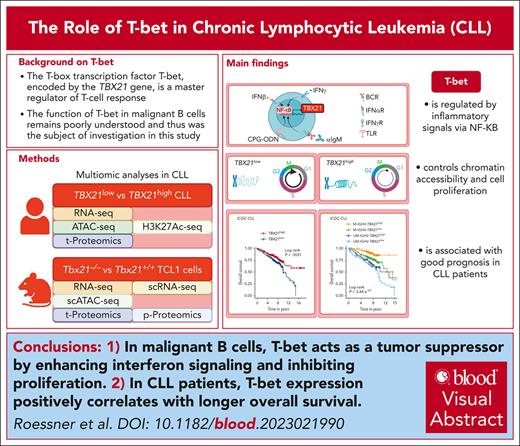
Key Points
T-bet acts as a tumor suppressor by enhancing interferon signaling and suppressing proliferation of malignant B cells.
T-bet expression in CLL cells is positively correlated with longer overall survival in patients with CLL.
Visual Abstract
The T-box transcription factor T-bet is known as a master regulator of the T-cell response but its role in malignant B cells has not been sufficiently explored. Here, we conducted single-cell resolved multi-omics analyses of malignant B cells from patients with chronic lymphocytic leukemia (CLL) and studied a CLL mouse model with a genetic knockout of Tbx21. We found that T-bet acts as a tumor suppressor in malignant B cells by decreasing their proliferation rate. NF-κB activity, induced by inflammatory signals provided by the microenvironment, triggered T-bet expression, which affected promoter-proximal and distal chromatin coaccessibility and controlled a specific gene signature by mainly suppressing transcription. Gene set enrichment analysis identified a positive regulation of interferon signaling and negative control of proliferation by T-bet. In line, we showed that T-bet represses cell cycling and is associated with longer overall survival of patients with CLL. Our study uncovered a novel tumor suppressive role of T-bet in malignant B cells via its regulation of inflammatory processes and cell cycling, which has implications for the stratification and therapy of patients with CLL. Linking T-bet activity to inflammation explains the good prognostic role of genetic alterations in the inflammatory signaling pathways in CLL.
Introduction
The T-box transcription factor (TF) T-bet encoded by TBX21 is well known for its role in the lineage commitment of CD4+ T helper cells, effector functions of CD8+ T cells, and differentiation of natural killer cells.1-3 But extensive literature attributes the important role of T-bet in B cells, mainly in the context of “age-associated” B cells (ABCs).4-6 First described in aging mice, ABCs also exist in healthy humans, in which they increase up to the age of 30 years, followed by stabilization of their frequency.6-8 Accumulation of ABCs has been observed during infections in which T-bet+ B cells contribute to protective immunity.6,9-13 In addition, an increase in ABC numbers is observed in patients with humoral autoimmune diseases, such as systemic lupus erythematosus, scleroderma, rheumatoid arthritis, Crohn disease, and Sjögren syndrome, as well as in adipose tissue during exacerbated metabolic disorders. Here, ABCs are linked to the production of autoreactive antibodies and are associated with worse clinical outcomes.5,6,8,14-22
The expression of T-bet in B cells is induced via the activation of several signaling pathways, including toll-like receptor (TLR), B-cell receptor (BCR), CD40, and cytokine receptor signaling. These signals are provided by the microenvironment of lymphoid tissues, for example, by bystander T cells.6,10,23-26 Using Tbx21-deficient B cells, the involvement of T-bet in immunoglobulin class switching and the generation of long-lived antibody-secreting B cells and their function in antiviral control was observed in several mouse models.9,10,26,27 Yang et al investigated a patient harboring a complete deficiency in T-bet and observed that T-bet is required for the generation of a CD11chigh subset of ABC-like B cells, and is dispensable for memory and plasma cell generation and antiviral control.26 T-bet expression was also detected in B-cell malignancies, including chronic lymphocytic leukemia (CLL), a disease of mature B cells with a highly heterogeneous course.28,29 However, its role and potential pathological function remain largely unexplored.
Here, we explored the role of T-bet in CLL using single-cell resolved multi-omics analyses of patient samples and a CLL mouse model with a Tbx21 knockout. We show that T-bet acts as a tumor suppressor in CLL by reducing proliferation and is associated with longer survival of patients.
Methods
Patient samples, mouse models, cell lines, and published data sets
Patient and healthy, age-matched control samples were obtained after approval of the study protocols by the local ethics committees according to the Declaration of Helsinki, and after obtaining informed consent of patients. Patients met standard diagnosis criteria for CLL. The details are provided in supplemental Table 1, available on the Blood website. The Eμ-TCL1 mouse model was the basis to generate Tbx21–/– TCL1 cells as previously described.30 A list of all cell lines is provided in supplemental Table 2, and an overview of the published data sets used in this study is provided in supplemental Table 3.
A detailed description of all methods is provided in the supplemental Methods.
Results
CLL cells have enhanced TBX21 expression
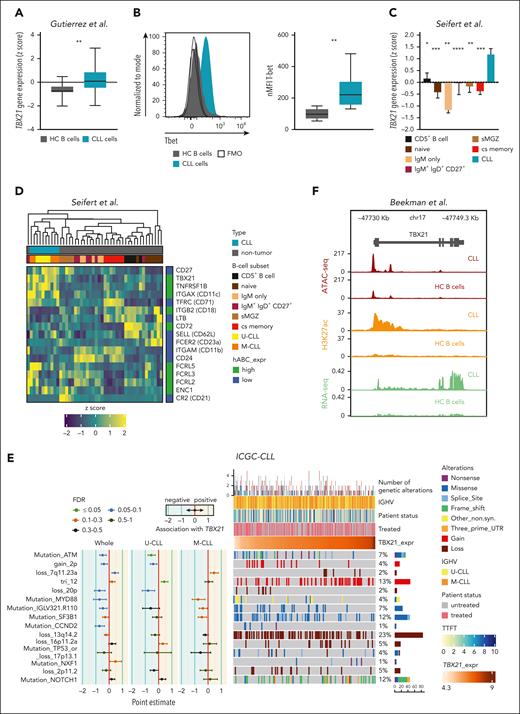
We first compared TBX21 expression levels in CLL cells and untransformed B cells of age-matched healthy controls, which revealed higher T-bet transcript and protein levels in CLL cells (Figure 1A-B). This was also the case when comparing CLL cells with several developmental states of untransformed B cells, including mature CD5+ and memory B cells, which have been suggested as the cellular origin of CLL (Figure 1C; supplemental Figure 1A),31,32 suggesting that high TBX21 expression in CLL cells is disease-specific.
TBX21 expression is higher in CLL cells than in B cells of healthy donors. (A) Gene expression of TBX21 in CLL cells (n = 41) and B cells from age-matched healthy controls (HC B cells; n = 11). P values were obtained by the unpaired t test. (B) Flow cytometric analysis of T-bet levels in CLL cells (n = 20) and B cells from age-matched healthy controls (n = 5). P values were obtained by the unpaired t test. (C) Gene expression of TBX21 in untransformed B-cell subsets (n = 5-7) and CLL cells (n = 10). Bar limits indicate mean expression and error bars indicate standard error of the mean. P values were obtained by the 1-way analysis of variance (ANOVA), controlling the false discovery rate (FDR) using the Benjamini-Hochberg (BH) method. (D) Expression of ABC marker genes in CLL cells (n = 10) and untransformed B-cell subsets (n = 5-7). High (green) and low (blue) expressions in the ABCs are depicted on the right. (E) Analysis of the association between TBX21 gene expression and the presence of specific driver genetic alterations. Point estimates with 95% confidence intervals were calculated for the whole CLL cohort and IGHV subtypes using 2-sided t tests and controlling the FDR using the BH method. The point estimates represent the difference between the mean TBX21 expression in individuals with CLL with and without each corresponding alteration. The point estimates were color-coded based on FDR. The OncoPrint shows the association of genetic driver alterations with higher or lower expression of TBX21, along with additional clinical information such as IGHV status, time to first treatment, and patient status (treated/untreated). Samples are ordered from lower to higher TBX21 gene expression. Monoclonal B lymphocytosis cases are excluded from this analysis. Genetic driver alterations are depicted using distinct colors corresponding to the alteration type. The number of samples with mutations, as well as the percentage of mutated samples over the whole cohort, is shown on the right. The analyzed data set consisted of gene expression microarray data from 364 CLL samples.33 (F) Chromatin landscape of TBX21 showing the median ATAC-seq, H3K27ac chromatin immunoprecipitation sequencing and positive-strand RNA-seq levels from 7 patients with CLL and 15 samples from 4 different B-cell subpopulations of healthy controls (naïve, germinal center, memory B cells, and plasma cells). ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
TBX21 expression is higher in CLL cells than in B cells of healthy donors. (A) Gene expression of TBX21 in CLL cells (n = 41) and B cells from age-matched healthy controls (HC B cells; n = 11). P values were obtained by the unpaired t test. (B) Flow cytometric analysis of T-bet levels in CLL cells (n = 20) and B cells from age-matched healthy controls (n = 5). P values were obtained by the unpaired t test. (C) Gene expression of TBX21 in untransformed B-cell subsets (n = 5-7) and CLL cells (n = 10). Bar limits indicate mean expression and error bars indicate standard error of the mean. P values were obtained by the 1-way analysis of variance (ANOVA), controlling the false discovery rate (FDR) using the Benjamini-Hochberg (BH) method. (D) Expression of ABC marker genes in CLL cells (n = 10) and untransformed B-cell subsets (n = 5-7). High (green) and low (blue) expressions in the ABCs are depicted on the right. (E) Analysis of the association between TBX21 gene expression and the presence of specific driver genetic alterations. Point estimates with 95% confidence intervals were calculated for the whole CLL cohort and IGHV subtypes using 2-sided t tests and controlling the FDR using the BH method. The point estimates represent the difference between the mean TBX21 expression in individuals with CLL with and without each corresponding alteration. The point estimates were color-coded based on FDR. The OncoPrint shows the association of genetic driver alterations with higher or lower expression of TBX21, along with additional clinical information such as IGHV status, time to first treatment, and patient status (treated/untreated). Samples are ordered from lower to higher TBX21 gene expression. Monoclonal B lymphocytosis cases are excluded from this analysis. Genetic driver alterations are depicted using distinct colors corresponding to the alteration type. The number of samples with mutations, as well as the percentage of mutated samples over the whole cohort, is shown on the right. The analyzed data set consisted of gene expression microarray data from 364 CLL samples.33 (F) Chromatin landscape of TBX21 showing the median ATAC-seq, H3K27ac chromatin immunoprecipitation sequencing and positive-strand RNA-seq levels from 7 patients with CLL and 15 samples from 4 different B-cell subpopulations of healthy controls (naïve, germinal center, memory B cells, and plasma cells). ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
We next explored potential similarities between CLL cells and ABCs by analyzing typical ABC marker genes,13,26,34 revealing a particularly high expression of TBX21, ITGAX (CD11c), and FCRL5, accompanied by low expression levels of MS4A1 (CD20) (Figure 1D) in CLL cells, suggesting a phenotypical overlap with ABCs.
We further compared TBX21 expression in genetically defined, prognostic subgroups of patients with CLL and did not observe major differences in cases with or without deletion of chromosome 13q (del13q) or somatic hypermutations of the IGHV gene locus, 2 commonly assessed prognostic features in CLL (Figure 1E). In contrast, patients with trisomy 12, associated with an intermediate prognosis,35 showed a significantly higher expression of TBX21, whereas in patients with ATM mutation, a driver of more aggressive disease,36TBX21 expression was significantly lower than in cases without these aberrations.
To infer whether the expression of TBX21 is epigenetically imprinted in CLL cells, we compared chromatin accessibility by assay for transposase-accessible chromatin (ATAC)-seq and H3K27-acetylation (H3K27ac) of the TBX21 gene locus in CLL vs healthy control B cells.37 This demonstrated that CLL cells not only show a higher transcriptional activity in TBX21 but also higher signs of epigenetic activation and chromatin accessibility in comparison to B cells or B-cell subsets from healthy controls (Figure 1F; supplemental Figure 1B-C).
Inflammatory signals drive TBX21 expression in CLL cells via NF-κB activity
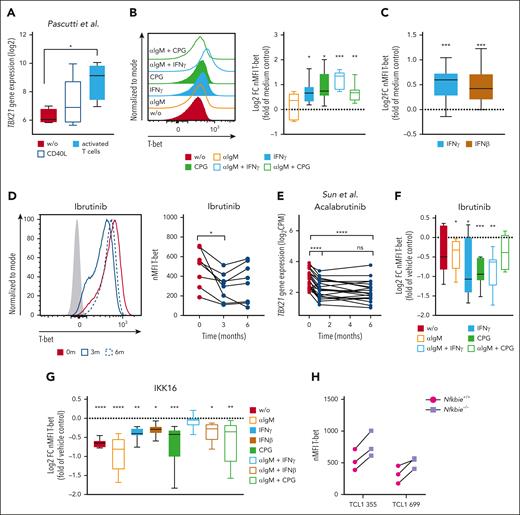
To explore the signals that induce TBX21 expression in CLL cells, we used data of CLL long-term cultures, including so-called nurse-like cells and T cells,38 as well as cocultures of CLL cells with in vitro–activated T cells.39,40 In both cultures, TBX21 expression was induced in the CLL cells (Figure 2A; supplemental Figure 2A). Through in vitro stimulation of CLL cells, we identified interferon gamma (IFN-γ), IFN-β, and CpG oligos as TLR9 ligands as TBX21-inducing signals, as well as combinations of these stimuli with BCR activation by αIgM (Figure 2B-C; supplemental Figure 2B-C). These data suggest that the inflammatory milieu in CLL is responsible for enhanced TBX21 expression in malignant B cells.41,42
Inflammatory signals from activated T cells drive TBX21 expression in CLL cells via NF-κB. (A) Log2-transformed TBX21 gene expression in CLL cells (n = 5) cultured alone, after coculture with CD40L-expressing fibroblasts, or in vitro–activated autologous T cells. P values were obtained by RM 1-way ANOVA and controlling the FDR using the BH method. (B-C) Flow cytometric analysis of T-bet levels in CLL cells. (B) CLL peripheral blood mononuclear cells (PBMCs) (n = 7) or (C) purified CLL cells (n = 11) were stimulated with various cytokines and combinations thereof. The induction of T-bet expression was compared with that in the medium control. P values were obtained by 1-sample t tests and Wilcoxon signed-rank tests and by controlling the FDR using the BH method. (D) Flow cytometric analysis of T-bet levels in CLL cells of patients before ibrutinib treatment and after 3 and 6 months of ibrutinib treatment (n = 8). P values were obtained by RM 1-way ANOVA and controlling the FDR using the BH method. (E) TBX21 gene expression in CLL cells of patients before acalabrutinib treatment and after 1 and 6 months of acalabrutinib treatment (n = 20). P values were obtained by RM 1-way ANOVA and controlling the FDR using the BH method. (F-G) Flow cytometric analysis of T-bet levels in CLL cells after stimulation of CLL PBMCs with various cytokines and combinations thereof. (F) Cells (n = 7) were stimulated in the presence of vehicle control or ibrutinib. Quantification displays log2FC in comparison to the vehicle control. P values were obtained by RM 1-way ANOVA and controlling the FDR using the BH method. (G) Purified CLL cells (n = 8) were stimulated in the presence of the vehicle control or the NF-κB inhibitor IKK-16. Quantification displays log2FC in comparison with vehicle control. P values were obtained using the Friedman test and controlling the FDR using the BH method. (H) T-bet levels of 2 individual TCL1 CLL clones harboring hyperactive NF-κB signaling (Nfkbie–/–) compared with WT controls, as analyzed by flow cytometry (n = 3 technical replicates). ns, not significant. ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Inflammatory signals from activated T cells drive TBX21 expression in CLL cells via NF-κB. (A) Log2-transformed TBX21 gene expression in CLL cells (n = 5) cultured alone, after coculture with CD40L-expressing fibroblasts, or in vitro–activated autologous T cells. P values were obtained by RM 1-way ANOVA and controlling the FDR using the BH method. (B-C) Flow cytometric analysis of T-bet levels in CLL cells. (B) CLL peripheral blood mononuclear cells (PBMCs) (n = 7) or (C) purified CLL cells (n = 11) were stimulated with various cytokines and combinations thereof. The induction of T-bet expression was compared with that in the medium control. P values were obtained by 1-sample t tests and Wilcoxon signed-rank tests and by controlling the FDR using the BH method. (D) Flow cytometric analysis of T-bet levels in CLL cells of patients before ibrutinib treatment and after 3 and 6 months of ibrutinib treatment (n = 8). P values were obtained by RM 1-way ANOVA and controlling the FDR using the BH method. (E) TBX21 gene expression in CLL cells of patients before acalabrutinib treatment and after 1 and 6 months of acalabrutinib treatment (n = 20). P values were obtained by RM 1-way ANOVA and controlling the FDR using the BH method. (F-G) Flow cytometric analysis of T-bet levels in CLL cells after stimulation of CLL PBMCs with various cytokines and combinations thereof. (F) Cells (n = 7) were stimulated in the presence of vehicle control or ibrutinib. Quantification displays log2FC in comparison to the vehicle control. P values were obtained by RM 1-way ANOVA and controlling the FDR using the BH method. (G) Purified CLL cells (n = 8) were stimulated in the presence of the vehicle control or the NF-κB inhibitor IKK-16. Quantification displays log2FC in comparison with vehicle control. P values were obtained using the Friedman test and controlling the FDR using the BH method. (H) T-bet levels of 2 individual TCL1 CLL clones harboring hyperactive NF-κB signaling (Nfkbie–/–) compared with WT controls, as analyzed by flow cytometry (n = 3 technical replicates). ns, not significant. ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
A common feature of IFN-, TLR-, and BCR-signaling is the subsequent activation of NF-κB. Therefore, we hypothesized that TBX21 expression in CLL is induced by NF-κB.43-45 In line, gene expression of TBX21 was reduced in patients with CLL during treatment with the Bruton tyrosine kinase (BTK) inhibitors ibrutinib46,47 and acalabrutinib,48 which among other signaling pathways are known to disrupt NF-κB activity46,49 (Figure 2D-E; supplemental Figure 2D). Furthermore, ibrutinib-mediated reduction in NF-κB signaling prevented inflammation-associated induction of T-bet in vitro (Figure 2F; supplemental Figure 2E). In contrast, neither a 7-day treatment of patients with CLL with the spleen tyrosine kinase inhibitor entospletinib (supplemental Figure 2F) affecting downstream protein kinase B (AKT) signaling50-52 nor a 48-hour treatment of CLL cells in vitro with the Src inhibitor dasatinib altered the expression of TBX21 (supplemental Figure 2G).53
Next, we measured NF-κB p65 phosphorylation after stimulation with microenvironmental factors to experimentally validate the dependency of TBX21 expression on NF-κB activity in CLL. As expected, CpG stimulation and the combination of αIgM and IFNγ showed increased total amounts of p65, and higher levels of p65 phosphorylation (supplemental Figure 2H-I). Inhibition of NF-κB signaling by IKK-16 reduced basal T-bet expression and prevented the induction of T-bet in response to NF-κB–dependent microenvironmental signals in CLL cells (Figure 2G). In line, Nfkbie–/– TCL1 leukemic cells, which are derived from the Eμ-TCL1 mouse model of CLL and harbor hyperactive NF-κB signaling,54 showed higher expression of T-bet than wild-type (WT) control cells (Figure 2H), confirming the importance of NF-κB in the induction of TBX21 expression.
T-bet activity can be assessed by its target gene expression signature
The regulatory activity of T-bet on transcriptional programs in T cells is well described;55 however, the transcriptional targets of T-bet in B cells are vastly unknown. Using the CRISPR/Cas9 approach, we generated Tbx21–/– TCL1 cells (supplemental Figure 3A),30 analyzed them by RNA sequencing (RNA-seq) and mass spectrometry (MS) in comparison to WT control cells, and identified differentially expressed genes (DEGs) and proteins (supplemental Tables 4-5), respectively. We further stratified the RNA-seq data of sorted CLL cells from patients according to their highest and lowest quartiles of TBX21 expression and analyzed DEGs in TBX21low vs TBX21high cells (supplemental Table 6).33 We then identified the overlap of all DEGs and proteins in these data sets with proteins obtained from the MS data of patients with CLL that correlated with T-bet levels (supplemental Table 7).56 This integrated multi-omics analysis generated a list of 104 genes that displayed a positive (55 genes) or negative (49 genes) correlation with TBX21 expression (supplemental Figure 3B-C; supplemental Table 8). Using this gene list, we explored whether T-bet activity was epigenetically imprinted in CLL by stratifying patients with CLL according to their H3K27ac levels at the TBX21 promoter, which correlated well with TBX21 gene expression (supplemental Figure 3D). We observed a clear correlation between differential gene expression and H3K27ac of T-bet–dependent genes in TBX21low vs TBX21high CLL cases, suggesting epigenetic imprinting of T-bet activity in CLL (supplemental Figure 3E). In addition, we independently validated our signature of T-bet–dependent genes in a published CLL data set, which confirmed that both the inducing and the repressing T-bet activity was higher in TBX21high CLL (supplemental Figure 3F).57
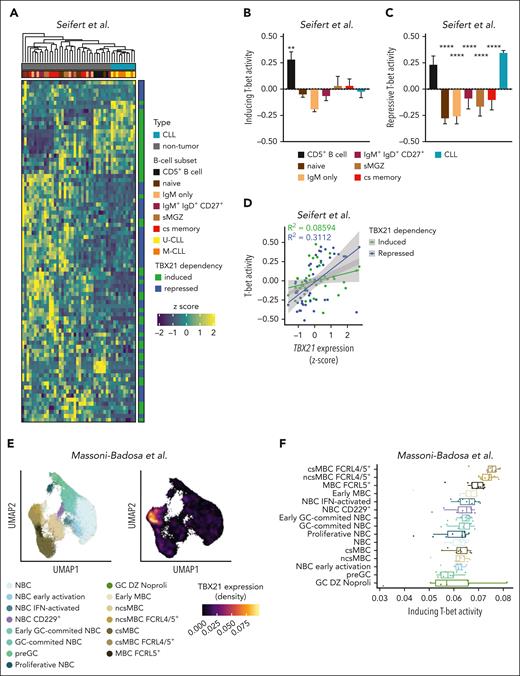
We next aimed to infer whether T-bet activity is specific to CLL cells or mirrored in B-cell subsets of healthy donors by comparing the expression of T-bet–dependent genes using published transcriptome data.31 Intriguingly, T-bet–dependent genes separated CLL samples from all other B-cell subsets; mature CD5+ B cells were most similar to CLL cells (Figure 3A). We further calculated the T-bet activity scores for induced and repressed genes separately. Although no major differences in the inducing activity of T-bet were observed (Figure 3B), the repressive activity of T-bet was the highest in CLL, followed by mature CD5+ B cells (Figure 3C). The correlation between T-bet activity scores and TBX21 gene expression revealed a strong correlation of repressive activity (Figure 3D), suggesting that T-bet acts as a silencing rather than activating TF in CLL. A single-cell omics study recently defined an atlas of B cells in the tonsils,58 including a cell subset annotated as FCRL4+/FCRL5+ memory B cells that showed similarities to ABCs.13,34,59,60 We hypothesized that this B-cell subset is controlled by TBX21 and investigated the T-bet activity scores across all B-cell subsets of the tonsil atlas. Notably, TBX21 expression was highest in FCRL4+/FCRL5+ memory B cells (Figure 3E; supplemental Figure 4A), which was accompanied by the highest T-bet activity according to the CLL–defined target gene signature (Figure 3F). To confirm the robustness of this gene signature, we independently calculated T-bet activity in these data using the recently published pySCENIC tool.61 This confirmed the highest T-bet activity in FCRL4+/FCRL5+ memory B cells (supplemental Figure 4B) and showed that the correlation between activity scores and TBX21 gene expression was similar in both approaches (supplemental Figure 4C-D). Notably, the CLL-defined T-bet activity was mainly driven by a gene module containing NOTCH1, IRF9, and RUNX3 in FCRL4+/FCRL5+ memory B cells (supplemental Figure 4E). Even though most T-bet–repressed genes in CLL also showed low expression in FCRL4+/FCRL5+ memory B cells, an exceptionally high expression of BHLHE41 was observed in this subset, highlighting the distinct regulatory activities of T-bet in malignant vs untransformed B cells.
T-bet has lineage-separating properties in CLL. (A) Expression of T-bet–dependent genes in untransformed B-cell subsets (n = 5-7) and CLL cells (n = 10). (B-C) Activity scores of T-bet were calculated based on (B) induced and (C) repressed genes individually for untransformed B-cell subsets and CLL cells. P values were obtained by 1-way ANOVA and controlling the FDR using the BH method. (D) Correlation between TBX21 gene expression and T-bet activity. P values were obtained using Pearson correlation testing. (E-F) Analysis of B-cell subsets in the human tonsil atlas with a representation of T-bet expression in different clusters. (F) T-bet activity scores in human tonsillar B cells calculated based on induced genes by T-bet in the CLL cells. P values were obtained using the Mann-Whitney test. ∗∗P < .01; ∗∗∗∗P < .0001.
T-bet has lineage-separating properties in CLL. (A) Expression of T-bet–dependent genes in untransformed B-cell subsets (n = 5-7) and CLL cells (n = 10). (B-C) Activity scores of T-bet were calculated based on (B) induced and (C) repressed genes individually for untransformed B-cell subsets and CLL cells. P values were obtained by 1-way ANOVA and controlling the FDR using the BH method. (D) Correlation between TBX21 gene expression and T-bet activity. P values were obtained using Pearson correlation testing. (E-F) Analysis of B-cell subsets in the human tonsil atlas with a representation of T-bet expression in different clusters. (F) T-bet activity scores in human tonsillar B cells calculated based on induced genes by T-bet in the CLL cells. P values were obtained using the Mann-Whitney test. ∗∗P < .01; ∗∗∗∗P < .0001.
In summary, we defined a T-bet–dependent gene expression signature in CLL cells that allowed us to robustly assess T-bet activity in CLL cells.
T-bet regulates transcription via suppression of long-range chromatin interactions
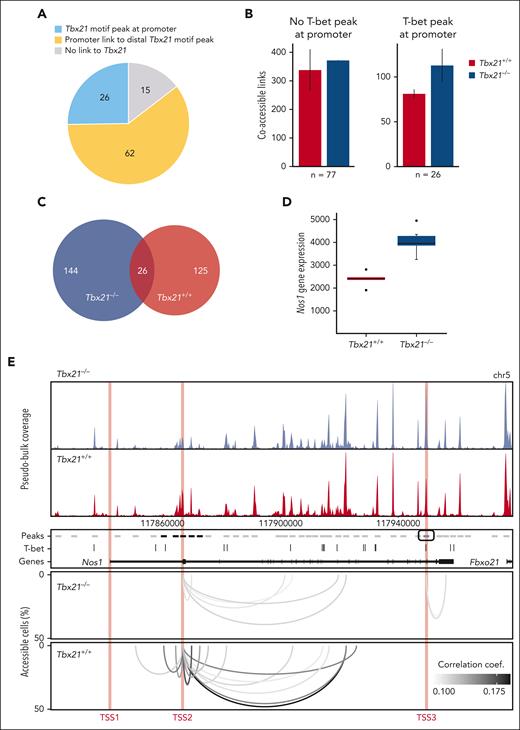
To dissect the mechanisms of transcription regulation by T-bet, we generated single-cell ATAC-seq (scATAC-seq) data for Tbx21+/+ vs Tbx21–/– TCL1 cells using our TurboATAC protocol.62 This protocol provides very high Transposase Tn5 integration efficacy and yielded a mean of ∼58 000 fragments per cell for a high coverage of regulatory elements. We analyzed the genomic location of accessible T-bet binding motifs associated with the T-bet–dependent gene set (supplemental Figures 3C and 5A-D). We identified T-bet binding motifs in 23% of all peaks from the pseudobulk scATAC-seq data, which were subsequently referred to as T-bet peaks. Using RWIRE software, we detected simultaneously accessible peaks in single cells between T-bet-dependent genes and T-bet peaks.63,64 Approximately 26% of T-bet-dependent genes contained an ATAC peak with a T-bet binding motif at their promoter (Figure 4A). These T-bet promoter peaks showed a higher number of coaccessible links to distal peaks in Tbx21–/– vs Tbx21+/+ TCL1 cells (Figure 4B). In contrast, there was no difference in the number of coaccessible links from T-bet–dependent gene promoters without T-bet peak between Tbx21–/– vs Tbx21+/+ TCL1 cells. We concluded that promoter-bound T-bet represses regulatory interactions with distal genomic loci in Tbx21+/+ TCL1 cells. In addition, we were able to link ∼60% of T-bet–dependent genes without a T-bet promoter peak to distal T-bet peaks by coaccessibility analysis of Tbx21–/– and Tbx21+/+ TCL1 cells (Figure 4A). The number of coaccessible links between T-bet-dependent genes and distal T-bet peaks increased in Tbx21–/– (n = 170) vs Tbx21+/+ TCL1 cells (n = 151; Figure 4C). Approximately 15% of the coaccessible links were detected in both Tbx21–/– and Tbx21+/+ TCL1 cells; however, most were unique to either Tbx21–/– or Tbx21+/+ TCL1 cells. These findings point to a rewiring of distal gene regulation in T-bet–dependent genes in Tbx21–/– TCL1 cells. For 15% of the T-bet–dependent genes, no regulatory link to accessible T-bet binding motifs was detected (Figure 4A), which could reflect genes regulated indirectly by T-bet. An example of transcription regulation by T-bet, which involves long-range interactions, is given for Nos1, which is negatively regulated by T-bet (Figure 4D-E, bulk RNA-seq). Different T-bet–dependent regulatory mechanisms appear to be active when comparing 1 kb regions around the 3 transcriptional start sites (TSSs) of Nos1, indicated as TSS1, TSS2, and TSS3 in Figure 4E, TSS1 is inaccessible in Tbx21–/– and Tbx21+/+ TCL1 cells and does not contain a T-bet binding motif. TSS2 is moderately accessible in Tbx21–/– and Tbx21+/+ TCL1 cells but does not contain a T-bet binding motif. It shows increased accessibility in Tbx21+/+ TCL1 cells with additional and enhanced coaccessible links from the promoter to the surrounding T-bet peaks. Thus, the lack of a T-bet binding motif at TSS2 is compensated for by putative repressive interactions with distal regulatory T-bet peaks in Tbx21+/+ TCL1 cells. TSS3 is highly accessible in both Tbx21–/– and Tbx21+/+ TCL1 cells and contains a T-bet motif. A coaccessible link to the downstream peak was found only in Tbx21–/– TCL1 cells. We concluded that the loss of repressive T-bet binding at TSS3 in Tbx21–/– TCL1 cells facilitates the formation of a regulatory link to a downstream peak.
T-bet regulates gene transcription mainly via distal chromatin coaccessibility in CLL cells. (A) T-bet–mediated regulation of T-bet-dependent genes. Promoter with an ATAC peak containing a T-bet binding motif, blue; coaccessible link of the promoter to the distal T-bet peak within 1 Mb, yellow; no link to the T-bet motif peak, gray. (B) Number of coaccessible links from T-bet–dependent gene promoters without (left) or with (right) ATAC peak with T-bet binding motif within a 1 Mb window in Tbx21–/– and Tbx21+/+ TCL1 cells. Whiskers represent the standard error of biological replicates (n = 2). (C) Overlap of coaccessible links from T-bet–dependent genes with distal T-bet peaks within 100 kb in Tbx21–/– and Tbx21+/+ TCL1 cells. The coaccessible links from the biological replicates were merged. (D) Gene expression of Nos1 in Tbx21–/– (n = 6) and Tbx21+/+ TCL1 cells (n = 5) from bulk RNA-seq data. (E) Coaccessibility in Tbx21–/– and Tbx21+/+ TCL1 cells at the T-bet-dependent gene Nos1 region. Browser tracks and coaccessible links from the biological replicates were merged. Top: browser tracks of pseudobulk chromatin accessibility from single cells. Middle: 2 kb regions around peaks from pseudobulk chromatin accessibility with no accessibility change (gray), significantly higher accessibility in Tbx21+/+ TCL1 cells (black), and significantly higher accessibility in Tbx21–/– TCL1 cells (blue); T-bet binding motif positions and gene annotation in black. Bottom: coaccessible links between peaks at Nos1 promoters and distal peaks in Tbx21–/– and Tbx21+/+ TCL1 cells. Promoters of Nos1 (1 kb around the TSS1-3) are marked in red.
T-bet regulates gene transcription mainly via distal chromatin coaccessibility in CLL cells. (A) T-bet–mediated regulation of T-bet-dependent genes. Promoter with an ATAC peak containing a T-bet binding motif, blue; coaccessible link of the promoter to the distal T-bet peak within 1 Mb, yellow; no link to the T-bet motif peak, gray. (B) Number of coaccessible links from T-bet–dependent gene promoters without (left) or with (right) ATAC peak with T-bet binding motif within a 1 Mb window in Tbx21–/– and Tbx21+/+ TCL1 cells. Whiskers represent the standard error of biological replicates (n = 2). (C) Overlap of coaccessible links from T-bet–dependent genes with distal T-bet peaks within 100 kb in Tbx21–/– and Tbx21+/+ TCL1 cells. The coaccessible links from the biological replicates were merged. (D) Gene expression of Nos1 in Tbx21–/– (n = 6) and Tbx21+/+ TCL1 cells (n = 5) from bulk RNA-seq data. (E) Coaccessibility in Tbx21–/– and Tbx21+/+ TCL1 cells at the T-bet-dependent gene Nos1 region. Browser tracks and coaccessible links from the biological replicates were merged. Top: browser tracks of pseudobulk chromatin accessibility from single cells. Middle: 2 kb regions around peaks from pseudobulk chromatin accessibility with no accessibility change (gray), significantly higher accessibility in Tbx21+/+ TCL1 cells (black), and significantly higher accessibility in Tbx21–/– TCL1 cells (blue); T-bet binding motif positions and gene annotation in black. Bottom: coaccessible links between peaks at Nos1 promoters and distal peaks in Tbx21–/– and Tbx21+/+ TCL1 cells. Promoters of Nos1 (1 kb around the TSS1-3) are marked in red.
In summary, T-bet controls the expression of most T-bet–dependent genes by binding to cis-regulatory elements and modulating long-range interactions that enhance the transcription of target genes, as inferred from coaccessibility analysis.
T-bet enhances IFN and represses cell cycle signatures in CLL
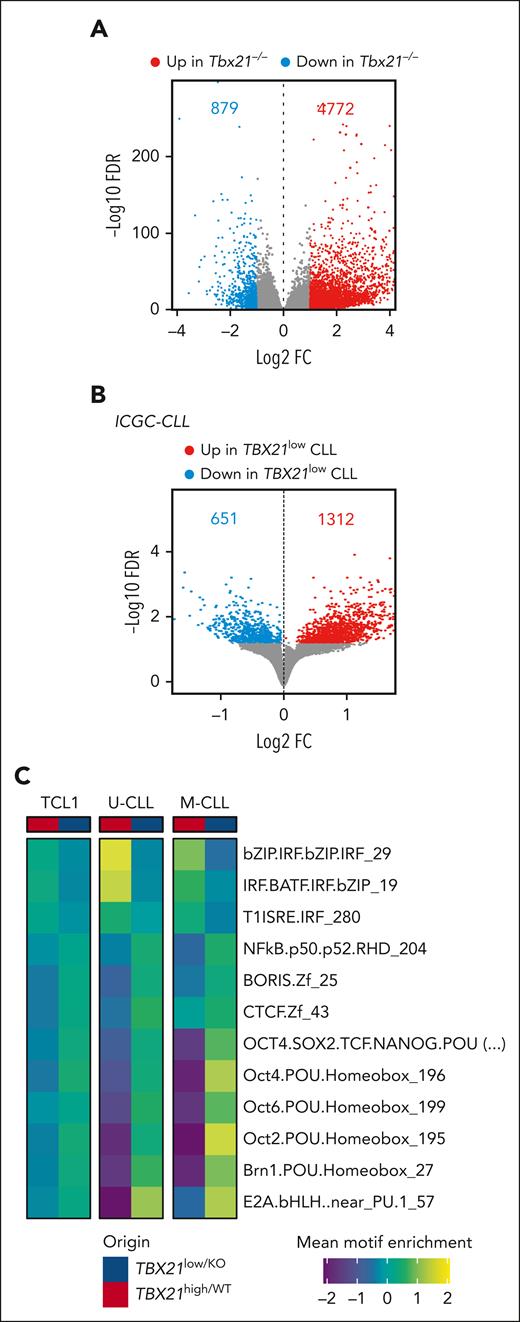
Next, we elucidated the function of TBX21 in CLL cells with a gene set enrichment analysis (GSEA) of published transcriptome data of human CLL cells and our RNA-seq data of Tbx21–/– vs Tbx21+/+ TCL1 cells. DEG sets (false discovery rate <0.05) were computed to identify TBX21-regulated pathways that are conserved across species. In total, 327 gene sets were differentially enriched in human and mouse, with 298 gene sets downregulated and 11 upregulated in TBX21low/KO cells (Figure 5A; supplemental Figure 6A). Interestingly, TBX21low/KO cells showed a lower abundance of IFN-associated pathways and an enrichment of the cell cycle signature (Figure 5A; supplemental Figure 6B). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using T-bet–correlated genes and proteins derived from published data in CLL56 revealed that T-bethigh CLL samples were enriched in B-cell, NF-κB, TLR, and TH17 pathways. In line with the GSEA results, the TBX21low/KO samples were enriched in cell cycle pathways (Figure 5B). To confirm the positive regulation of IFN pathway activity by T-bet, we compared the expression of IFN-stimulated genes (ISGs) in Tbx21–/– and Tbx21+/+ TCL1 cells. At steady state, we noted a higher expression of many ISGs in Tbx21+/+ than in Tbx21–/– TCL1 cells in our RNA-seq data (Figure 5C). Stimulation of these cells with IFNβ in vitro resulted in stronger induction of ISGs in Tbx21+/+ than in Tbx21–/– cells (Figure 5D), confirming the positive regulation of IFN signaling in CLL by T-bet.
T-bet is required for interferon signaling in CLL cells. (A) GSEA of RNA-seq of Tbx21–/– (n = 6) vs Tbx21+/+ (n = 6) TCL1 cells and TBX21low vs TBX21high CLL cells was performed, and commonly regulated gene sets are depicted. (B) KEGG pathway analysis of RNA-seq and MS data of T-betlow vs T-bethigh CLL cells. (C) Basal expression of ISGs in Tbx21–/– compared with Tbx21+/+ TCL1 cells, as analyzed by RNA-seq. (D) Purified Tbx21–/– and Tbx21+/+ TCL1 cells were stimulated in vitro with IFNβ. Log2FC of ISG expression in comparison with the medium control, as analyzed by quantitative reverse transcription polymerase chain reaction. P values were obtained by multiple t tests and controlling the FDR using the BH method. NES, normalized enrichment score. ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
T-bet is required for interferon signaling in CLL cells. (A) GSEA of RNA-seq of Tbx21–/– (n = 6) vs Tbx21+/+ (n = 6) TCL1 cells and TBX21low vs TBX21high CLL cells was performed, and commonly regulated gene sets are depicted. (B) KEGG pathway analysis of RNA-seq and MS data of T-betlow vs T-bethigh CLL cells. (C) Basal expression of ISGs in Tbx21–/– compared with Tbx21+/+ TCL1 cells, as analyzed by RNA-seq. (D) Purified Tbx21–/– and Tbx21+/+ TCL1 cells were stimulated in vitro with IFNβ. Log2FC of ISG expression in comparison with the medium control, as analyzed by quantitative reverse transcription polymerase chain reaction. P values were obtained by multiple t tests and controlling the FDR using the BH method. NES, normalized enrichment score. ∗P ≤ .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
We further characterized the effects of T-bet on chromatin accessibility and T-bet-controlled TFs using our scATAC-seq of Tbx21–/– vs Tbx21+/+ TCL1 cells and ATAC-seq data from TBX21low vs TBX21high patients with CLL. This showed that TBX21low/KO cells harbored a higher accessibility of multiple chromatin sites (+500% in TCL1 CLL and +200% in human CLL cells; Figure 6A-B). Motif enrichment analyses inferred differential TF activities in Tbx21–/– vs Tbx21+/+ TCL1 cells (supplemental Figure 7A) and TBX21low vs TBX21high CLL cells from patients with IGHV-unmutated CLL and mutated CLL (supplemental Figure 7B). This analysis confirmed the most repressive transcriptional activity of T-bet in CLL. Overlap of differential binding motif enrichment of TCL1, unmutated-CLL, and mutated-CLL cells (supplemental Figure 7C-D) identified an enrichment of 3 and a depletion of 9 binding motifs in TBX21high compared with TBX21low cells (Figure 6C; supplemental Figure 7D). Interferon regulatory factor (IRF), IRF and basic leucine zipper ATF-like transcription factor (BATF) coregulation and type-1 IFN-sensitive response element (T1ISRE) motifs were enriched in TBX21high cells (Figure 6C), which is in line with our results above showing that T-bet induces IFN signaling. In contrast, the motifs of NF-κB p50, brother of the regulator of imprinted sites (BORIS), CCCTC-binding factor (CTCF), B-cell TF E2A, and members of the Pit-Oct-Unc (POU) TF family were enriched in TBX21low cells. These TFs were shown to prevent spontaneous apoptosis in CLL65 and enhance the proliferation of B cells,66 which is in line with the negative correlation of T-bet with the cell cycle observed by GSEA and KEGG analysis (Figure 5A-B).
T-bet acts as a silencing TF in CLL cells. (A-B) Differential chromatin accessibility in (A) Tbx21–/– vs Tbx21+/+ TCL1 cells analyzed by scATAC-seq and (B) TBX21low vs TBX21high CLL cells analyzed by ATAC-seq (FDR 0.05). The numbers of up- and downregulated peaks are indicated. (C) Motif enrichment analysis of the ATAC-seq data was performed individually for U-CLL, M-CLL, and TCL1 cells. Commonly enriched motifs are displayed.
T-bet acts as a silencing TF in CLL cells. (A-B) Differential chromatin accessibility in (A) Tbx21–/– vs Tbx21+/+ TCL1 cells analyzed by scATAC-seq and (B) TBX21low vs TBX21high CLL cells analyzed by ATAC-seq (FDR 0.05). The numbers of up- and downregulated peaks are indicated. (C) Motif enrichment analysis of the ATAC-seq data was performed individually for U-CLL, M-CLL, and TCL1 cells. Commonly enriched motifs are displayed.
In summary, our data showed that T-bet maintains an inflammatory program, particularly type 1 IFN signaling, and represses cell cycle signatures in CLL cells.
T-bet suppresses cell proliferation
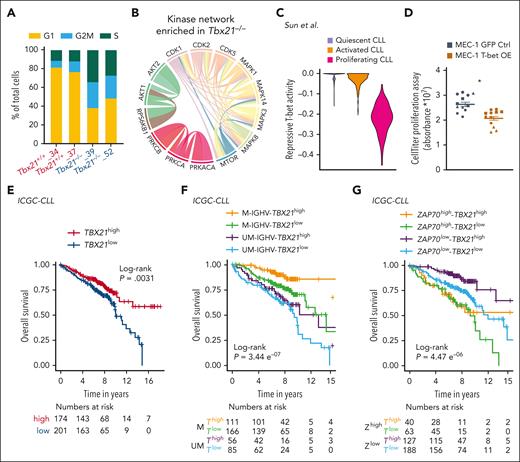
To validate the observed association between T-bet and cell cycle activity, we performed single-cell RNA sequencing (scRNA-seq) of Tbx21–/– vs Tbx21+/+ TCL1 cells. First, we detected a higher fraction of Tbx21–/– cells in the G2/M or S phase than Tbx21+/+ cells (Figure 7A; supplemental Figure 8). Second, by analyzing KI-67 expression in TCL1 cells isolated from the peritoneal cavity, bone marrow, and spleen of leukemic mice, we observed a higher frequency of KI-67+ cells in Tbx21–/– compared with control mice (supplemental Figure 9A). Third, phosphoproteomic analysis of Tbx21–/– vs Tbx21+/+ TCL1 cells revealed an overrepresentation of AKT and protein kinase C networks, as well as an enrichment of the CMGC kinase group (including cyclin-dependent kinases (CDK), mitogen-activated protein kinases (MAPK), glycogen synthase kinases, and CDK-like kinases) in Tbx21–/– cells (Figure 7B; supplemental Figure 9B-C). The latter included important mediators of cell cycle progression, such as CDK1, CDK2, and CDK5, which showed higher activity in the absence of T-bet. In support of this, 13 of the total 104 T-bet–dependent genes, all of which were repressed by T-bet, were associated with cell cycling (supplemental Figure 3C), including Bub1, which is involved in G1/S phase entry and cell cycle progression.67
T-bet represses cell cycling in CLL cells and predicts a good outcome. (A) scRNA-seq of Tbx21–/– and Tbx21+/+ TCL1 cells was performed and single cells were annotated according to their cell cycle phase. (B) Phospho-specific MS analysis of Tbx21–/– and Tbx21+/+ TCL1 cells was performed and kinase networks enriched in Tbx21–/– TCL1 cells are displayed. (C) The T-bet activity score based on repressed genes was calculated from scRNA-seq data of CLL lymph node samples annotated according to their proliferation status. (D) MEC-1 cell lines with inducible overexpression of T-bet or green fluorescent protein (GFP) as control were generated. Using doxycycline, overexpression was induced and expansion of GFP+ control cells and T-bet-overexpressing MEC-1 cells was analyzed using the CellTiter Glo proliferation assay (n = 4 biological replicates á 3 technical replicates). P values were obtained by unpaired t test of the means of biological replicates. (E-G) RNA-seq of patient samples with CLL was performed at diagnosis. Patients with CLL were stratified according to TBX21 mRNA abundance using the maximally selected rank statistics-based cutoff. (E) The OS of TBX21high vs TBX21low patients with CLL was analyzed. (F) Patients with CLL were stratified according to TBX21 mRNA abundance using the maximally selected rank statistics-based cutoff and their IGHV mutational status (M = mutated vs UM = unmutated). The OS of M-TBX21high, M-TBX21low, UM-TBX21high, and UM-TBX21low patients with CLL was analyzed. (G) Patients with CLL were stratified according to TBX21 mRNA abundance using the maximally selected rank and statistics-based cutoff and their ZAP70 gene expression level (ZAP70high vs ZAP70low). The OS of ZAP70high-TBX21high, ZAP70high-TBX21low, ZAP70low-TBX21high, and ZAP70low-TBX21low patients with CLL was analyzed. P values were obtained by log-rank testing. mRNA, messenger RNA. ∗P ≤ .05.
T-bet represses cell cycling in CLL cells and predicts a good outcome. (A) scRNA-seq of Tbx21–/– and Tbx21+/+ TCL1 cells was performed and single cells were annotated according to their cell cycle phase. (B) Phospho-specific MS analysis of Tbx21–/– and Tbx21+/+ TCL1 cells was performed and kinase networks enriched in Tbx21–/– TCL1 cells are displayed. (C) The T-bet activity score based on repressed genes was calculated from scRNA-seq data of CLL lymph node samples annotated according to their proliferation status. (D) MEC-1 cell lines with inducible overexpression of T-bet or green fluorescent protein (GFP) as control were generated. Using doxycycline, overexpression was induced and expansion of GFP+ control cells and T-bet-overexpressing MEC-1 cells was analyzed using the CellTiter Glo proliferation assay (n = 4 biological replicates á 3 technical replicates). P values were obtained by unpaired t test of the means of biological replicates. (E-G) RNA-seq of patient samples with CLL was performed at diagnosis. Patients with CLL were stratified according to TBX21 mRNA abundance using the maximally selected rank statistics-based cutoff. (E) The OS of TBX21high vs TBX21low patients with CLL was analyzed. (F) Patients with CLL were stratified according to TBX21 mRNA abundance using the maximally selected rank statistics-based cutoff and their IGHV mutational status (M = mutated vs UM = unmutated). The OS of M-TBX21high, M-TBX21low, UM-TBX21high, and UM-TBX21low patients with CLL was analyzed. (G) Patients with CLL were stratified according to TBX21 mRNA abundance using the maximally selected rank and statistics-based cutoff and their ZAP70 gene expression level (ZAP70high vs ZAP70low). The OS of ZAP70high-TBX21high, ZAP70high-TBX21low, ZAP70low-TBX21high, and ZAP70low-TBX21low patients with CLL was analyzed. P values were obtained by log-rank testing. mRNA, messenger RNA. ∗P ≤ .05.
Next, we investigated published scRNA-seq data of CLL lymph nodes, in which CLL cells have been annotated to be either quiescent, activated, or proliferating.68 Using our list of T-bet–dependent genes, we observed the lowest repressive T-bet activity in proliferating CLL cells (Figure 7C; supplemental Figure 9D), which is in line with the proposed repressive activity of T-bet for cell cycle activity. Analysis of published data from cells of patients with monoclonal B lymphocytosis, a precursor state of CLL with a yearly progression rate of 1% to 2% of cases,69 revealed no differences in T-bet activity before and after progression to CLL (supplemental Figure 10A), suggesting no major differences in cell cycle activity at the stage of disease progression.70
Finally, to experimentally validate the regulation of the cell cycle by T-bet, we generated a CLL-like MEC-1 cell line and 2 B-cell lymphoma cell lines, OCI-Ly7, and U-2940, with inducible T-bet expression. Overexpression of T-bet upon doxycycline treatment (supplemental Figure 10B-C) resulted in lower expansion and proliferation rates in comparison to either green fluorescent protein–transduced or nontreated control cells (Figure 7D; supplemental Figure 10 D-E). Thus, T-bet plays a central role in suppressing the proliferation of malignant B cells.
In summary, the omics data analyses of TCL1 and human CLL cells, followed by functional assays showed that T-bet activity inhibits malignant B-cell proliferation.
T-bet expression is a marker of good prognosis for CLL
A subset of patients with CLL suffer from disease transformation into the more aggressive Richter syndrome (RS), which often resembles diffuse large B-cell lymphoma (DLBCL) and is associated with a poor outcome with a short overall survival (OS) of less than a year. Recently, a novel mouse model mimicking the transformation of CLL cells into RS cells was published.71 Using this model, we observed higher expression of Tbx21 in CLL vs RS cells (supplemental Figure 11A). In addition, analysis of scRNA-seq data of paired CLL cells and transformed RS cells of 4 patients revealed the lowest repressive T-bet activity in a distinct cluster of proliferating RS cells (supplemental Figure 11B).72 Accordingly, comparing patient samples of CLL and other B-cell non-Hodgkin lymphoma entities revealed the highest repressive T-bet activity in CLL and the lowest in DLBCL (supplemental Figure 11C), which is in line with our observation that T-bet activity limits the proliferation of malignant B cells.
We then assessed the prognostic impact of T-bet in CLL and stratified patients according to their T-bet expression levels acquired by RNA-seq or MS in high- and low-expressing groups. Patients with CLL with high gene expression and protein levels of T-bet showed better outcomes, specifically longer time to treatment and OS, compared with cases with low expression levels (Figure 7E; supplemental Figure 12A-D).33,56,57 The longer treatment-free and OS of patients with CLL with high T-bet expression was independent of their IGHV mutational status or ZAP70 gene expression, which was confirmed by multivariate analysis (Figure 7F-G; supplemental Figure 13A-D).
Altogether, our data identified T-bet as a novel prognostic marker for CLL. Mechanistically, this can be explained by T-bet driving inflammatory processes via IFN signaling and limiting the proliferation of malignant B cells. These novel findings have implications for the stratification and therapy of patients with CLL and likely other B-cell non-Hodgkin lymphoma.
Discussion
Our study provides evidence for a so far unexplored role of T-bet in CLL and provides insights into its transcriptional regulation and activity, as well as its prognostic role. We showed higher T-bet expression in CLL cells than in B cells from healthy donors, and its induction by multiple factors within the tumor microenvironment via NF-κB and likely other pathways. We observed that T-bet acts mostly as a transcription repressor and that its activity maintains IFN signaling and represses the cycling of CLL cells. Therefore, targeting T-bet or its associated pathways might serve as a novel treatment strategy for CLL.
Because T-bet expression is a feature of most ABCs,5 we compared the phenotypic and functional properties of CLL cells and ABCs to assess whether CLL cells might originate from this B-cell subset. Similarly, as in ABCs, T-bet expression is induced in CLL cells by inflammatory signals present in the micromilieu of CLL through BCR, TLR, and IFN stimulation.6,24,73 In addition, we detected the expression of several ABC marker genes in CLL but not in nonmalignant B cells. CLL cells are suggested to be derived from self-reactive B-cell precursors; however, they do not secrete autoantibodies, which is a typical feature of ABC.6,74,75 Furthermore, although T-bet expression in ABCs is regulated by STAT signaling, we identified NF-κB as a key mediator of T-bet expression in CLL cells.24 Thus, the regulatory properties of T-bet induction in malignant and nonmalignant B cells are likely distinct. To ultimately assess whether CLL cells originate from ABCs, further cell-of-origin modeling comparing different subsets of CD11c+ untransformed B cells and CLL cells will be necessary.
Our in vitro and in vivo data showed that BTK inhibitors reduced T-bet expression in CLL cells, which is in line with the negative effect of these drugs on NF-κB activity.46,49 However, the overall clinical efficacy of BTK inhibition is clearly independent of the altered expression or activity of T-bet.
Type I IFN in the tumor microenvironment is known to suppress tumor growth, but in CLL, the response to IFN differs between the good and bad prognostic subgroups. In low-risk patients, IFN signaling is associated with growth arrest, whereas in aggressive CLL, IFN promotes tumor growth.76 This might be explained by recent findings suggesting that aggressive forms of CLL are hypersensitive to autocrine IFN signaling.77 In light of our findings that T-bet induces IFN signaling and limits proliferation, this implies that the chronic inflammatory micromilieu that is mediated by T-bet activity in CLL, is a characteristic of indolent disease with a low proliferative rate, in contrast to low T-bet activity in aggressive lymphomas such as RS. Notably, clinical IFNα treatment showed overall limited efficacy, but the best responses were noted in a subset of previously untreated, early-stage CLL,78 which possibly could resemble patients with CLL with high T-bet activity.
Our bulk ATAC-seq analysis of a large cohort of patients with CLL combined with scATAC-seq analysis of the CLL mouse model revealed a remarkable reduction in chromatin accessibility and subsequent gene expression by T-bet. This repressive activity of T-bet is in line with published data showing that T-bet represses specific gene programs in B cells.27 Our findings show that T-bet has multiple modes of action to regulate target gene expression, including direct promoter binding, and also controls the regulatory long-range interactions of enhancers. Our finding that T-bet deficiency is associated with an enrichment of binding sites for the POU TF family is in contrast with observations in human CD21low ABC-like B cells that express T-bet and are enriched for POU-binding sites in open chromatin regions,8 highlighting the difference between ABCs and CLL cells. Members of the POU TF family are required for the progression of the cell cycle.66,79 In accordance, the enhanced activity of POU TFs in TBX21low/KO CLL cells was associated with a higher expression of cell cycle-associated gene signatures and higher cell proliferation rates. This is in line with, and might explain, the reduced OS of patients with CLL with lower T-bet expression. Moreover, Penter et al noted a reduced activity of POU TF family members in patients with CLL in comparison to the more aggressive RS,80 which is consistent with our finding of reduced T-bet activity in RS and in more aggressive types of lymphoma such as DLBCL.
In summary, we showed that T-bet is induced in CLL cells by microenvironmental signals present in lymphoid tissues via NF-κB signaling. It acts as a tumor suppressor by maintaining IFN signaling and repressing the cell cycle. As a consequence, T-bet expression levels are positively correlated with longer survival of patients with CLL, which has implications for clinical applications.
Toward this goal, specific immune stimulatory agents that lead to the induction of T-bet in CLL cells need to be evaluated in preclinical models. Notably, such compounds likely also have a positive impact on other immune cells, thereby improving cancer-directed immune responses, which could result in an additive multitarget therapeutic efficacy.
Acknowledgments
The authors are grateful to Verena Kalter, Norman Mack, and Sibylle Ohl for their very valuable technical assistance, and to Ka Hou Man for sharing experimental expertise. The authors thank Dan Landau, who provided raw RNA sequencing data of ibrutinib-treated patients with chronic lymphocytic leukemia. The authors thank the entire team of the Proteomics Core Facility of the DKFZ for support with the proteomic sample analysis, and the Core Facility Cytometry of the Medical Faculty at Ulm University for providing technical and expert support. The visual abstract was created with BioRender.com.
P.M.R. has been funded by a fellowship from the German Cancer Research Center Clinician Scientist Program, supported by the Dieter Morszeck Foundation. M.C. and H.B. have been supported by the DKFZ International PhD Program Fellowship. This project was funded by the Deutsche Krebshilfe (M. Seiffert). T.N. was supported by the Bundesministerium für Bildung und Forschung (grant agreement no. 161L0212E). D.G.E. received funding from the Italian Foundation for Cancer Research (Fondazione AIRC) project IG 2020, ID. 24566. S.S. and C. Schneider were supported by SFB1074 subproject B1, and K.R. by subproject Z1 of the German Research Foundation (DFG). The used instrumentation of the Core Facility Cytometry is funded by DFG (grants 162388165 and 68236468). C. Sun and A.W. are supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Authorship
Contribution: P.M.R. conceptualized the study, designed and performed experiments, analyzed and interpreted data, generated figures, and wrote the manuscript; I.S. analyzed and interpreted data, generated figures, and wrote parts of the manuscript; V.C. analyzed and interpreted data, generated figures; R.J. and H.B. performed experiments, analyzed data, and generated figures; R.M.-B., P.B., and T.R. performed the bioinformatics analyses and generated figures; J.B. and H.B. performed the experiments and analyzed the data; L.A., M.C., S.C., A.M.V., M. Sivina, M.M., and S.R.-R. performed experiments; A.B. provided experimental materials; S.A.H., M.Z., C. Sun, H.K., T.N, P.-M.B., F.C., E.t.H., M. Schneider, and D.H. performed bioinformatics analyses or proteome analyses; D.Y.Y., J.K., A.V.D., M.B., K.H., C. Schneider, S.S., and A.W. provided patient samples, clinical information, or data sets; J.P.M. provided expertise as well as materials for single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq); J.A.B., D.G.E., and P.L. provided oversight and logistic and budget support for the generation of experimental materials; S.D. provided patient samples and clinical information, access to patient data, and intellectual feedback to the manuscript; J.I.M.-S. and K.R. provided oversight, budget support, intellectual feedback, and wrote parts of the manuscript; M. Seiffert supervised the study, provided logistic and budget support, and wrote the manuscript; and all the authors read, reviewed, and revised the manuscript.
Conflict-of-interest disclosure: C.S. received research funding from Genmab. A.V.D. received consulting fees from AbbVie, AstraZeneca, BeiGene, Bristol Meyers Squibb, Genentech, Genmab, Incyte, Janssen, Lilly Oncology, MEI Pharma, Nurix, Oncovalent, Pharmacyclics, and TG Therapeutics; and has ongoing research funding from AbbVie, AstraZeneca, Bayer Oncology, Bristol Meyers Squibb, Cyclacel, Lilly Oncology, MEI Pharma, Nurix, and Takeda Oncology. A.W. received research support from Pharmacyclics LLC, an AbbVie Company, Acerta Pharma, a member of the AstraZeneca group, Merck, Nurix, Verastem, and Genmab. J.A.B. received research funding from Pharmacyclics LLC and BeiGene; served on the advisory board for Janssen, Gilead, TG Therapeutics, Pharmacyclics LLC, BeiGene, and Novartis. The remaining authors declare no competing financial interests.
Correspondence: Martina Seiffert, Molecular Genetics, German Cancer Research Center, Im Neuenheimer Feld 280, Heidelberg 69120, Germany; email: m.seiffert@dkfz.de.
References
Author notes
Bulk gene expression data were deposited in the ArrayExpress repository of the European Bioinformatics Institute (https://www.ebi.ac.uk/biostudies/arrayexpress/studies; accession number E-MTAB-13030). The scRNA-seq and scATAC-seq data are available from the Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/) under (accession number GSE234226). Previously published sequencing data that were used in the analysis are listed in supplemental Table 3. GEO data (accession number GSE22529,81 GSE36907,31 and GSE5057239) were analyzed and derived from R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). Gene expression of Tbx21 in a mouse model of matched CLL and Richter syndrome (GSE186137) was analyzed after the transfer of cells in wild-type animals.71 Custom code used is available from GitHub at https://github.com/RippeLab/RWire-IFN, https://github.com/tnaake/TBET_in_CLL, and https://github.com/massonix/Tbet_in_CLL.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal