Key Points
Next-generation sequencing–based IG/TR profiling enables also clonal evolution analysis and detection of nonmalignant clones.
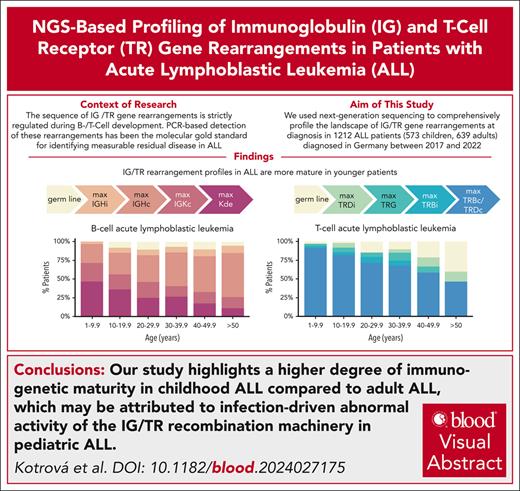
Next-generation sequencing–based IG/TR rearrangement profiling provides hints toward a higher immunogenetic maturity in pediatric ALL.
Visual Abstract
We comprehensively profiled the landscape of immunoglobulin (IG) and T-cell receptor (TR) rearrangements at diagnosis in 1212 patients with acute lymphoblastic leukemia (ALL; 573 children and 639 adults) diagnosed in Germany between 2017 and 2022. Our study revealed a significant age-related decrease in immunogenetic maturation, where IG κ rearrangements in B-ALL and complete TR β/δ rearrangements in T-ALL, hallmarks of maturity, were more frequent in pediatric patients compared to adults (B-ALL: 68.7% vs 39.0%, P < 2.2e-16; T-ALL: 85.7% vs 67.3%, P = 6.7e-03). Compared to adults, children had a higher average number of IG/TR markers per patient (6 vs 4; P = 2.5e-38) and markedly fewer lacked these markers (0.5% compared to 6.7%). IG heavy chain clonal evolution was most pronounced among pro-B-ALL cases (60.9%), with the V-to-DJ mechanism driving pro-B evolution (78.6%), whereas V-replacement dominated other immunophenotypes. Furthermore, expanded accompanying T-cell clones of unknown significance in B-ALL increased with age. This next-generation sequencing-based study offers an unprecedented characterization of IG/TR rearrangement patterns across ALL subtypes and age groups. It highlights the higher immunogenetic maturity in children, which may be explained by the infection-driven abnormal activity of the IG/TR recombination machinery in pediatric ALL.
Introduction
For >3 decades, polymerase chain reaction (PCR)–based detection of immunoglobulin (IG) and T-cell receptor (TR) gene rearrangements has been the molecular gold standard for identifying measurable residual disease (MRD) in acute lymphoblastic leukemia (ALL).1 A major development in this field was the introduction of amplicon-based next-generation sequencing (NGS) assays for the characterization of clonally rearranged IG/TR genes by the EuroClonality-NGS working group in 2019.2 This innovation has gained traction in laboratories worldwide, surpassing traditional Sanger sequencing–based marker screening in both efficiency and capability.
To date, no comprehensive overview exists on the prevalence of leukemia-associated IG/TR gene rearrangement types in ALL across different immunophenotypes, genetic subtypes, and age groups. Existing reports using low-throughput methods often fail to cover all IG/TR rearrangement types or to provide a comprehensive age-related perspective.3-6 Notably, the IG/TR immunogenotype of B-cell precursor ALL (B-ALL) is reported to be more immature and stable in adults compared with children, suggesting that the immunogenetic maturation grade (IMG) might be prognostically relevant.6 NGS-based analysis offers significant advantages, such as higher sensitivity for detecting subclones and accompanying clones,7 and high throughput for assessing clone relatedness and clonal evolution (ClEvo).8,9 Two mechanisms driving immunoglobulin heavy chain (IGH) ClEvo in B-ALL (ongoing V-DJ recombination and V replacement) are associated with maturation arrest at different stages of B-cell development.8 In addition, data indicate that accompanying T-cell clones might have an impact on ALL treatment outcomes.7
Study design
We examined 1212 patients with ALL, including 573 children (aged 1-17.99 years) and 639 adults (aged 18-56 years), diagnosed in Germany between 2017 and 2022 (for patient characteristics, see supplemental Table 1, available on the Blood website). Immunophenotypes were centrally assessed for all patients and evaluated according to the European Group for the Immunological Classification of Leukemia recommendations10 to ensure comparability. Molecular subtypes were assessed from RNA sequencing data for adult patients; and from fluorescence in situ hybridization, array comparative genomic hybridization, and targeted RNA sequencing11 for children.
All patients underwent routine IG/TR-marker screening using the EuroClonality-NGS amplicon assay.2 For pediatric cases, intronRSS-Kde (κ deleting element) rearrangements were analyzed in a separate PCR tube, whereas in adults all immunoglobulin κ chain (IGK) rearrangements were amplified in 1 PCR tube (we excluded this class from our analysis to avoid bias).
To improve the specificity of distinguishing leukemia-associated rearrangements from the physiological background, an algorithm named Marksman was implemented, as described previously7 and outlined in the supplemental Methods.
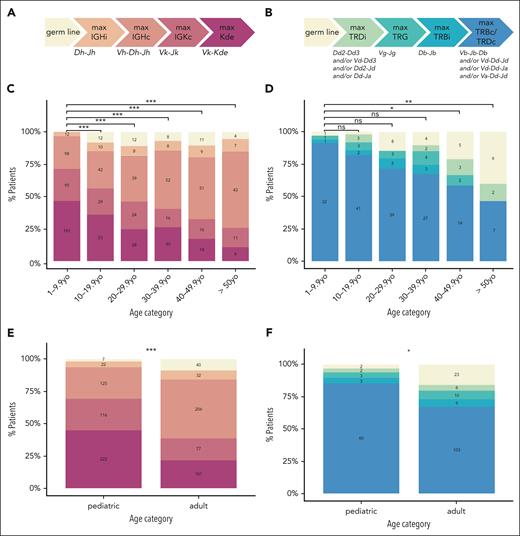
For each patient, we defined an IMG, based on the IG- and TR rearrangement profile for B-ALL12 and T-ALL,13-15 respectively. Briefly, we ordered the IG/TR rearrangement classes according to their sequence of appearance during B-/T-cell development. The most mature rearrangement determined each patient’s IMG (Figures 1A-B).
IMG based on IG (left) and TR (right) rearrangements in B-ALL and T-ALL cases. The most mature rearrangement identified for each patient determines the IMG for that individual. The IMG and corresponding rearrangements are ordered (left to right) by their appearance during the physiological B-cell (A) and T-cell (B) development. (C-D) The distribution by age of the IG-based IMG in patients with B-ALL (C) and the TR-based IMG in patients with T-ALL (D) is shown. (E-F) The distribution in pediatric and adult patients of the IG-based IMG in patients with B-ALL (E) and the TR-based IMG in patients with T-ALL (F) is illustrated. Patient numbers are shown within each bar. P values were corrected for multiple testing using the Bonferroni method. Statistical significance is indicated as follows: ns, P > .05; ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001. c, complete; i, incomplete; max, maximum; ns, not significant; TRG, T-cell receptor γ rearrangement; yo, years old.
IMG based on IG (left) and TR (right) rearrangements in B-ALL and T-ALL cases. The most mature rearrangement identified for each patient determines the IMG for that individual. The IMG and corresponding rearrangements are ordered (left to right) by their appearance during the physiological B-cell (A) and T-cell (B) development. (C-D) The distribution by age of the IG-based IMG in patients with B-ALL (C) and the TR-based IMG in patients with T-ALL (D) is shown. (E-F) The distribution in pediatric and adult patients of the IG-based IMG in patients with B-ALL (E) and the TR-based IMG in patients with T-ALL (F) is illustrated. Patient numbers are shown within each bar. P values were corrected for multiple testing using the Bonferroni method. Statistical significance is indicated as follows: ns, P > .05; ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001. c, complete; i, incomplete; max, maximum; ns, not significant; TRG, T-cell receptor γ rearrangement; yo, years old.
Only patients with at least 1 leukemic IGH-VJ or T-cell receptor β (TRB)–VJ rearrangement were further analyzed for the presence of ClEvo at the IGH or TRB locus. Rearrangements were considered leukemia related if they were either evolving or assigned as dominant by Marksman. The analysis was performed as described previously.8,9 Briefly, clonotypes sharing the same “DNJ-stem” sequence were considered clonally related and further analyzed, determining whether the stem was stable or evolving, and identifying the ClEvo mechanism (V-DJ or V replacement).
We further investigated the presence of expanded accompanying T-cell clones in B-ALL samples. These were identified by the presence of complete T-cell receptor δ (TRD) rearrangements or complete TRB rearrangements with a TRBJ1 segment comprising ≥5% reads, because such rearrangements are virtually never found in B-ALL cases.5-7
All patients provided written informed consent in accordance with the Declaration of Helsinki. This study was approved by the institutional review board of the Christian-Albrechts-University in Kiel (ethics approval number D416/21).
Results and discussion
In physiological lymphocytes, the IG/TR loci rearrangement process follows a strict chronological order (Figure 1A-B). Consequently, IG/TR rearrangements could provide valuable insights into the cell stage at which malignant transformation occurred. However, because the recombination machinery may remain active in malignant cells arrested at early developmental stages, ClEvo can occur within the leukemic clone. The character of ClEvo is also cell-stage specific.8,9
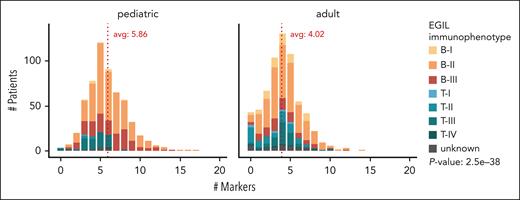
Our data indicate that mature immunogenotypes are more frequent in younger patients. Children more frequently exhibit rearrangements occurring at later stages of B-/T-cell development. Figure 1C-D illustrates that the percentage of patients with IGK (B-ALL) and complete TRB/TRD (T-ALL) rearrangements declines with age. Specifically, 338 of 492 (68.7%) pediatric patients with B-ALL and 60 of 70 (85.7%) pediatric patients with T-ALL carried an IGK or complete TRB and/or TRD rearrangement, respectively. In contrast, only 178 of 456 (39.0%) adult patients with B-ALL and 103 of 153 (67.3%) patients with T-ALL carried these rearrangements (B-ALL, P < 2.2e-16; T-ALL, P = 6.7e-03; Figure 1E-F). Although there seems to be a certain immunogenotype-immunophenotype correlation (supplemental Figure 1), this trend of more mature immunogenotypes in children persists within patients exhibiting the same immunophenotype (supplemental Figure 2) and even when patients are categorized by molecular ALL subtype (supplemental Figure 3). Furthermore, this trend also remains statistically significant after excluding ETV6::RUNX1+ and hyperdiploid cases (the molecular classes that differ most in frequency between children and adults) from our cohort. In addition, children have a higher average number of IG/TR markers (n = 5.86) than adults (n = 4.02; P = 2.5e-38; Figure 2). Only 3 of 573 pediatric patients (0.5%, including 2 with T-ALL and 1 with an unknown immunophenotype) vs 43 of 639 adult patients (6.7%, including 14 with B-ALL, 23 with T-ALL, and 6 with an unknown immunophenotype) lack IG/TR markers. Higher IG/TR-based immunogenetic maturity of pediatric B-ALL is consistent with a better prognosis of this disease in childhood and may reflect a distinct origin of the disease in children and adults. Pediatric ALL is widely believed to result from an abnormal immune response to common infections16 coupled with the concurrent expression of activation-induced cytidine deaminase and the recombination-activating gene enzymes under strong inflammatory stimuli.17 In contrast, the mechanism in older patients likely involves the gradual accumulation of mutations over time within the context of lymphoid clonal hematopoiesis.18
Total number of clonal IG/TR rearrangements per patient for pediatric and adult patients with ALL. The x-axis represents the number of clonal rearrangements per patient, whereas the y-axis shows the patient count. Pediatric patients are displayed on the left and adult patients on the right. Different colors indicate immunophenotypes as defined by European Group for the Immunological Classification of Leukemia (EGIL). The difference in the number of markers per patient between children and adults is highly significant (P = 2.5e-38). Avg, average.
Total number of clonal IG/TR rearrangements per patient for pediatric and adult patients with ALL. The x-axis represents the number of clonal rearrangements per patient, whereas the y-axis shows the patient count. Pediatric patients are displayed on the left and adult patients on the right. Different colors indicate immunophenotypes as defined by European Group for the Immunological Classification of Leukemia (EGIL). The difference in the number of markers per patient between children and adults is highly significant (P = 2.5e-38). Avg, average.
Illegitimate crosslineage TR rearrangements are rare in normal B cells and mature B-cell malignancies. However, they are common in B-ALL, in which they result from the ongoing and aberrant activity of the V(D)J recombinase system. We have observed that the percentage of patients with B-ALL with crosslineage TR rearrangements decreases with age (supplemental Figure 4A). Specifically, 459 of 492 (93.3%) pediatric patients showed crosslineage rearrangements, compared with 360 of 456 (78.9%) adult patients (supplemental Figure 4B; P = 2.3e-10). The lower frequency of crosslineage TR rearrangements in immature B-ALL (pro–B-ALL) compared with CD10+ B-ALL has been previously documented.3,19 These findings are also consistent with the data presented in supplemental Figure 4C, demonstrating the highest prevalence of crosslineage TR rearrangements in patients with a B-II immunophenotype. One possible explanation could be the epigenetically regulated limited accessibility of TR loci in pro-B cells.20 Our data also reveal a lower frequency of TR rearrangements in KMT2A-rearranged and TCF3::PBX1 B-ALL cases (supplemental Figure 5), which is in line with published data.19,21 In TCF3::PBX1 cases, the deregulated E2A function was suggested to be responsible for the absence of TR rearrangements.21
The ability to detect IG/TR ClEvo and its mechanism is another advantage of NGS over low-throughput methods (supplemental Figure 6). IGH ClEvo was observed in 147 of 455 (32.3%) pediatric and 121 of 409 (29.6%) adult B-ALL cases (P = .36). Our data indicate that ClEvo of the IGH locus is most prevalent in B-I ALL (42/69 [60.9%]), compared with 154 of 582 (26.5%) in B-II and 72 of 213 (33.8%) B-III cases (supplemental Figure 6A). Consistent with published data, we confirmed that the V-DJ recombination mechanism drives pro-B-cell evolution, whereas V replacement predominates in other immunophenotypes (V-DJ detected in 33/42 [78.6%] B-I, 79/147 [53.7%] B-II, and 31/72 [43.1%] B-III cases).8 This is probably related to the higher proportion of KMT2A-rearranged leukemias among the B-I cases, in which the IGH ClEvo is mostly driven by the V-DJ mechanism (supplemental Figure 7). Notably, IGH rearrangements appear to drive ClEvo more frequently than TRB rearrangements: TRB ClEvo was detected in 47 of 156 (30.1%) pediatric and 44 of 281 (15.7%; P = .001) adult B-ALL cases and in 5 of 57 (8.8%) pediatric and 9 of 131 (6.9%; P = .79) adult T-ALL cases (supplemental Figure 6C-F). The higher frequency of TRB ClEvo in pediatric patients may be explained by the concurrent expression of activation-induced cytidine deaminase and the recombination-activating gene enzymes in the presence of strong inflammatory stimuli, which can lead to renewed somatic recombination and/or mutation activity in their (pre)leukemic cells.16,17 Consistent with this, 21 of 46 pediatric patients (45.7%) with TRB ClEvo are aged between 2 and 5 years, an age range that closely aligns with the typical age distribution of ALL and common childhood infectious diseases. This finding might have significant clinical implications, because ClEvo could lead to the loss of MRD markers, potentially resulting in false-negative MRD results.
Our data also reveal a higher frequency of expanded accompanying T-cell clones in older patients (supplemental Figure 8); only 9 of 386 (2.3%) and 1 of 386 (0.3%) patients aged 1 to 9.9 years carried at least 1 accompanying clonal TRD and TRB rearrangement, respectively. In contrast, 26 of 73 (35.6%) and 21 of 73 (28.8%) patients aged >50 years carried at least 1 accompanying TRD or TRB rearrangement, respectively. This is probably attributable to the antigen-driven clonal T-cell expansion, thymic involution, and the associated shrinkage of the T-cell repertoire in older individuals.22,23 Such clones may possess antileukemic properties, because better treatment responses in ALL patients with expanded γδ T-cell clones have been reported7 but may also be leukemia-reactive24 or represent a premalignant condition.25 This finding underscores the need for caution to avoid misinterpreting accompanying rearrangements as MRD markers, particularly in the adult population.
Finally, we provide a comprehensive overview of the prevalence of rearrangement classes across different immunophenotypes and age groups (supplemental Figure 9). Although previous studies have attempted to overview leukemic IG/TR rearrangements,3-6 none of them has included such a substantial number of patients across such a wide range of age groups. Furthermore, low-throughput methods can only offer a limited perspective on the rearrangement profiles because of their limited sensitivity and readout. Our findings are consistent with previously published low-throughput data3-6 and establish a valuable reference point for future NGS-based research on IG/TR rearrangements in ALL.
Acknowledgments
The authors thank the ALL-BFM and German Multicenter Study Group for Adult ALL trial centers, patients and their guardians, physicians, and all participating hospitals for patient recruitment, care, and logistics.
This work was partly supported by the Deutsche José Carreras Leukämie-Stiftung (grants DJCLS R 15/11 and DJCLS 06R/2019 [M. Szczepanowski, M.B.]), the Deutsche Forschungsgemeinschaft (German Research Foundation; project number 444949889; Clinical Research Unit CATCH ALL KFO 5010/1 [S.B., G. Chitadze, G. Cario, M. Schrappe, L.B., C.D.B., M.B.]), and the Clinician Scientist Program in Evolutionary Medicine (project number 413490537 [G. Chitadze]).
Authorship
Contribution: M.K., C. Pott, M.R., N.G., C.D.B., G. Cario, M. Schrappe, J.A., R.K., and M.B. designed the research; M.K., C. Proske, N.D., A.L., B.K., J.K., S.B., S.K., M. Szczepanowski, W.W., Z.A., G. Chitadze, A.B., L.B., S.S., and R.K. analyzed and interpreted data; J.J.M.v.D. interpreted the data; M.K. performed statistical analysis and wrote the manuscript; and all authors reviewed the manuscript and approved its final version.
Conflict-of-interest disclosure: S.S. has received speaker honoraria from the Akademie für Infektionsmedizin e.V., Amgen, CSI Hamburg, Pfizer, and SERB SAS; has served as consultant or advisory board member for Amgen, Pfizer, and SERB SAS; and has received travel grants from Amgen, Pfizer, and SERB SAS. G. Cario received travel support from Jazz Pharma. The remaining authors declare no competing financial interests.
Correspondence: Monika Brüggemann, Unit for Hematological Diagnostics, Medical Department II, University Medical Center Schleswig-Holstein, Campus Kiel, Langer Segen 8-10, 24105 Kiel, Germany; email: m.brueggemann@med2.uni-kiel.de.
References
Author notes
Data are available on request from the corresponding author, Monika Brüggemann (m.brueggemann@med2.uni-kiel.de).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal