“Multilineage” vs “lymphoid-only” BCR::ABL1 involvement and distinct cooperating events determine gene expression in BCR::ABL1-positive ALL.
Outcome with recent GMALL protocols is similar for BCR::ABL1 lineage clusters, but inferior for an IKZF1-/- enriched “lymphoid” subcluster.
Visual Abstract
Distinct diagnostic entities within BCR::ABL1-positive acute lymphoblastic leukemia (ALL) are currently defined by the International Consensus Classification of myeloid neoplasms and acute leukemias (ICC): “lymphoid only”, with BCR::ABL1 observed exclusively in lymphatic precursors, vs “multilineage”, where BCR::ABL1 is also present in other hematopoietic lineages. Here, we analyzed transcriptomes of 327 BCR::ABL1-positive patients with ALL (age, 2-84 years; median, 46 years) and identified 2 main gene expression clusters reproducible across 4 independent patient cohorts. Fluorescence in situ hybridization analysis of fluorescence-activated cell-sorted hematopoietic compartments showed distinct BCR::ABL1 involvement in myeloid cells for these clusters (n = 18/18 vs n = 3/16 patients; P < .001), indicating that a multilineage or lymphoid BCR::ABL1 subtype can be inferred from gene expression. Further subclusters grouped samples according to cooperating genomic events (multilineage: HBS1L deletion or monosomy 7; lymphoid: IKZF1-/- or CDKN2A/PAX5 deletions/hyperdiploidy). A novel HSB1L transcript was highly specific for BCR::ABL1 multilineage cases independent of HBS1L genomic aberrations. Treatment on current German Multicenter Study Group for Adult ALL (GMALL) protocols resulted in comparable disease-free survival (DFS) for multilineage vs lymphoid cluster patients (3-year DFS: 70% vs 61%; P = .530; n = 91). However, the IKZF1-/- enriched lymphoid subcluster was associated with inferior DFS, whereas hyperdiploid cases showed a superior outcome. Thus, gene expression clusters define underlying developmental trajectories and distinct patterns of cooperating events in BCR::ABL1-positive ALL with prognostic relevance.
Introduction
BCR::ABL1-positive acute lymphoblastic leukemia (ALL) is a high-risk disease subtype in children and adults,1,2 treated with tyrosine-kinase inhibitors (TKIs) combined with dose-reduced chemotherapy regimens, including allogenic stem cell transplantation (SCT).2,3 Combinations of TKIs and the bispecific antibody blinatumomab provide further promising results in clinical trials.4,5 However, dependency on lineage-specific targets poses the risk of lineage infidelity as a resistance mechanism in particular for lineage-restricted immunotherapies.6 We and others described 2 developmental trajectories in BCR::ABL1-positive ALL with either a lymphoid or a stem cell origin.6-9 The ICC classification of myeloid neoplasms and acute leukemia defines BCR::ABL1-positive ALL accordingly as “lymphoid only” or “multilineage,”10 but diagnostic standards are not yet established. In addition, the leukemia-intrinsic biology of these subtypes and their clinical phenotype remain poorly understood.
Study design
We analyzed 327 BCR::ABL1-positive ALL patient samples (age, 2-84 years; median, 46 years) from 4 cohorts11-14 by transcriptomic (RNA sequencing [RNA-Seq]15: n = 327) and genomic profiling (whole-genome sequencing12: n = 61/single-nucleotide polymorphism array16: n = 102) and BCR::ABL1 fluorescence in situ hybridization on fluorescence-activated cell-sorted hematopoietic populations6 (supplemental Table 1 [available on the Blood website]). Machine learning–based gene expression analysis15 was used to define in our GMALL cohort BCR::ABL1-ALL gene expression clusters, reproducibly validated in 3 external cohorts.12-14 Long-read RNA-Seq17 and single-cell assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-Seq)/RNA-Seq (10× Genomics, Pleasanton, CA) were used to confirm cluster-specific gene isoform expressions. Immunophenotyping and measurement of minimal residual disease were performed in central reference laboratories.18,19 Clinical outcome of BCR::ABL1-positive ALL clusters was analyzed in 98 evaluable adult patients treated according to GMALL protocols with dose-reduced chemotherapy induction combined with imatinib, followed by consolidation I with continuous imatinib treatment and indication for allogeneic SCT in first complete remission (CR)3 (supplemental Appendix).
The GMALL studies (NCT02872987, NCT02881086) reported here were approved by the ethical committees of the participating centers, and patients gave written informed consent for biological research.
Results and discussion
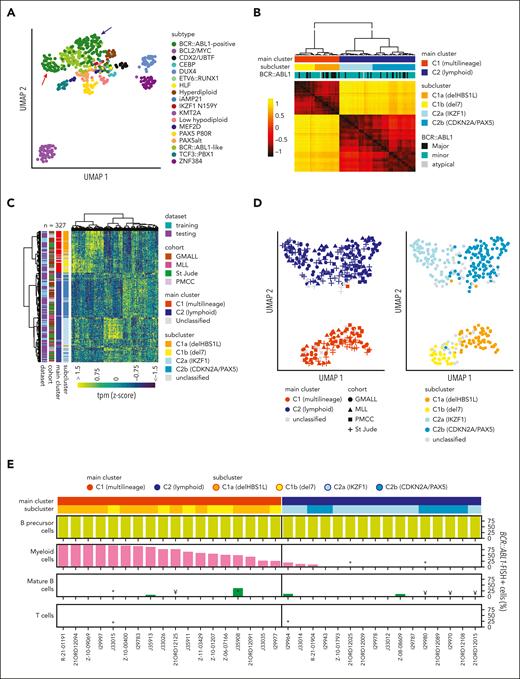
Transcriptomic profiles of B-cell precursor ALL (BCP-ALL) define molecular disease subtypes.15 Unsupervised analysis of subtype-specific gene expression in our GMALL cohort (n = 493) revealed 2 distinct main clusters within BCR::ABL1-positive ALL (Figure 1A). For validation, we obtained sample-to-sample distances from systematic variation of uniform manifold approximation and projection (UMAP) parameters on gene expression profiles of BCR::ABL1-positive GMALL samples (supplemental Figure 1). Unsupervised clustering of these distances confirmed 2 main clusters (C1, C2) and 2 subclusters (C1a, C1b, C2a, and C2b) within each of the main clusters (Figure 1B), which could be robustly predicted by a machine learning–based classifier (supplemental Figure 2). We used the underlying gene set definition (n = 331 genes; supplemental Tables 2-6) for unsupervised clustering of an aggregated data set of all cohorts (n = 32711-14; Figure 1C), which confirmed the cluster separation (Figure 1D). Differential gene expression analysis provided complex transcriptomic signatures for the 2 main and 4 subclusters (supplemental Figure 3; supplemental Tables 7-11), indicating underlying biological differences. BCR::ABL1 fluorescence in situ hybridization on diagnostic samples from 34 adult patients (supplemental Figure 4) based on fluorescence-activated cell-sorted6 hematopoietic compartments (myeloid, lymphoid precursor, and mature B/T cells) identified BCR::ABL1 in 28% to 99% of myeloid cells in 18 of 18 samples from C1 (therefore termed: multilineage), whereas 13 of 16 samples from C2 showed BCR::ABL1 exclusively in lymphoid precursors or mature B cells (termed lymphoid; P < .001; Figure 1E; supplemental Table 12). Thus, ICC-defined BCR::ABL1 multilineage vs lymphoid-only ALL subtypes are characterized by clearly distinct gene expression profiles.
Developmental trajectories of BCR::ABL1-positive ALL can be determined by gene expression. (A) Uniform manifold approximation and projection (UMAP) plot shows unsupervised clustering of 493 BCP-ALL patients (GMALL study group) based on 2802 genes previously established15 for allocation to 21 molecular disease subtypes. A total of 18 subtypes represented in this adult cohort are shown. Arrows indicate separation of BCR::ABL1-positive patients into 2 distinct clusters. (B) BCR::ABL1-positive samples from this cohort (n = 113) were reanalyzed by UMAP analysis with systematic variation of 30 setting combinations for the parameters “min_dist” and “n_neighbors” (supplemental Figure 1). Sample-to-sample distances for each setting were calculated, z-transformed, and averaged. Hierarchical clustering of the averaged distances is shown. To define the final number of clusters, the dendrogram was progressively split at each junction and the integrity of the resulting clusters was determined using machine learning (SVM linear). When the predictability (Cohen κ) of a cluster decreased below 0.8, no further cluster splitting was performed (for details, see supplemental Figure 2). This resulted in 2 main clusters (C1 and C2) with 4 subclusters (C1a, C1b, C2a, C2b), which could be reliably predicted. (C) To test whether similar clusters were present in other cohorts, 2 machine learning classifiers (1 for the 2 main clusters and 1 for the 4 subclusters) were trained on the basis of 178 and 331 LASSO genes, respectively, derived from the GMALL discovery cohort (supplemental Tables 2-6). Gene expression data from validation cohorts (Munich Leukemia Laboratry (MLL), n = 61; St. Jude Children's Research Hospital, n = 104; and Princess Margaret Cancer Centre (PMCC), n = 49) were used for hierarchical clustering together with the GMALL reference cohort after batch correction. Newly established classifiers were used for sample allocation to the 2 main and 4 subclusters (supplemental Table 1), which are shown in the annotation. (D) UMAP plots obtained from the data in panel C, showing the classifier predictions for the main clusters (left) and subclusters (right). (E) Bone marrow/peripheral blood samples at first diagnosis of ALL were fluorescence-activated cell sorted into hematopoietic compartments on cover slides and used for BCR::ABL1 fluorescence in situ hybridization (FISH) (supplemental Figure 4). Bars depict the frequency of BCR::ABL1-positive cells in the corresponding compartments: myeloid cells (CD45lowCD19−CD10-CD34+/−CD13/33+), mature B cells (CD45highCD19+CD10−CD20+), T cells (CD45highCD19-CD3+CD16/65−), or B lymphoid precursor/ALL cells (CD45lowCD19+CD10+; in 1 case with pro-B immunophenotype, ALL cells were only identified by CD45lowCD19+). FISH signal constellations and distribution in analyzed cells are detailed in the supplemental Table 12. Note: ∗less than 100 cells analyzed, ¥less than 50 cells analyzed.
Developmental trajectories of BCR::ABL1-positive ALL can be determined by gene expression. (A) Uniform manifold approximation and projection (UMAP) plot shows unsupervised clustering of 493 BCP-ALL patients (GMALL study group) based on 2802 genes previously established15 for allocation to 21 molecular disease subtypes. A total of 18 subtypes represented in this adult cohort are shown. Arrows indicate separation of BCR::ABL1-positive patients into 2 distinct clusters. (B) BCR::ABL1-positive samples from this cohort (n = 113) were reanalyzed by UMAP analysis with systematic variation of 30 setting combinations for the parameters “min_dist” and “n_neighbors” (supplemental Figure 1). Sample-to-sample distances for each setting were calculated, z-transformed, and averaged. Hierarchical clustering of the averaged distances is shown. To define the final number of clusters, the dendrogram was progressively split at each junction and the integrity of the resulting clusters was determined using machine learning (SVM linear). When the predictability (Cohen κ) of a cluster decreased below 0.8, no further cluster splitting was performed (for details, see supplemental Figure 2). This resulted in 2 main clusters (C1 and C2) with 4 subclusters (C1a, C1b, C2a, C2b), which could be reliably predicted. (C) To test whether similar clusters were present in other cohorts, 2 machine learning classifiers (1 for the 2 main clusters and 1 for the 4 subclusters) were trained on the basis of 178 and 331 LASSO genes, respectively, derived from the GMALL discovery cohort (supplemental Tables 2-6). Gene expression data from validation cohorts (Munich Leukemia Laboratry (MLL), n = 61; St. Jude Children's Research Hospital, n = 104; and Princess Margaret Cancer Centre (PMCC), n = 49) were used for hierarchical clustering together with the GMALL reference cohort after batch correction. Newly established classifiers were used for sample allocation to the 2 main and 4 subclusters (supplemental Table 1), which are shown in the annotation. (D) UMAP plots obtained from the data in panel C, showing the classifier predictions for the main clusters (left) and subclusters (right). (E) Bone marrow/peripheral blood samples at first diagnosis of ALL were fluorescence-activated cell sorted into hematopoietic compartments on cover slides and used for BCR::ABL1 fluorescence in situ hybridization (FISH) (supplemental Figure 4). Bars depict the frequency of BCR::ABL1-positive cells in the corresponding compartments: myeloid cells (CD45lowCD19−CD10-CD34+/−CD13/33+), mature B cells (CD45highCD19+CD10−CD20+), T cells (CD45highCD19-CD3+CD16/65−), or B lymphoid precursor/ALL cells (CD45lowCD19+CD10+; in 1 case with pro-B immunophenotype, ALL cells were only identified by CD45lowCD19+). FISH signal constellations and distribution in analyzed cells are detailed in the supplemental Table 12. Note: ∗less than 100 cells analyzed, ¥less than 50 cells analyzed.
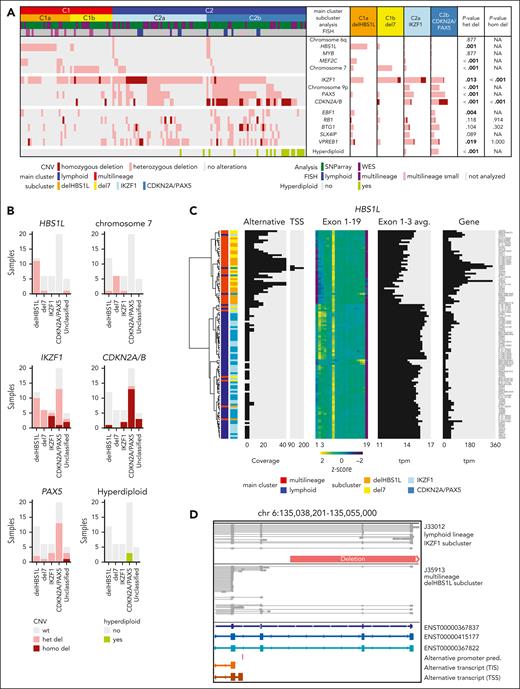
To identify the underpinnings of the subclusters, we obtained genomic profiles of 149 cases of the aggregated cohort (Figure 2A; supplemental Figure 5). Focal deletions in HBS1-like translational GTPase (HBS1L) were exclusively observed in the multilineage cluster C1 with strong enrichment for subcluster C1a (termed “delHBS1L”; P = .001), whereas monosomy 7 was strongly enriched in C1b (“del7”; P < .001). The lymphoid subcluster C2a was enriched for homozygous deletions in IKZF1 (“IKZF1”; P < .001), whereas C2b was enriched for homozygous CDKN2A/B deletions (P < .001), PAX5 deletions (P < .001), and/or hyperdiploidy (“CDKN2A/PAX5”; P < .001). Validation on an external cohort14 confirmed this enrichment pattern of genomic aberrations (Figure 2B), indicating that the 4 gene expression subclusters represent distinct patterns of underlying genomic aberrations.
Cooperating genomic events define gene expression subclusters, including alternative HBS1L isoform expression in delHBS1L and del7 as novel candidate. (A) Distribution of recurrent copy number variants (CNVs) in the 4 BCR::ABL1-positive ALL subclusters. CNVs were assessed in samples with subcluster allocation (GMALL: ground truth; MLL: predictions, excluding n = 14 samples that remained "unclassified" by machine learning classifier for the 2- and/or the 4-cluster definition) by whole-genome sequencing (WGS) (n = 47) or single-nucleotide polymorphism (SNP) array (n = 102) and validated by fluorescence in situ hybridization (FISH), polymerase chain reaction (PCR), and/or multiplex ligation-dependent probe amplification (MLPA). The identified recurrent HBS1L deletion harbored the same breakpoints in all samples as identified by WGS (chr6:135,044,863-135,116,862; GRCh38hg38), including the HBS1L promoter and exon 1 to 2. Bars represent the percentage of BCR::ABL1-positive cases with a given alteration within each category. Associations between delHBS1L vs del7 vs IKZF1 vs CDKN2A/PAX5 were assessed by χ2 or Fisher exact test (P values below the significance level of 0.05 are depicted in bold). For detailed statistic please refer to supplemental Figure 5 and supplemental Appendix. (B) Subcluster-specific patterns of genomic aberrations were validated in the PMCC cohort (n = 49) using subcluster allocations obtained from a machine learning classifier trained on the GMALL cohort and the published14 genomic aberration profile of these samples. (C) Hierarchical clustering was performed using HBS1L alternative transcription start side expression (TSS; chr6: 135,040,344-135,040,447), HBS1L exon use, and HBS1L total gene expression in 113 GMALL samples. In addition, the average expression on HBS1L exons 1 to 3 is shown. (D) Direct long-read RNA-sequencing reads of HBS1L region between exons 1 and 4 are shown for 1 lymphoid and 1 multilineage BCR::ABL1-positive sample. The predicted alternative promoter in the intronic region between exon 3 and 4 is depicted in red. The orange bar shows the identified genomic deletion in HBS1L. A more detailed overview of the alternative HBS1L transcript and confirmation of the alternative TSS by single-cell ATAC-Seq is shown in supplemental Figures 7 and 8.)
Cooperating genomic events define gene expression subclusters, including alternative HBS1L isoform expression in delHBS1L and del7 as novel candidate. (A) Distribution of recurrent copy number variants (CNVs) in the 4 BCR::ABL1-positive ALL subclusters. CNVs were assessed in samples with subcluster allocation (GMALL: ground truth; MLL: predictions, excluding n = 14 samples that remained "unclassified" by machine learning classifier for the 2- and/or the 4-cluster definition) by whole-genome sequencing (WGS) (n = 47) or single-nucleotide polymorphism (SNP) array (n = 102) and validated by fluorescence in situ hybridization (FISH), polymerase chain reaction (PCR), and/or multiplex ligation-dependent probe amplification (MLPA). The identified recurrent HBS1L deletion harbored the same breakpoints in all samples as identified by WGS (chr6:135,044,863-135,116,862; GRCh38hg38), including the HBS1L promoter and exon 1 to 2. Bars represent the percentage of BCR::ABL1-positive cases with a given alteration within each category. Associations between delHBS1L vs del7 vs IKZF1 vs CDKN2A/PAX5 were assessed by χ2 or Fisher exact test (P values below the significance level of 0.05 are depicted in bold). For detailed statistic please refer to supplemental Figure 5 and supplemental Appendix. (B) Subcluster-specific patterns of genomic aberrations were validated in the PMCC cohort (n = 49) using subcluster allocations obtained from a machine learning classifier trained on the GMALL cohort and the published14 genomic aberration profile of these samples. (C) Hierarchical clustering was performed using HBS1L alternative transcription start side expression (TSS; chr6: 135,040,344-135,040,447), HBS1L exon use, and HBS1L total gene expression in 113 GMALL samples. In addition, the average expression on HBS1L exons 1 to 3 is shown. (D) Direct long-read RNA-sequencing reads of HBS1L region between exons 1 and 4 are shown for 1 lymphoid and 1 multilineage BCR::ABL1-positive sample. The predicted alternative promoter in the intronic region between exon 3 and 4 is depicted in red. The orange bar shows the identified genomic deletion in HBS1L. A more detailed overview of the alternative HBS1L transcript and confirmation of the alternative TSS by single-cell ATAC-Seq is shown in supplemental Figures 7 and 8.)
Interestingly, BCR::ABL1 multilineage cluster samples showed an increased HBS1L gene expression but reduced exon 1 to 3 use (Figure 2C). This pattern was absent in BCR::ABL1-negative ALL and healthy lymphoid progenitors (supplemental Figure 6). Long-read RNA-Seq confirmed a previously not described HBS1L transcript (HBS1Lalt) in delHBS1L and del7 cases initiated from a putative transcription start side in HBS1L intron 3 (Figure 2C,D; supplemental Figure 7). Single-cell ATAC-Seq revealed an open chromatin region at the putative HBS1Lalt promoter in 2 del7 cases, which was absent in 1 lymphoid BCR::ABL1 case (supplemental Figure 8). Besides, no significantly enriched genomic aberrations were identified in the proximity of HBS1L (whole-genome sequencing; chromosome 6:134,800,000-135,310,000, GRCh38), which could explain HSB1Lalt expression by disruption of regulatory elements. Expression of HBS1Lalt due to independent mechanisms, promoter loss (delHBS1L) or epigenetic/transcriptional regulations (del7), suggests HBS1Lalt as a novel cooperating event in multilineage BCR::ABL1-positive ALL.
Our subtype definition extends a recent description of transcriptomic classes of BCR::ABL1-positive ALL by Kim et al.14 In direct comparison, the published Early-Pro subtype corresponds to our multilineage definition (supplemental Figure 9). Within the multilineage cluster, we additionally identified subclusters delHBS1L and del7, which were confirmed also on the published data (Figure 1C; supplemental Figure 9A). The published Inter-Pro and Late-Pro definitions correspond to our IKZF1 and CDKN2A/PAX5 subclusters, respectively. However, our IKFZ1 cluster definition included more samples with heterozygous IKZF1 deletions compared with “Inter-Pro,” which was exclusively restricted to samples with biallelic IKZF1 loss in the published data. Unsupervised clustering of the aggregated cohorts using the published14 gene set definition grouped together samples classified IKZF1 according to our definition, independent of the genomic IKZF1 deletion status, suggesting that epigenetic regulations might contribute to a shared transcriptomic profile in cases without biallelic IKFZ1 loss (supplemental Figure 9B). Thus, validation across independent analyses specifies 4 molecular subtypes of BCR::ABL1-positive ALL.
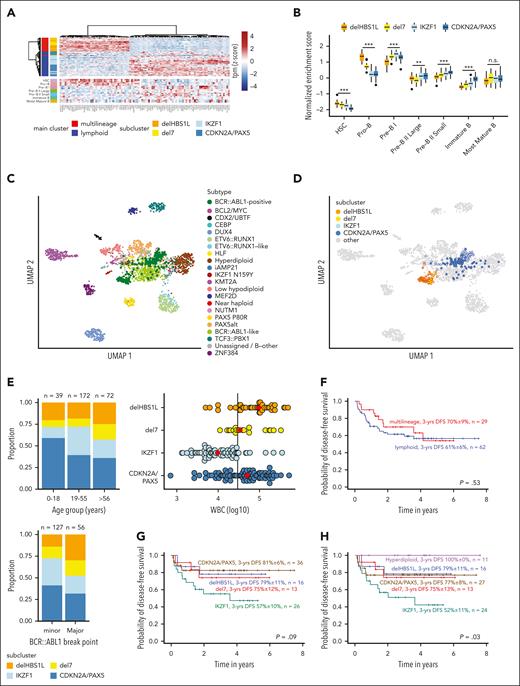
To further evaluate lineage-specific phenotypes, we analyzed diagnostic BCP-ALL immunophenotypes and compared gene expression with normal lymphopoiesis. More frequent myeloid coexpression (CD13, CD33) was observed in the multilineage subclusters, whereas lymphoid markers (CD20, CD22) were higher expressed in the lymphoid subclusters (supplemental Figure 10). Top differentially expressed genes between multilineage and lymphoid main cluster (Figure 3A) and single-sample gene set enrichment analyses for normal B lymphopoiesis stages15 (Figure 3B; supplemental Figure 11) revealed a closer proximity to pro-B cells in multilineage and to pre-B I cells in lymphoid cluster samples, confirming distinct underlying developmental trajectories. Analysis of involved signaling pathways confirmed functional independence of the BCR::ABL1 clusters with strong enrichment for Hippo and Hedgehog pathways in IKZF1 and CDKN2A/PAX5, respectively (supplemental Figure 12).
Proximity to more immature lymphopoiesis stages defines multilineage BCR::ABL1-positive ALL, which has a similar outcome as lymphoid BCR::ABL1-positive ALL. (A) Differential gene expression analysis between multilineage and lymphoid BCR::ABL1-positive ALL was performed using 1-way analysis of variance (supplemental Table 7). The 100 most significantly differentially expressed genes were used for hierarchical clustering of BCR::ABL1-positive samples (upper panel). The expression heat map in the lower panel shows gene expression of the same genes in healthy B-cell progenitors.15 (B) ALLCatchR15 single-sample enrichment scores20 for samples from the 4 subclusters are shown using gene set definitions of normal B lymphopoiesis. (C-D) Uniform manifold approximation and projection (UMAP) plots showing gene expression data of 2567 patients with BCP-ALL, previously aggregated from 3 cohorts15 and including now the 2 major (C) and 4 subcluster (D) BCR::ABL1 groups. The UMAP plots are based on the 3058 genes defined for BCP-ALL subtypes15 and BCR::ABL1 clusters. The updated version of ALLCatchR, providing molecular subtype allocation to BCP-ALL subtypes, including the novel BCR::ABL1 clusters, is available online (https://github.com/ThomasBeder/ALLCatchR_bcrabl1). (E) The distribution of age groups (upper left), BCR::ABL1 break points (lower left), and white blood cell counts (WBC, upper right) at initial diagnosis are shown for subclusters of the aggregated data set. The solid line in the dot plot showing WBC distribution represents a WBC of 30 000/μL, and red diamonds are the medians. Corresponding data and statistical analyses are provided in supplemental Tables 1,13, and 14. (F-H) DFS recorded at a median of 3 years for 91 GMALL BCR::ABL1-positive patients treated according to GMALL protocols with dose-reduced chemotherapy induction combined with imatinib, followed by consolidation I with continuous imatinib treatment and indication for allogeneic stem cell transplantation in first complete remission is shown. Kaplan-Meier analysis was used to calculate survival probabilities, and differences were assessed by log-rank test.
Proximity to more immature lymphopoiesis stages defines multilineage BCR::ABL1-positive ALL, which has a similar outcome as lymphoid BCR::ABL1-positive ALL. (A) Differential gene expression analysis between multilineage and lymphoid BCR::ABL1-positive ALL was performed using 1-way analysis of variance (supplemental Table 7). The 100 most significantly differentially expressed genes were used for hierarchical clustering of BCR::ABL1-positive samples (upper panel). The expression heat map in the lower panel shows gene expression of the same genes in healthy B-cell progenitors.15 (B) ALLCatchR15 single-sample enrichment scores20 for samples from the 4 subclusters are shown using gene set definitions of normal B lymphopoiesis. (C-D) Uniform manifold approximation and projection (UMAP) plots showing gene expression data of 2567 patients with BCP-ALL, previously aggregated from 3 cohorts15 and including now the 2 major (C) and 4 subcluster (D) BCR::ABL1 groups. The UMAP plots are based on the 3058 genes defined for BCP-ALL subtypes15 and BCR::ABL1 clusters. The updated version of ALLCatchR, providing molecular subtype allocation to BCP-ALL subtypes, including the novel BCR::ABL1 clusters, is available online (https://github.com/ThomasBeder/ALLCatchR_bcrabl1). (E) The distribution of age groups (upper left), BCR::ABL1 break points (lower left), and white blood cell counts (WBC, upper right) at initial diagnosis are shown for subclusters of the aggregated data set. The solid line in the dot plot showing WBC distribution represents a WBC of 30 000/μL, and red diamonds are the medians. Corresponding data and statistical analyses are provided in supplemental Tables 1,13, and 14. (F-H) DFS recorded at a median of 3 years for 91 GMALL BCR::ABL1-positive patients treated according to GMALL protocols with dose-reduced chemotherapy induction combined with imatinib, followed by consolidation I with continuous imatinib treatment and indication for allogeneic stem cell transplantation in first complete remission is shown. Kaplan-Meier analysis was used to calculate survival probabilities, and differences were assessed by log-rank test.
Integration of gene set definitions for the new molecular BCR::ABL1 clusters with our previous definitions for 21 molecular subtypes provided a clear separation in a large BCP-ALL data set15 (Figure 3C,D). Therefore, we implemented a predictive model for the novel BCR::ABL1 clusters in ALLCatchR15 to facilitate independent validation for routine diagnostic application (https://github.com/ThomasBeder/ALLCatchR_bcrabl1).
Integrated analysis across all cohorts revealed further clinical phenotypes of the BCR::ABL1 clusters (Figure 3E; supplemental Tables 13 and 14). We observed a predominance of the CDKN2A/PAX5 subcluster in pediatric patients despite equal distribution of multilineage and lymphoid cases in children and younger adults. Elderly patients were more frequently classified as multilineage with an increase of del7 cases. Patients in the multilineage cluster more frequently harbored BCR::ABL1 major breaks with a significant increase of BCR exon14 involvement in delHBS1L. White blood cell counts at diagnosis also differed between the novel BCR::ABL1 clusters (highest in delHBS1L and CDKN2A/PAX5), together indicating that the novel cluster definitions also represent distinct clinical profiles.
We analyzed the clinical implications of BCR::ABL1 subtypes in our homogeneously treated adult patient cohort (n = 98; first diagnosis 2014-2021) in the context of recent GMALL multicenter protocols including imatinib combined with adapted chemotherapy, minimal residual disease monitoring, and allogeneic SCT in first CR. The SCT rate was 86% in this cohort and 85% in the most recent GMALL trial 08/2013.3 Seven patients were aged >55 years and treated according to less intensive protocols. We observed fewer complete molecular responders in the multilineage cluster compared with the lymphoid cluster (supplemental Figure 13). Disease-free survival (DFS) probabilities were uniformly high in both subtypes (3-year DFS multilineage vs lymphoid, 70% ± 9% vs 61% ± 6%; P = .530; n = 91 evaluable patients; Figure 3F), in line with recent reports.9,21 Analysis of the subclusters revealed an inferior outcome for the IKZF1 cluster, whereas DFS was comparable in the remaining clusters (3-year DFS: delHBS1L, 79% ± 11%; del7, 75% ± 12%; IKZF1, 57% ± 10%; CDKN2A/PAX5, 81% ± 6%; P = .090; Figure 3G). Hyperdiploid cases had an excellent outcome (DFS: 100% ± 0%; Figure 3H), extending previous reports.22,23 DFS probabilities were comparable to overall survival (data not shown). Kim et al14 observed heterogeneous outcomes of BCR::ABL1 subtypes with a poor 5-year overall survival of 33% in “Early-Pro” cases, which correspond to our multilineage definition. These adult patients were diagnosed between 1992 and 2019 and were treated mostly with protocols including imatinib or other TKIs. SCT in first CR was performed in only 32% to 39% of cases, suggesting that higher transplantation rates might equalize biological differences between BCR::ABL1 subtypes. Future trials need to evaluate the role of BCR::ABL1 subtypes in context of chemotherapy-free treatments relying on immunotherapy targets. Our analyses provide a novel framework integrating developmental trajectories and defined genomic patterns in BCR::ABL1-positive ALL based on shared transcriptomic regulations to subclassify BCR::ABL1-positive ALL into distinct biological and clinically relevant entities.
Acknowledgments
The authors gratefully acknowledge support for fluorescence-activated cell sorting performed by Evelin Kerner and Christian Peters at CYTO Kiel–Cytometry facility, Institute of Immunology (head: Alexander Scheffold). This study was conducted using data provided by the Ontario Institute for Cancer Research, which is funded by the Government of Ontario.
This study was funded in part by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) project number 444949889 (KFO 5010/1 Clinical Research Unit “CATCH ALL” to L. Bastian, A.M.H., G. Chitadze, M.N., M.B., G. Cario, and C.D.B.) and project number 413490537 (Clinician Scientist Program in Evolutionary Medicine to B.-T.H.) and Deutsche José Carreras Leukämie-Stiftung (DJCLS 06R/2019 to M.B.). The GMALL trial 07/03 and the GMALL trial 08/13 were funded by Deutsche Krebshilfe. C.R., B.B., and F.-J.M. were supported by the German Federal Ministry for Research and Education (BMBF IntraEpiGliom, FKZ 13GW0347C, FKZ 13GW0347D). B.B. and F.-J.M. were partly supported by the DFG under Germany’s Excellence Strategy–EXC 22167-390884018 and by an intramural grant of the University Comprehensive Cancer Center Schleswig-Holstein (UCCSH Twinning 2022).
The views expressed in the publication are those of the authors and do not necessarily reflect those of the Ontario Institute for Cancer Research or the Government of Ontario.
Authorship
Contribution: L. Bastian, T.B., M.J.B., M.B., and C.D.B. designed the study and performed research; L. Bastian and T.B. developed the classifier; T.B., M.J.B., S.B., W.W., N.W., B.-T.H., A.M.H., D.D.G., L. Bartsch, and J.Z. established bioinformatic workflows and performed data analyses; L. Bastian, M.N., G. Cario, C.H., H.P., N.G., M.B., and C.D.B. supervised the study; L. Bastian, M.J.B., L. Bartsch, K.I., M.D., and M.W. performed single-nucleotide polymorphism array and multiplex ligation-dependent probe amplification analyses; L. Bastian, M.J.B., S.B., M.D., G. Chitadze, and L.H. established and performed fluorescence-activated cell sorting and BCR::ABL1 fluorescence in situ hybridization analysis; B.B., C.R., and F.-J.M. performed and analyzed nanopore long-read RNA sequencing; W.W. and C.H. contributed gene expression and whole-genome data and validations; H.P. and N.G. provided clinical data of the GMALL study group and performed outcome analysis; M.N., F.S., W.F., B.S., M.S., C.F., and S.S. treated patients and provided samples and clinical and diagnostic data, supporting the study; L. Bastian, T.B., M.J.B., and C.D.B. wrote the manuscript; and all authors revised and approved the final version of the manuscript.
Conflict-of-interest disclosure: M.B. is contracted to perform research for Affimed, Amgen, and Regeneron; is a member of the advisory boards of Amgen and Incyte; and is on the speaker bureaus of Amgen, Janssen, Pfizer, and Roche. W.F. recieved personal fees and nonfinancial support from AbbVie; received grants, personal fees, and nonfinancial support from Amgen and Pfizer; received personal fees from Jazz Pharmaceuticals, Celgene, Morphosys, Ariad/Incyte, stem line therapeutics Daiichi Sankyo, Apis, Otsuka, and Servier outside the submitted work; has a patent issued for Amgen; and received support for medical writing from Amgen, Pfizer, and AbbVie. C.H. is part owner of the Munich Leukemia Laboratory. The remaining authors declare no competing financial interests.
Correspondence: Lorenz Bastian, Medical Department II, Hematology and Oncology, Universitätsklinikum Schleswig-Holstein, Campus Kiel, Arnold-Heller-Str 3, Kiel 24105, Germany; email: lorenz.bastian@uksh.de.
References
Author notes
L. Bastian, T.B., and M.J.B. contributed equally to this study.
RNASeq raw data of BCR::ABL1 samples from the German Multicenter Study Group for Adult Acute Lymphoblastic Leukemia (GMALL) can be obtained from EGAS00001006107. RNASeq raw from FACS sorted healthy B cell progenitors can be obtained from EGAS00001007305. Other RNASeq and WGS data obtained from cooperation partners should be requested from the corresponding author, Lorenz Bastian (lorenz.bastian@uksh.de).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal