep3B cells under hypoxia with the addition of 10−4 mol/L L-NMMA or cGMP, and EMSA was performed under the same conditions as described in Figure 8. The addition of cGMP inhibited the L-NMMA–induced binding activity of GATA-2 in a dose-dependent manner (data not shown). The effects of L-NMMA on the binding activity of HIF-1, HNF-4, chicken ovalbumin upstream promoter–transcription factor 1 (COUP-TF1), nuclear factor (NF)-κB were measured by EMSA. L-NMMA did not alter the binding activity of any of these transcription factors (data not shown).
GATA-2 messenger RNA expression was induced by L-NMMA
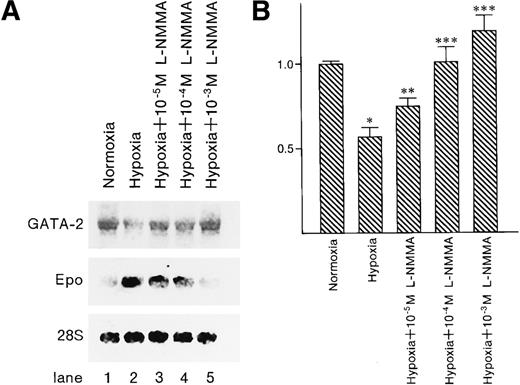
To examine whether GATA-2 and Epo messenger RNA (mRNA) expression levels were affected by the addition of L-NMMA, Northern blot analysis was performed. Northern blot analysis showed hypoxia-induced Epo mRNA expression; however, the addition of L-NMMA inhibited this induction of Epo mRNA (Figure10A, middle panel). In contrast, hypoxia reduced GATA-2 mRNA expression, whereas L-NMMA induced the expression of GATA-2 mRNA (Figure 10A, upper panel, and B). The 28S used as a control revealed a constant level of mRNA expression from the cells incubated under normoxia or hypoxia and with or without L-NMMA (Figure 10A, lower panel).
Effect of L-NMMA on the expression of GATA-2 and Epo mRNA.
(A) Northern blot analysis was performed using 20 μg of RNA from Hep3B cells incubated under conditions of normoxia (21% O2) (lane 1), conditions of hypoxia (1% O2) (lane 2), hypoxia with 10−5 mol/L L-NMMA (lane 3), hypoxia with 10−4 mol/L L-NMMA (lane 4), and hypoxia with 10−3 mol/L L-NMMA (lane 5) for 8 hours. Upper, middle, and lower panels show GATA-2, Epo, and 28S mRNA, respectively. (B) Quantitative differences in GATA-2 mRNA levels were evaluated by molecular imager FX. Abundance of GATA-2 mRNA is expressed as units of densitometry relative to normoxia. Three separate experiments were performed (n = 3). *Significance compared with normoxia,P < .01. **Significance compared with hypoxia,P < .05. ***Significance compared with hypoxia,P < .01.
Effect of L-NMMA on the expression of GATA-2 and Epo mRNA.
(A) Northern blot analysis was performed using 20 μg of RNA from Hep3B cells incubated under conditions of normoxia (21% O2) (lane 1), conditions of hypoxia (1% O2) (lane 2), hypoxia with 10−5 mol/L L-NMMA (lane 3), hypoxia with 10−4 mol/L L-NMMA (lane 4), and hypoxia with 10−3 mol/L L-NMMA (lane 5) for 8 hours. Upper, middle, and lower panels show GATA-2, Epo, and 28S mRNA, respectively. (B) Quantitative differences in GATA-2 mRNA levels were evaluated by molecular imager FX. Abundance of GATA-2 mRNA is expressed as units of densitometry relative to normoxia. Three separate experiments were performed (n = 3). *Significance compared with normoxia,P < .01. **Significance compared with hypoxia,P < .05. ***Significance compared with hypoxia,P < .01.
Discussion
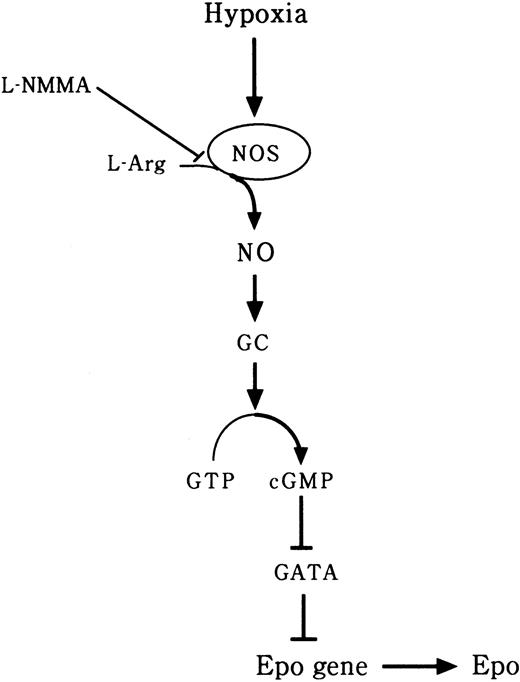
It was reported that L-NAME, also an NOS inhibitor, significantly blunted interferon-γ (IFN-γ) induction of Epo, nitrite, and cGMP in Hep3B cells.22 Furthermore, L-NAME significantly decreased hypoxia-induced elevations of Epo mRNA and cGMP in an isolated perfused rat kidney system.22 It has been suggested that NO plays an important role in the oxygen-sensing mechanism in Epo production.22 However, the effect of an NOS inhibitor on the transcription factors of the Epo gene has not yet been clarified. NO is known to be an endothelium-derived relaxing factor28 and has been found to play an important role in the physiology of the cardiovascular, immune, and nervous systems.23,29 The biosynthesis of NO involves the oxidation of l-arginine by NOS.30,31 NO stimulates a soluble GC, resulting in an increase in cGMP levels in cells.23 The mRNA of nNOS is abundantly expressed in the kidney inner medulla and macula densa cells.32 We hypothesized that NO produced by cNOS, which behaves as an intracellular or extracellular messenger, activates GC and, through stimulation of cGMP, consequently up-regulates Epo gene expression in peritubular cells. However, in chronic renal failure, we suspect that an NOS inhibitor such as L-NMMA suppresses Epogene expression through inhibition of NO production. In the present study, we found that L-NMMA decreased the expression of NO and cGMP and increased the expression of GATA-2 mRNA and the level of GATA-2 binding activity, thereby inhibiting Epo promoter activity and causing a decrease in the expression of Epo protein (Figure 11).
The predialysis concentration of L-NMMA in patients with chronic renal failure is approximately 10−5 mol/L.8 This concentration of L-NMMA significantly inhibited Epo protein production in Hep3B cells (Figure 2). However, a higher concentration of L-NAME in comparison to the serum concentration of L-NMMA in chronic renal failure was required for the inhibition of Epo production in vivo (Figure 5). There are 2 possible reasons for this. (1) A high local concentration of L-NMMA has a physiologic role in the kidney, liver, and pancreas. An inappropriately high concentration of L-NMMA due to renal failure may affect Epo production in peritubular cells despite its low serum concentration. (2) Not only L-NMMA but also asymmetric dimethylarginine (ADMA), which is also increased in patients with chronic renal failure,33 causes a dose-dependent inhibition of NOS.34
In the present study, the effect of L-NMMA on GATA was investigated in detail. The effects of L-NMMA on HIF-1, HNF-4, COUP-TF1, NF-κB were assessed by EMSA and Epo promoter assay. L-NMMA did not alter the binding activity of HIF-1, HNF-4, COUP-TF1, or NF-κB at all. Neither EMSA nor Epo reporter gene transfection experiments showed any effect of L-NMMA on Epo enhancer activity. L-NMMA did not affect Luc activity when GATA in the Epo promoter was mutated (Pm7). Recently, Kimura and coworkers reported that L-NAME did not interfere with hypoxia-induced vascular endothelial growth factor (VEGF) promoter (HIF-1 binding site) activation.35 This result is compatible with our data. These results strongly suggest that L-NMMA affects GATA-2 binding.
The effect of NO on VEGF expression via HIF-1 is controversial. Some recent reports show an inhibitory effect of NO on VEGF expression. Huang and colleagues36 and Sogawa and associates37 have demonstrated that sodium nitroprusside (SNP, NO donor) suppresses hypoxia-induced VEGF gene activation and HIF-1 binding activity. SNP inhibits the hypoxic induction of the VEGF gene in a dose-dependent manner, in contrast to the effects S-nitroso-N-acetyl-D,l-penicillamine (SNAP) and 3-(2-hydroxy-1-(1-methylethyl)-2-nitrosohydrazino)-1-propanamine (NOC5) (another NO donor) as shown by Kimura and colleagues.35This concentration is clearly due to the specific nature of SNP. As to the effect of NO on Epo, Fisher and coworkers reported that serum levels of Epo in hypoxic polycythemic mice were significantly increased after injections of 200 μg/kg SNP.22 Furthermore, cGMP levels in hypoxic Hep3B cells were also elevated. SNP (10 and 100 μmol/L) and NO (2 μmol/L) increased cGMP levels in Hep3B cells.22 These results are compatible with our data and strongly suggest that L-NMMA inhibits Epo production via the GATA transcription factor.
Further analysis of the GATA-2 expression level and NO and cGMP from patients with renal anemia is necessary to clarify the details of the pathogenesis of renal anemia.
Acknowledgments
We thank D. L. Galson for providing the Pwt, Pm7, Δ18pXP2, and Δ18m7pXP2 plasmids. We also thank M. Nakamura and A. Yamazaki for their expert technical assistance.
Supported by grants-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan, and the Chugai Foundation, Tokyo, Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Shigehiko Imagawa, Division of Hematology, Institute of Clinical Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan; e-mail: simagawa@md.tsukuba.ac.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal