In this issue of Blood, Bastian et al1 report that Philadelphia chromosome-positive acute lymphoblastic leukemias (Ph+ ALLs) are more heterogeneous than previously thought. The authors identified subtypes with distinct transcriptomic and genomic profiles, which correlate with multilineage or lymphoid-only BCR::ABL1 involvement and have distinct clinical phenotypes.
It has been 5 decades since Janet Rowley’s groundbreaking discovery of the translocation t(9;22) and its association with chronic myeloid leukemia (CML) and Ph+ ALL. The identification of the BCR::ABL1 fusion gene marked a turning point in understanding these hematologic malignancies, leading to the development of targeted therapies that transformed the landscape of leukemia treatment.2
Since then, it has been assumed that disease heterogeneity within BCR::ABL1-driven malignancies, specifically CML or de novo Ph+ ALL, largely depended on different types of BCR::ABL1 fusions: the major breakpoint fusion producing the p210 isoform associated with CML occurred in an hematopoietic stem cell, and the minor fusion producing p190 was confined to the B-cell precursor compartment.3 Although a subset of Ph+ ALL presented with major fusion, there was a persistent doubt as to whether those cases represented authentic de novo Ph+ ALL or CML blast crisis with an unrecognized chronic stage. Recent studies challenged this view by demonstrating BCR::ABL1 multilineage involvement in a subset of Ph+ ALL, regardless of the fusion breakpoint.4-6 This dichotomy within Ph+ ALL, namely “multilineage” or “lymphoid-only” has been recently recognized in the International Consensus Classification of acute leukemias.7 However, little is known regarding the distinctive biology of these subtypes and their associated clinical and prognostic features. Moreover, there is currently no consensus on the appropriate methodology to classify patients into these subtypes.
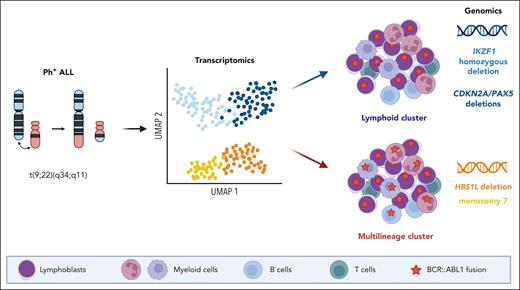
In this study, Bastian et al characterized a large cohort of Ph+ ALL using comprehensive transcriptomic and genomic profiling. They identified 2 transcriptomic clusters and showed that they segregated cases with or without multilineage involvement. Each cluster was further explored and found to be associated with distinct genomic patterns, namely HBS1L deletion and monosomy 7 for the multilineage clusters and IKZF1 homozygous deletion and CDKN2A/PAX5 deletions for the lymphoid-only clusters (see figure). Moreover, multilineage subtypes displayed a gene expression pattern mirroring the normal pro-B stage, and lymphoid subtypes were closer to pre-B stages. Finally, they analyzed the prognostic implications of molecular subtypes of Ph+ ALL within a cohort of adults treated homogeneously in German multicenter ALL protocols. They observed an equivalent prognosis for the multilineage as compared with the lymphoid-only Ph+ ALL groups, but an inferior outcome for the IKZF1 lymphoid cluster.
Gene expression profiling in Ph+ ALL delineates 2 distinct groups that segregate cases with or without multilineage involvement, referred as multilineage or lymphoid clusters, respectively. These groups are associated with distinct genomic features including HBS1L deletion and monosomy 7 for the multilineage cluster and IKZF1 homozygous deletion and CDKN2A/PAX5 deletions for the lymphoid cluster. UMAP, uniform manifold approximation and projection.
Gene expression profiling in Ph+ ALL delineates 2 distinct groups that segregate cases with or without multilineage involvement, referred as multilineage or lymphoid clusters, respectively. These groups are associated with distinct genomic features including HBS1L deletion and monosomy 7 for the multilineage cluster and IKZF1 homozygous deletion and CDKN2A/PAX5 deletions for the lymphoid cluster. UMAP, uniform manifold approximation and projection.
This study significantly expands a recent study by Kim et al, which also described distinct molecular subtypes of Ph+ ALL.8 However, although these studies appear to be largely similar at the transcriptomic and genomic levels, there are noticeable differences in the findings and conclusions. First, Bastian et al propose that the cell of origin, either multipotent or lymphoid, may determine the molecular subtype, in contrast to Kim et al who could not observe an association between molecular subtypes and multilineage involvement and consequently assumed that BCR::ABL1 occurs in all cases in a multipotent stem cell. This point will require further study. Second, there are differences between the studies in the prognosis of molecular subtypes, with the HBS1L subtype associated with worse outcomes in Kim et al but good survival rates (3-year disease-free survival 79%) in the present study. Although these variations may be attributed to different treatment regimens, validation in other cohorts would be necessary to establish robust conclusions and integrate these new markers into clinical practice.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal