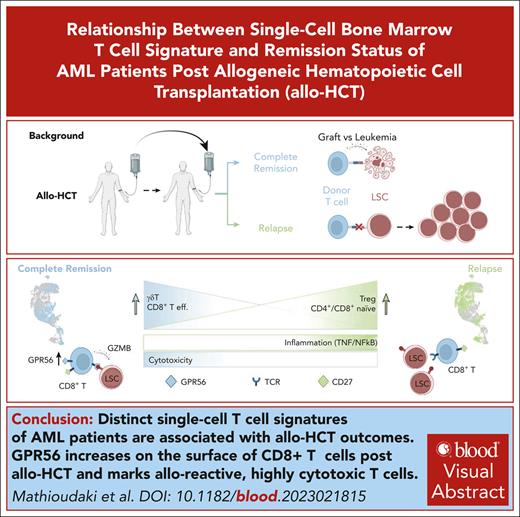
Distinct scRNA-seq signatures characterize BM T cells of patients with AML in remission and with relapse after allo-HCT.
GPR56 increases on the surface of CD8+ T cells after allo-HCT and marks allo-reactive, highly cytotoxic T cells recognizing non-self.
Visual Abstract
Acute myeloid leukemia (AML) is a hematologic malignancy for which allogeneic hematopoietic cell transplantation (allo-HCT) often remains the only curative therapeutic approach. However, incapability of T cells to recognize and eliminate residual leukemia stem cells might lead to an insufficient graft-versus-leukemia (GVL) effect and relapse. Here, we performed single-cell RNA-sequencing (scRNA-seq) on bone marrow (BM) T lymphocytes and CD34+ cells of 6 patients with AML 100 days after allo-HCT to identify T-cell signatures associated with either imminent relapse (REL) or durable complete remission (CR). We observed a higher frequency of cytotoxic CD8+ effector and gamma delta (γδ) T cells in CR vs REL samples. Pseudotime and gene regulatory network analyses revealed that CR CD8+ T cells were more advanced in maturation and had a stronger cytotoxicity signature, whereas REL samples were characterized by inflammatory tumor necrosis factor/NF-κB signaling and an immunosuppressive milieu. We identified ADGRG1/GPR56 as a surface marker enriched in CR CD8+ T cells and confirmed in a CD33-directed chimeric antigen receptor T cell/AML coculture model that GPR56 becomes upregulated on T cells upon antigen encounter and elimination of AML cells. We show that GPR56 continuously increases at the protein level on CD8+ T cells after allo-HCT and confirm faster interferon gamma (IFN-γ) secretion upon re-exposure to matched, but not unmatched, recipient AML cells in the GPR56+ vs GPR56– CD8+ T-cell fraction. Together, our data provide a single-cell reference map of BM–derived T cells after allo-HCT and propose GPR56 expression dynamics as a surrogate for antigen encounter after allo-HCT.
Introduction
Acute myeloid leukemia (AML) is a malignant hematopoietic disorder in which immature myeloid precursors accumulate in bone marrow (BM) and blood. Although clinical trials exploring novel strategies such as leukemic stem cell (LSC)–directed therapies or chimeric antigen receptor (CAR) T-cell (CAR-T) treatment1 are underway, allogeneic hematopoietic cell transplantation (allo-HCT) is the best established immunotherapy in AML. After allo-HCT, donor hematopoietic stem and progenitor cells (HSPCs) repopulate the BM to regenerate the hematopoietic system, whereas donor T cells in the graft can recognize and target residual leukemic cells (graft-versus-leukemia [GVL] effect) as well as the patients’ healthy cells (graft-versus-host disease [GVHD]).2 Both effects are partially interconnected, because mild chronic GVHD (cGVHD) not requiring restart of immunosuppression is associated with better outcome than absence of cGVHD.3 In line, exhaustion of effector memory T cells might be associated with relapse in patients with AML after allo-HCT,4 and increased levels of regulatory T cells (Tregs) have been linked to worse prognosis.5,6
The necessity for immunosuppression during the first months, which hampers GVL effects, makes early relapse after transplantation a major challenge. Besides salvage chemotherapy, donor lymphocyte infusions represent a key component of therapy options once relapse has occurred, but their efficacy is highly variable, and we currently lack biomarkers for their antileukemic potential.7 Furthermore, AML-specific characteristics such as high-risk genetic aberrations contribute to immune-escape mechanisms,8 for example, by downregulating major histocompatibility complex class II molecules on the surface or upregulating inhibitory ligands such as PD-L1, thus making some patients more susceptible to relapse than others.9,10 The substantial heterogeneity in outcome even within supposedly similar genetic AML groups, suggests the contribution of other factors, such as donor cell characteristics and immune cell composition at the time of relapse. Indeed, high CD8+ and high CD34+ cell doses in the graft have been associated with better survival in AML.11,12 Yet, little is known about how donor T cell and HSPC composition change after transplantation. Moreover, most T-cell studies, including single-cell RNA-sequencing (scRNA-seq), focused on peripheral blood, whereas GVL happens predominantly in the BM, in which it is heavily influenced by surrounding healthy HSPCs, stroma cells, and residual LSCs.
Here, we provide a holistic view of the BM T-cell repertoire to distinguish T cells of relapsed patients with failed early GVL from T cells of patients with long-term remission. Our study provides a reference map of single-cell BM-derived T cells and links higher frequencies of BM γδΤ and a specific class of antigen-experienced CD8+ effector T cells with remission after allo-HCT. We identify the adhesion G-protein–coupled receptor GPR56 as a dynamic biomarker in T-cell reactivity after transplantation, which we validate with in vitro CAR-T/HL-60 and primary AML T-cell coculture assays mimicking GVL, as well as on protein level by flow cytometry on a larger cohort of patients.
Methods
Patient samples
BM samples were collected from adult patients after obtaining written informed consent in accordance with the Declaration of Helsinki. The local ethics board of Heidelberg University approved the project. For sample and patient details supplemental Table 1 (available on the Blood website) and supplemental Methods. For scRNA-seq profiling, BM collected 100 days after allo-HCT from 6 patients was used. Three patients had a borderline blast burden of 5% at sampling and developed full blown relapse despite reduction of immunosuppression within 150 days after allo-HCT (failure of early GVL, termed relapse [REL]); 3 patients remained in complete remission (CR) until last follow-up (at least 12 months) despite minimal residual disease positivity or incomplete donor chimerism at sampling.
scRNA library preparation, sequencing, and analysis
Frozen ficoll-processed primary BM samples were sorted into CD34+ and CD3+ fractions (supplemental Figure 1 for sorting strategy). 10× Genomics single-cell 3′ Gene Expression v3 assay was performed according to the manufacturer’s protocol CG000204, and libraries were sequenced with Illumina NextSeq 500. The raw scRNA-seq data were preprocessed using Cell Ranger Single Cell Software Suite provided by 10× Genomics. All scripts for scRNA-seq downstream analysis are available at: https://github.com/ZauggGroup/scBM_alloSCT.
CAR-T cell assay
CD33.CAR-T cells from healthy donors were generated as follows: peripheral blood mononuclear cells (PBMCs) were activated with anti-CD3/anti-CD28 antibodies (Biolegend, San Diego, CA) and transduced with a third-generation retroviral vector (SFG) supplied by the CAGT, Baylor College, Houston, TX, followed by CAR-T cell expansion as previously described.13
Flow cytometry and cell sorting
Patient samples were assessed for surface expression of CD3, CD4, CD8, CD27, CD33, CD34, CD45RA, CD56, CCR7, and GPR56 (supplemental Methods). Data were acquired using BD LSRII and Canto flow cytometers and analyzed using BD FACS Diva 8.0/9.0 and Flowjo X (Treestar Inc) software. Sorting was performed on FACSAria Fusion and BD FACSAria II sorters. Caspase 3-FITC was used as a viability marker.
ELISpot assay
GPR56+ and GPR56– CD8+ T cells were sorted from PBMCs of patients with AML after allo-HCT in remission. Cells were then cocultured with matched AML blasts for 24 hours, and interferon gamma (IFN-γ) production was measured using enzyme-linked immunosorbent spot (ELISpot) assay (MabTech, catalog no. 3420-4HPW-2). For sample and patient details, see supplemental Table 1 and supplemental Methods.
See supplemental Methods for further details.
Results
Characterization of BM T cells after allo-HCT
To define the BM landscape of donor T cells of patients with AML 100 days after allo-HCT, we sorted CD3+ T cells from 3 patients in CR and 3 with REL and performed scRNA-seq (Figure 1A; sample characteristics in supplemental Table 1). To ensure that both groups had a healthy donor-derived HSPC compartment at the time of analysis, we also sorted and sequenced the corresponding CD34+ cells. After quality control, we retained 4038 cells from patients in CR and 4461 cells from those with REL (average number of cells per patient, 1415; supplemental Figure 2). Genotype deconvolution was used to detect 6 genetically unique individuals (supplemental Figure 3; supplemental Methods).
Single-cell characterization of BM HSPCs and T cells from patients with REL and those with CR. (A) Overview of the experimental setup. (B) Uniform manifold approximation and projection (UMAP) of 8492 post-QC cells representing BM HSPCs and T cells from 6 patients with AML on day 100 after allo-HCT. Cells are colored according to cell type. HSPCs included myeloid/lymphoid progenitors (MLP), B-cell precursors (preB), T-cell precursors (pre/proT), megakaryocyte-erythroid progenitors (MEP), neutrophil progenitors (NP), monocyte-dendritic progenitors (MDP), late monocytic precursors/monocytes (MP/mono), and dendritic cells (pDCs, cDCs). (C-D) Scaled expression (C) of marker genes and TF activity (D) are shown for each cell type cluster in panel B as heat maps. Values are averaged across all cells in the cluster. TF activity is obtained from SCENIC (“Methods”). (E-G) UMAP as shown in panel B indicating the normalized gene expression of selected genes, with the range of normalized gene expression indicated in the parenthesis of each UMAP. QC, quality control. The BM biopsy illustration in the lower part of panel A was adapted from the original work of Cancer Research UK under a CC BY-SA 4.0 license.
Single-cell characterization of BM HSPCs and T cells from patients with REL and those with CR. (A) Overview of the experimental setup. (B) Uniform manifold approximation and projection (UMAP) of 8492 post-QC cells representing BM HSPCs and T cells from 6 patients with AML on day 100 after allo-HCT. Cells are colored according to cell type. HSPCs included myeloid/lymphoid progenitors (MLP), B-cell precursors (preB), T-cell precursors (pre/proT), megakaryocyte-erythroid progenitors (MEP), neutrophil progenitors (NP), monocyte-dendritic progenitors (MDP), late monocytic precursors/monocytes (MP/mono), and dendritic cells (pDCs, cDCs). (C-D) Scaled expression (C) of marker genes and TF activity (D) are shown for each cell type cluster in panel B as heat maps. Values are averaged across all cells in the cluster. TF activity is obtained from SCENIC (“Methods”). (E-G) UMAP as shown in panel B indicating the normalized gene expression of selected genes, with the range of normalized gene expression indicated in the parenthesis of each UMAP. QC, quality control. The BM biopsy illustration in the lower part of panel A was adapted from the original work of Cancer Research UK under a CC BY-SA 4.0 license.
We grouped the cells using Louvain clustering and annotated the clusters based on the expression of known marker genes into 9 HSPC clusters including all expected HSPC populations,14 8 CD8+, 5 CD4+, and 2 unconventional T-cell clusters (Figure 1B-G; supplemental Figure 4A; supplemental Table 2). The CD8+ clusters segregated into the following: CD8+ naive cells (CD8+ NV, expressing CCR7 and SELL); CD8+ cells expressing the transcription factor (TF) HOBIT/ZNF683, hereafter called “CD8+ hobit”; CD8+ effector 1 and 2 (CD8+ eff. 1: CD160+; CD8+ eff. 2: CD160–; expressing NKG7 and GZMB); CD8+ memory (mem.) 1, 2, and 3, characterized by expression of GZMK and lower levels of cytotoxic genes compared with CD8+ effector; and CD8+ IFN (enriched for IFN-γ pathway genes). The CD4+ T cells included CD4+ naive (CD4+ NV), Tregs, CD4+ IFN, CD4+ memory cells (CD4+ mem.), and T helper 17 (Th17). Moreover, we identified gamma-delta (γδ) T cells and mucosal-associated invariant T cells (MAIT) consistent with previous BM scRNA data sets (Figure 1B-G).15
Altered BM T-cell composition is associated with remission status
After confirming that both groups, patients with REL and CR, were successfully engrafted with healthy donor-derived CD34+ at the time of sequencing (supplemental Figure 4B-C), we focused subsequent analyses on the T-cell compartment as the main driver of GVL effects (reviewed in Bleakley and Riddell20) to assess whether the BM T-cell composition was associated with either sustained remission or imminent relapse.
Although all T-cell types were present in patients in CR and those with REL (supplemental Figure 4B,D), their abundances significantly differed between the 2 groups (Figure 2A-C; supplemental Figure 4E). CD4+ NV, Tregs, CD8+ NV, and CD8+ hobit, as well as the 2 interferon clusters (CD4+ IFN and CD8+ IFN) were enriched in patients with REL, whereas samples from those achieving CR were enriched in CD8+ eff. 1, the more mature CD8+ mem. 2 and 3, and γδT cells (Figure 2C; supplemental Table 3). Comparable patterns were observed when assessing the population abundances for each sample individually for most cell populations, and we only labeled those as significant that were consistent among individual donors (supplemental Figure 4E). Since we aimed to identify T-cell subsets associated with GVL activity, we subsequently focused our analyses on the CD8 compartment, which contained all CR-associated subsets.
Altered BM T-cell composition is associated with the remission status. (A) UMAP highlighting CR and REL cells (green, REL; blue, CR). (B) Absolute numbers of cells across CD3+ T-cell types. The different colors indicate REL (green) and CR (blue) samples. (C) Differential abundance per cell type, within the CD3+ population. The bars represent the log2 odds ratios (Fisher exact test, P value after Bonferroni correction; n.s., not significant; ∗∗∗P < .0001). (D) (Left) Scaled expression (z-score) of publicly available cellular indexing of transcriptome and epitopes (CITE)-seq data. Features (x-axis) with antibody (AB) suffix indicate that the measurement was performed on protein level. (Right) Percentage of cells per cluster (x-axis) that map to public reference clusters (y-axis). Bold cluster names indicate CD8+ TEMRA subsets (CD45RA+ CCR7–). (E) Comparison of the CD8+ pseudotime (calculated with Monocle3) between CR and REL using naive CD8+ as the starting population. Box plot (top) and density plot (bottom) depicting the pseudotime of CD8+ cells in CR (blue) and REL (green). Difference between CR and REL was assessed using t test. (F) Heat map depicting the scaled expression across pseudotime of selected effector, memory, and exhaustion genes in CD8+ cells. (G) Diffusion maps for the CD8+ cells colored according to the inferred pseudotime using Monocle3 split per condition (top) and based on the clusters (bottom).
Altered BM T-cell composition is associated with the remission status. (A) UMAP highlighting CR and REL cells (green, REL; blue, CR). (B) Absolute numbers of cells across CD3+ T-cell types. The different colors indicate REL (green) and CR (blue) samples. (C) Differential abundance per cell type, within the CD3+ population. The bars represent the log2 odds ratios (Fisher exact test, P value after Bonferroni correction; n.s., not significant; ∗∗∗P < .0001). (D) (Left) Scaled expression (z-score) of publicly available cellular indexing of transcriptome and epitopes (CITE)-seq data. Features (x-axis) with antibody (AB) suffix indicate that the measurement was performed on protein level. (Right) Percentage of cells per cluster (x-axis) that map to public reference clusters (y-axis). Bold cluster names indicate CD8+ TEMRA subsets (CD45RA+ CCR7–). (E) Comparison of the CD8+ pseudotime (calculated with Monocle3) between CR and REL using naive CD8+ as the starting population. Box plot (top) and density plot (bottom) depicting the pseudotime of CD8+ cells in CR (blue) and REL (green). Difference between CR and REL was assessed using t test. (F) Heat map depicting the scaled expression across pseudotime of selected effector, memory, and exhaustion genes in CD8+ cells. (G) Diffusion maps for the CD8+ cells colored according to the inferred pseudotime using Monocle3 split per condition (top) and based on the clusters (bottom).
To further characterize the CD8+ populations, we integrated our data with public cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) data of PBMCs,21 in which T-cell populations were defined by both RNA and surface protein expression. This enabled mapping of CD8+ hobit and CD8+ mem. 1 clusters to CD45RA–CCR7+ central memory (TCM) and CD45RA–CCR7– effector memory (TEM) cells, respectively (Figure 2D). The CD8+ eff. 1 and 2, as well as CD8+ mem. 2 and 3 corresponded to subsets of CD45RA+CCR7– effector memory cells (TEMRA), which differed in their expression levels for CD45RA and the cytotoxic molecules GZMB, GZMK, and GNLY (Figure 2D).
To better reflect the dynamics of T-cell maturation, which is a continuum from the naive to the effector memory state, we inferred pseudotime of CD8+ T-cell maturation using Monocle3,22 a computational approach, which orders cells along a differentiation trajectory, here T-cell maturation, with the naive CD8 T cells as starting population (Figure 2E-G; supplemental Figure 5A-C). We found that REL cells were significantly less advanced in pseudotime (Figure 2E,G).
CD8+ T cells in patients in CR are characterized by high expression of cytotoxicity-related genes
We next asked which genes and regulatory networks are associated with the differences in T-cell subset composition. Because differentiation processes are typically driven by TFs, we inferred gene regulatory networks using SCENIC16 (supplemental Methods; supplemental Figure 7A-D). We first aimed to identify global gene regulatory network differences in the 2 major types of T-cell lineages, CD4+ and CD8+. We defined TFs within CD4+ and CD8+ clusters as differentially active in CR vs REL, when their regulons were enriched among differentially expressed genes (DEGs; 422 and 539 respectively; log2FC > 1, adjusted P value < 0.05 after Bonferroni correction; supplemental Table 4; supplemental Figure 6A-C). This revealed 14 differentially active TFs (Fisher test; adjusted P value < 0.05 after false discovery rate correction; Figure 3A; supplemental Tables 5-6; supplemental Figure 8A). Many TFs with enriched activity in the REL samples were shared between CD4+ and CD8+ cells, yet with different target genes (supplemental Figure 8B), which is in line with our previous observations of cell type–specific regulons of commonly expressed TFs.23 These shared TFs included activator protein 1 TFs (FOS/JUN), REL from the NF-κB family, as well as CREM and NFE2L2/NRF2, which have been linked to an immunosuppressive tumor microenvironment and CD8+ exhaustion, respectively.24,25 TBX21 was the only TF whose regulon was enriched among the genes upregulated in CR CD8+ cells and contained mostly effector-associated genes (Figure 3B).
CD8+ T cells in REL samples have lower cytotoxic potential. (A) Differential TF activity analysis using SCENIC. Heat map indicating log2 odds ratios; only significantly enriched TFs are colored (Fisher exact test false discovery rate < 0.05) (left). Characterization of target genes based on known gene sets (right). Assignment of target genes to known functions was performed using publicly available gene sets (supplemental Methods). The colored bars represent the fraction of target genes per TF, which belong to the different gene sets (IFN, IFN response; Activation, Immune cell activation; TNF, TNF signaling). (B) CD8+ T gene regulatory network (GRN) of exemplary differentially active TFs (TBX21, REL, and FOS) and their target DEGs. (C) UMAP highlighting the T-cell clusters that belong to the CD8+ effector memory (EM) cells and were used for the differential expression analysis. (D) Heat map depicting scaled expression (z-score) across all 6 samples of 235 DEGs (CR, 144 genes; REL, 91 genes) in CD8+ EM clusters. Analysis was performed using the MAST algorithm (log2FC > 0.5; adjusted P value [p.adj] < 0.05 after Bonferroni correction). (E) Gene ontology and hallmark enrichment analysis on the DEGs from D. Terms were selected from the top-enriched terms. Full list provided in supplemental Table 8. (F) UMAP indicating the normalized gene expression of ADGRG1/GPR56. (G) GPR56 and CD27 expression across pseudotime of CD8+ cells, split per condition. The bar plots below indicate the percentages of GPR56+ and CD27+ cells in REL and CR samples when considering all CD8+ clusters. (H) Percentage of GPR56 positive cells in the indicated fractions determined by flow cytometry in the same 6 samples used for scRNA-seq. (Top left cartoon) Gating strategy used to identify naive, central memory (TCM), EM (TEM), and CD45RA+ EM (TEMRA) cells using CCR7 and CD45RA. P values were calculated using Student t test. (I) Volcano plot illustrating the DEGs between GPR56+ (purple) and GPR56– (orange) CD8+ TEM cells of patients in CR. The y-axis represents P value after Bonferroni correction (p.adj), and points were colored according to absolute log2FC > 0.5 and P.adj < .05 (purple/orange). (J) Box plots illustrating the percentage of GZMB+ (top) and PRF1+ (bottom) T cells in the GPR56+ and GPR56− fractions assessed by intracellular flow cytometry. Connected points indicate fractions originating from the same sample. P value was calculated using paired Wilcoxon test, n = 10.
CD8+ T cells in REL samples have lower cytotoxic potential. (A) Differential TF activity analysis using SCENIC. Heat map indicating log2 odds ratios; only significantly enriched TFs are colored (Fisher exact test false discovery rate < 0.05) (left). Characterization of target genes based on known gene sets (right). Assignment of target genes to known functions was performed using publicly available gene sets (supplemental Methods). The colored bars represent the fraction of target genes per TF, which belong to the different gene sets (IFN, IFN response; Activation, Immune cell activation; TNF, TNF signaling). (B) CD8+ T gene regulatory network (GRN) of exemplary differentially active TFs (TBX21, REL, and FOS) and their target DEGs. (C) UMAP highlighting the T-cell clusters that belong to the CD8+ effector memory (EM) cells and were used for the differential expression analysis. (D) Heat map depicting scaled expression (z-score) across all 6 samples of 235 DEGs (CR, 144 genes; REL, 91 genes) in CD8+ EM clusters. Analysis was performed using the MAST algorithm (log2FC > 0.5; adjusted P value [p.adj] < 0.05 after Bonferroni correction). (E) Gene ontology and hallmark enrichment analysis on the DEGs from D. Terms were selected from the top-enriched terms. Full list provided in supplemental Table 8. (F) UMAP indicating the normalized gene expression of ADGRG1/GPR56. (G) GPR56 and CD27 expression across pseudotime of CD8+ cells, split per condition. The bar plots below indicate the percentages of GPR56+ and CD27+ cells in REL and CR samples when considering all CD8+ clusters. (H) Percentage of GPR56 positive cells in the indicated fractions determined by flow cytometry in the same 6 samples used for scRNA-seq. (Top left cartoon) Gating strategy used to identify naive, central memory (TCM), EM (TEM), and CD45RA+ EM (TEMRA) cells using CCR7 and CD45RA. P values were calculated using Student t test. (I) Volcano plot illustrating the DEGs between GPR56+ (purple) and GPR56– (orange) CD8+ TEM cells of patients in CR. The y-axis represents P value after Bonferroni correction (p.adj), and points were colored according to absolute log2FC > 0.5 and P.adj < .05 (purple/orange). (J) Box plots illustrating the percentage of GZMB+ (top) and PRF1+ (bottom) T cells in the GPR56+ and GPR56− fractions assessed by intracellular flow cytometry. Connected points indicate fractions originating from the same sample. P value was calculated using paired Wilcoxon test, n = 10.
We next explored DEGs in the CD8+ TEM compartment (CD8 eff. 1 and 2 and CD8 mem. 1-3) in more depth, since the effector-memory compartment was enriched in CR samples (Figure 3C-E; supplemental Tables 7-8). Genes upregulated in REL showed increased expression of CXCR4, associated with homing of naive T cells to the BM,26 and CD27, a TNF receptor associated with early T-cell activation27 (Figure 3D), and were enriched in NF-κB–dependent TNF-α signaling and IL-2 production (Figure 3E). Notably, CXCR4 and other genes were also upregulated in REL CD8+ naive and MAIT cells (supplemental Figure 9 A-B), pointing toward a common mechanism driving gene expression programs in REL CD8+ subsets. CR samples were enriched in RAS protein signaling, cell killing, and immune cell activation processes and comprised TBX21 targets including the known cytotoxic genes GZMB and KLRG1 but also the surface marker and adhesion GPCR ADGRG1/GPR56 (Figure 3B-E; supplemental Tables 7-8). Given its well-known function as stemness marker and self-renewal regulator in AML,28 we explored its expression pattern in T-cell subsets in more detail. GPR56 was predominantly expressed in the TEM compartment, but a trend toward higher GPR56 expression in CR vs REL samples was also observed in other clusters such as CD4+ and γδ T cells (supplemental Figure 10D; supplemental Table 7). Its expression in CD8+ cells increased with pseudotime opposite to CD27 (Figure 3F-G), and it was coexpressed with genes associated with effector functions, such as NKG7, GZMB, and PRF1 but not with exhaustion markers such as PCDC1 (PD-1) and CTLA4 (supplemental Figure 10A-C). We used flow cytometry to detect surface protein expression in the 6 samples and confirmed increasing fractions of GPR56+ cells during maturation of CD8+ from naive to TEM and TEMRA cells, as well as higher GPR56+ fractions in CR vs REL samples (Figure 3H).
To gain functional insight into the role of GPR56 in the TEM compartment (effector and memory clusters), in which it is predominantly expressed (Figure 3F), we determined a GPR56+ gene signature through differential expression analysis of the GPR56+ vs GPR56– TEM in CR samples. The signature comprised the cytotoxicity genes NKG7, GZMB, GZMH, KLRD1, GNLY, and PRF1, suggesting that GPR56 marks a specific subpopulation of cytotoxic T cells (Figure 3I; supplemental Figure 10E; supplemental Table 9). Accordingly, cytotoxicity-related gene ontology terms (“leukocyte-mediated cytotoxicity” and “cell killing”) were enriched in the GPR56+ signature (supplemental Figure 10F; supplemental Table 10). Intracellular flow cytometry analysis of PBMCs from 10 AML patients in CR (supplemental Table 1) confirmed significantly higher levels of the central cytotoxic response molecules PRF1 and GZMB on protein level in GPR56+ vs GPR56– CD8+ T cells (Figure 3I; paired Wilcoxon test, P < .01; supplemental Figure 11). Of note, there was no complete overlap between GPR56 and known activation and exhaustion molecules, suggesting that GPR56 has a nonredundant role with these markers (CD44, PD-1, CD107a, and CD69; supplemental Figure 12).
CAR-T cells dynamically upregulate GPR56 upon target cell elimination
Increased GPR56 expression on maturing CD8+ T cells suggested that GPR56 expression may reflect “non-self” recognition and, therefore, a higher GVL potential in CR vs REL samples. To test this, we used CAR-T cells as an artificial but well-defined effector-target cell model. CAR-T cells mimic GVL, as their T-cell receptors are genetically engineered to specifically recognize a defined (leukemic) target and eliminate cells expressing this target. We cocultured CD33-directed CAR-T cells engineered from activated T cells of 4 healthy donors together with the AML cell line HL-60, in which CD33 was knocked out29 (HL60 KO) vs HL-60 with preserved CD33 expression (scrambled CRISPR control, HL60 WT) (outlined in Figure 4A).
GPR56 is dynamically upregulated by CAR-T cells upon target recognition. (A) Schematic visualizing the experimental setup: Peripheral blood mononucleated cells (PBMCs) from 4 healthy donors were first activated, activated T cells (ATCs) were then transduced with a retroviral vector comprising a CD33.CAR construct. On day 15 of production, CAR-T cells were coincubated with the AML cell line HL60, expressing CD33 on the surface, or with HL60 cells, in which CD33 was knocked out using CRISPR/Cas9 (HL60 CD33 KO). (B) Representative FACS plots showing CD27 and GPR56 expression on CAR-T cells after activation and transduction, but without contact to leukemia cells (upper left), after 5-day coculture with CD33+ HL60 (upper right), after coculture with HL60 CD33 KO cells (lower left), and on nontransduced cells after coculture with HL60 CD33+ cells (lower right). Note that GPR56 upregulation occurs exclusively when CAR-T cells carrying the CD33.CAR were incubated with HL60 CD33+ cells. This suggests that GPR56 upregulation occurs only upon antigen recognition by the T cell receptor. (C) Statistical analysis of the experiment shown in 4B. ∗∗∗ P < .0005. (D) Percentage of GPR56+CD27+ fractions (blue) and CD15+ HL60 cells (orange) in the 4 individual donors during the 5 serial 5-day challenges. (E) (Top) Experimental setup of sorting experiment; CAR-T cells were challenged once with HL60 for 5 days. Then, cultures were sorted for CD8+ and the following 4 quadrants: GPR56+CD27–, GPR56+CD27+, GPR56–CD27+, and GPR56–CD27–. Subsequently, equal numbers of sorted cells were re-exposed to HL60 cells for 5 days and subsequently analyzed for surface marker expression. All fractions were capable of eliminating HL60 WT cells except for 1 culture with double-negative cells (data not shown), confirming that all fractions contained the CAR construct. (Lower left) Representative FACS plots of the sorted CAR-T cells after re-exposure to target cells. The label above the plots indicates the originally sorted phenotype. (Lower right) Stacked bar graph showing the distribution of the 4 quadrants in the 4 different conditions. Mean and standard deviation of the 4 donors are shown. The x-axis labels indicate the originally sorted fraction. Colors of bars indicate the output phenotype according to the legend. nt, nontransduced; wt, wild type.
GPR56 is dynamically upregulated by CAR-T cells upon target recognition. (A) Schematic visualizing the experimental setup: Peripheral blood mononucleated cells (PBMCs) from 4 healthy donors were first activated, activated T cells (ATCs) were then transduced with a retroviral vector comprising a CD33.CAR construct. On day 15 of production, CAR-T cells were coincubated with the AML cell line HL60, expressing CD33 on the surface, or with HL60 cells, in which CD33 was knocked out using CRISPR/Cas9 (HL60 CD33 KO). (B) Representative FACS plots showing CD27 and GPR56 expression on CAR-T cells after activation and transduction, but without contact to leukemia cells (upper left), after 5-day coculture with CD33+ HL60 (upper right), after coculture with HL60 CD33 KO cells (lower left), and on nontransduced cells after coculture with HL60 CD33+ cells (lower right). Note that GPR56 upregulation occurs exclusively when CAR-T cells carrying the CD33.CAR were incubated with HL60 CD33+ cells. This suggests that GPR56 upregulation occurs only upon antigen recognition by the T cell receptor. (C) Statistical analysis of the experiment shown in 4B. ∗∗∗ P < .0005. (D) Percentage of GPR56+CD27+ fractions (blue) and CD15+ HL60 cells (orange) in the 4 individual donors during the 5 serial 5-day challenges. (E) (Top) Experimental setup of sorting experiment; CAR-T cells were challenged once with HL60 for 5 days. Then, cultures were sorted for CD8+ and the following 4 quadrants: GPR56+CD27–, GPR56+CD27+, GPR56–CD27+, and GPR56–CD27–. Subsequently, equal numbers of sorted cells were re-exposed to HL60 cells for 5 days and subsequently analyzed for surface marker expression. All fractions were capable of eliminating HL60 WT cells except for 1 culture with double-negative cells (data not shown), confirming that all fractions contained the CAR construct. (Lower left) Representative FACS plots of the sorted CAR-T cells after re-exposure to target cells. The label above the plots indicates the originally sorted phenotype. (Lower right) Stacked bar graph showing the distribution of the 4 quadrants in the 4 different conditions. Mean and standard deviation of the 4 donors are shown. The x-axis labels indicate the originally sorted fraction. Colors of bars indicate the output phenotype according to the legend. nt, nontransduced; wt, wild type.
CAR-T cells maintained their AML killing potential for up to 5 serial challenges confirming enduring functionality. Given our focus on CD8+ T cells, we used donors with predominant CD8+ CAR T-cell expansion (supplemental Figure 13A). Already during the first challenge, GPR56 was upregulated on CD8+ CD33.CAR-T cells exposed to CD33+ HL-60 WT cells; but neither on those exposed to HL60 KO cells lacking CD33 expression, nor on nontransduced activated T cells exposed to HL60 WT cells (Figure 4B,C; supplemental Figure 13B). Furthermore, nonspecific T-cell stimulation by addition of anti-CD3 alone or with anti-CD28 antibodies did not cause GPR56 upregulation in PBMCs (supplemental Figure 13C), and we observed only marginal GPR56 upregulation during CAR-T production, that is, activation and transduction (Figure 4B-C). Together, this suggested that T-cell receptor antigen encounter promotes strong GPR56 upregulation. The GPR56+ fraction segregated into a CD27+ and CD27– fraction. Although the GPR56+CD27– fraction was more variable, the GPR56+CD27+ fraction continuously increased during the serial challenges and dropped when CAR-Ts stopped expanding (Figure 4D; supplemental Figure 13D), indicating that the proliferative capacity was highest in the GPR56+CD27+ fraction after contact with target cells.
To analyze whether the phenotype was reversible, we sorted CD8+ CD33.CAR-Ts from challenge 2 as outlined in Figure 4E and re-exposed the sorted fractions to HL60 WT cells. All fractions were capable of eliminating HL60 WT cells except for 1 culture with double-negative cells (data not shown), confirming that all fractions contained the CAR construct. When comparing the phenotypes of the sorted cells after target cell exposure, we observed that GPR56+CD27+ cells had the greatest capability to regenerate all 4 quadrants (largest plasticity), followed by the GPR56+CD27– fraction, whereas the 2 GPR56– fractions mostly maintained their GPR56– phenotypes (Figure 4E). In conclusion, this effector-target CAR-T/HL-60 model confirmed that GPR56 upregulation occurs upon antigen encounter and suggested that a reciprocal shift between GPR56+CD27+ and GPR56+CD27– cells is possible.
GPR56 increases on the surface of CD8+ T cells after allo-HCT and marks allo-reactive cytotoxic T cells
To relate the observations of GPR56 dynamics from the CAR-T cell model with the allo-HCT setting, we collected 338 BM aspirates from a cohort of 139 AML patients (outlined in supplemental Figure 14; supplemental Table 1). To determine the transplantation effect on GPR56 expression, we compared BM from patients who never underwent allo-HCT with BM of patients prior to and after allo-HCT. The fraction of GPR56+ cells in the CD8+ TEM, CD8+ TEMRA, and CD4+ TEMRA compartments were significantly higher after allo-HCT, whereas the overall percentages of CD8+ TEM and TEMRA within the BM did not differ (Figure 5A; supplemental Figure 15A-B). In contrast, there was no significant increase in the fraction of GPR56+ cells on double negative CD3+CD4–CD8– and CD3–CD56+ natural killer (NK) cells between patients before and after transplantation (supplemental Figure 15C).
GPR56 increases after non-self recognition by allo-reactive T cells after allo-HCT. (A) Percentage of CD8+ TEM in BM (left) and percentage of GPR56+ on CD8+ TEM (right). Numbers below plots show the median percentage. Numbers between groups of patients without (no-Allo), before (pre-Allo), and after (post-Allo) allo-HCT indicate the P values (unpaired Wilcoxon test). Note that the total fraction of CD8+ TEM in BM does not significantly differ between the 3 groups. Box plots showing medians and quartiles, each dot represents an individual sample. (B) Representative FACS plots showing CD27 and GPR56 expression on CD3+CD8+ cells in healthy BM (left), in a patient with low GPR56 upregulation (patient 1 [middle]), and a patient with a dominant GPR56+CD27– fraction (patient 2 [right]). (C) Time course of percentage of GPR56–CD27+ (blue), GPR56+CD27+ (purple), and GPR56+CD27– (pink) in the CD8+ compartment in patients with CR after allo-HCT. Numbers above the box plots indicate the median percentages. Box plots represent medians, quartiles, and outliers. (D) Proposed model of CD8+ T-cell phenotype switch after allo-HCT. (E) Percentage of GPR56+ on CD8+ TEMRA in recipients with CMV IgG negativity (left) and positivity (right). Numbers above box plots indicate the median percentages. Box plots represent medians, quartiles, and outliers. (F) Time course of the percentage of the indicated cell types in patient GXW165. As indicated, CMV status was positive for the recipient and negative for the donor (CMV R/D pos/neg); donor sex was male. The text below the x-axis provides clinical information on the course of the disease. Note the drop in chimerism, neutrophils, and GPR56 positivity accompanied by FACS MRD positivity around day +100. IS was reduced, and the patient developed skin GVHD requiring steroids, which was accompanied by a steady increase in GPR56+, re-establishment of full donor chimerism, and CR. (G) Schematic illustrating the experimental strategy for measuring IFN-γ production using ELISpot. GPR56+ and GPR56– CD8+ T cells were sorted using FACS from PBMCs of 6 patients with AML in CR after allo-HCT. Sorted populations were then cocultured for 24 hours with primary AML blasts from initial diagnosis of the same patients. ELISpot assay was performed to detect and quantify IFN-γ production by the T cells in response to AML blasts. (H) Representative wells from the ELISpot assay after coculturing primary AML blasts with GPR56– (orange) and GPR56+ (purple) CD8+ T cells for 24 hours. (I) ELISpot results showing the numbers of spots generated in the GPR56+ and GPR56– T-cell fractions of 6 patients with AML in remission within 24 hours of contact with matched AML blasts. P value was calculated using paired Wilcoxon test. Individual points indicate biological replicates (mean across technical replicates) and connected points indicate fractions originating from the same sample. Chim, donor chimerism; IgG, immunoglobulin G; IS, under immunosuppression; MRD, minimal residual disease.
GPR56 increases after non-self recognition by allo-reactive T cells after allo-HCT. (A) Percentage of CD8+ TEM in BM (left) and percentage of GPR56+ on CD8+ TEM (right). Numbers below plots show the median percentage. Numbers between groups of patients without (no-Allo), before (pre-Allo), and after (post-Allo) allo-HCT indicate the P values (unpaired Wilcoxon test). Note that the total fraction of CD8+ TEM in BM does not significantly differ between the 3 groups. Box plots showing medians and quartiles, each dot represents an individual sample. (B) Representative FACS plots showing CD27 and GPR56 expression on CD3+CD8+ cells in healthy BM (left), in a patient with low GPR56 upregulation (patient 1 [middle]), and a patient with a dominant GPR56+CD27– fraction (patient 2 [right]). (C) Time course of percentage of GPR56–CD27+ (blue), GPR56+CD27+ (purple), and GPR56+CD27– (pink) in the CD8+ compartment in patients with CR after allo-HCT. Numbers above the box plots indicate the median percentages. Box plots represent medians, quartiles, and outliers. (D) Proposed model of CD8+ T-cell phenotype switch after allo-HCT. (E) Percentage of GPR56+ on CD8+ TEMRA in recipients with CMV IgG negativity (left) and positivity (right). Numbers above box plots indicate the median percentages. Box plots represent medians, quartiles, and outliers. (F) Time course of the percentage of the indicated cell types in patient GXW165. As indicated, CMV status was positive for the recipient and negative for the donor (CMV R/D pos/neg); donor sex was male. The text below the x-axis provides clinical information on the course of the disease. Note the drop in chimerism, neutrophils, and GPR56 positivity accompanied by FACS MRD positivity around day +100. IS was reduced, and the patient developed skin GVHD requiring steroids, which was accompanied by a steady increase in GPR56+, re-establishment of full donor chimerism, and CR. (G) Schematic illustrating the experimental strategy for measuring IFN-γ production using ELISpot. GPR56+ and GPR56– CD8+ T cells were sorted using FACS from PBMCs of 6 patients with AML in CR after allo-HCT. Sorted populations were then cocultured for 24 hours with primary AML blasts from initial diagnosis of the same patients. ELISpot assay was performed to detect and quantify IFN-γ production by the T cells in response to AML blasts. (H) Representative wells from the ELISpot assay after coculturing primary AML blasts with GPR56– (orange) and GPR56+ (purple) CD8+ T cells for 24 hours. (I) ELISpot results showing the numbers of spots generated in the GPR56+ and GPR56– T-cell fractions of 6 patients with AML in remission within 24 hours of contact with matched AML blasts. P value was calculated using paired Wilcoxon test. Individual points indicate biological replicates (mean across technical replicates) and connected points indicate fractions originating from the same sample. Chim, donor chimerism; IgG, immunoglobulin G; IS, under immunosuppression; MRD, minimal residual disease.
Combined GPR56 and CD27 staining revealed a highly variable CD27/GPR56 pattern (Figure 5B). Stratifying the available CR samples by collection time point, we noticed a continuous decrease of the GPR56–CD27+ fraction and a transient increase followed by a decrease of the double positive fraction (Figure 5C-D). The GPR56+CD27– fraction continuously increased, reaching a median of ∼50% 1 year after allo-HCT compared with 10% in patients who did not receive transplantation (Figure 5C). No specific pattern of the GPR56+ fraction over time was observed on NK cells and CD3+CD4–CD8– cells (supplemental Figure 15D). These observations suggested a phenotype switch of CD8+ T cells after allo-antigen encounter (Figure 5D).
Antigen encounter after allo-HCT is not restricted to leukemia antigens but also occurs during infections or viral reactivation. We identified the cytomegalovirus (CMV) serostatus of the recipient as a major contributor to high GPR56 expression on T cells (Figure 5E). Importantly, the dynamic increase in GPR56 expression on TEMRA and TEM after allo-HCT was detectable in both, patients with CMV sero-positivity and those with sero-negativity, but GPR56 expression levels were generally higher in patients with CMV sero-positivity at all time points (Figure 5E; supplemental Figure 15E, upper panel). In contrast, the percentage of GPR56 on NK cells was independent of CMV status (supplemental Figure 15E, lower panel). Individual examples of 5 patients (3 with CR and 2 REL) for whom samples from multiple time points were available confirmed intrapatient dynamics of GPR56 expression after allo-HCT (Figure 5F; supplemental Figure 16A). When comparing the patients succumbing to relapse soon after sampling with those who stayed in CR, we observed that the GPR56+ fraction within CD8+ and CD8+ TEMRA cells was significantly lower in patients with imminent relapse than those in CR (n = 25; P = .01 for CD8+ and n = 23; P = .006 for CD8+ TEMRA; supplemental Figure 16B). Given the small cohort size, this observation needs to be followed up with large prospective cohorts.
To corroborate that GPR56 upregulation marks allo-reactive T cells, we measured IFN-γ production in vitro using an IFN-γ ELISpot assay, in which GPR56+ and GPR56– T cells sorted from PBMCs of 6 patients in CR were exposed to matched primary AML blasts from initial diagnosis (Figure 5G; supplemental Figure 17A). Within only 24 hours, the GPR56+ fractions showed stronger IFN-γ secretion in all 6 cocultures compared to the GPR56– fractions (Figure 5H, I; paired Wilcoxon; P = .031; supplemental Figure 17B; supplemental Table 11). No IFN-γ secretion was observed within 24 hours when coculturing GPR56+ T cells with nonmatched AML blasts (supplemental Figure 17C), supporting that IFN-γ secretion upon contact with matched recipient cells in this assay indicated T-cell memory of previous allo-antigen encounters. Together, these observations establish dynamic upregulation of GPR56 on CD8+ T cells as a novel hallmark of the allo-transplant setting and suggest GPR56 as a dynamic marker of antigen-experienced cytotoxic T cells, which might help monitor “non-self” recognition after allo-HCT.
Discussion
Allo-HCT in AML constitutes a highly efficient immunotherapy, but identification of GVL activity in individual patients is challenging. In this study, we combined scRNA-seq of BM CD3+ T cells and CD34+ HSPCs from day +100 BM aspirates of patients with AML who stayed in CR or suffered relapse shortly after sampling with a range of functional assays (CAR-T cell experiments, flow cytometry, ELISpot). This allowed us to delineate compositional, transcriptional, and functional characteristics of T cells that might be associated with outcome.
Among donor-derived T-cell subsets, cytotoxic cells (CD8+ T effector memory subsets and γδΤ cells) were enriched in CR samples. Higher γδΤ cell content in the grafts has been associated with better outcome after allo-HCT,30 and clinical trials with γδΤ cells as salvage therapy are ongoing (eg, NCT03790072), but their safety and efficacy remain to be determined.
We identified relapse-associated T-cell signatures defined by inflammatory/TNF-α (FOS/JUN31,32) and NF-κB signaling, which has been associated with dysfunctional T cells in renal cell carcinoma,33 and an immunosuppressive microenvironment (CREM24). The proinflammatory TFs were more active in REL samples in all cell types (CD4+ and CD8+), suggesting a general proinflammatory milieu in the BM that may hamper GVL.
Furthermore, we identified TBX21 activity as a hallmark of CR, and its regulon was enriched for cytotoxicity genes and also comprised the adhesion GPCR GPR56. Although GPR56 is considered inhibitory in NK cells,34 its function in T cells is less clear.35,36 We previously identified GPR56 as an LSC marker in AML37 that promotes self-renewal of stem cells. In human NK cells, GPR56 seems to inhibit NK cell function, and its expression is driven by the TF HOBIT/ZNF683.34 Here, we show that GPR56 and HOBIT are coexpressed with NKG7 in the pseudotime analysis and in CD8+ memory 2 cells, which were enriched in CR. Because HOBIT has been linked to lifelong, periodic challenges38 and considering the role of GPR56 in stemness of HSCs, it is possible that GPR56 is part of the HOBIT network that drives long-term memory in T cells. In support of such a model, we show here that GPR56+ T cells recognize and react to patients’ AML cells faster upon recontact than their GPR56– counterparts. Moreover, there is concomitant GPR56 and HOBIT upregulation in lifelong CMV-specific T cells.38,39 Further functional T-cell studies are thus warranted to dissect the role of GPR56 in T-cell memory.
Here, we provide evidence that (1) GPR56 expression on T cells occurs only upon antigen encounter, (2) the GPR56+ compartment segregates into a more proliferative CD27+ and a less proliferative CD27– fraction, (3) both fractions can regenerate each other upon re-challenge, (4) the CMV-serostatus highly influences the GPR56 baseline level of T cells, and (5) GPR56 marks an allo-reactive, highly cytotoxic T-cell population that recognizes the patients’ original AML blasts. Assuming that donor-derived T cells recognize both healthy and leukemic “non-self” antigens, higher GPR56 expression after allo-HCT may thus indicate a higher likelihood of recognizing and eliminating residual LSCs. Larger and prospective cohorts will be required to establish GPR56 as an early biomarker for therapy response. Ultimately, knowledge about the clues that foster GPR56 upregulation and those responsible for the inflammatory, immunosuppressive milieu found in REL samples might help design small molecules that drive GVL while preventing relapse.
Together, our study establishes a reference map of donor-derived BM T cells after allo-HCT and proposes GPR56 dynamics as a surrogate for the extent of “non-self” recognition after allo-HCT.
Acknowledgments
The authors thank V. Eckstein of the FACS Core Facility of Medical Department V, Heidelberg University Hospital, Heidelberg, Germany, for support with flow cytometry experiments, D. Ordonez of EMBL FACS Core Facility for assistance with FACS sorting for single cell RNA sequencing (scRNAseq), and V. Benes and members of EMBL Genomics Core Facility for support with scRNAseq. The authors thank N. Servaas for the discussions about T-cell biology. The authors thank R. Kruhmann, C. Holitsch, and S. Leonhardt for assistance with patient sample preparation, the members of the GMP laboratory of Medical Department V, Heidelberg University Hospital, for assistance with CAR T-cell experiments. The authors thank the team of the Biomaterial bank of Medical Department V, Heidelberg University Hospital, and all donors for providing primary human cells. Schematics were created using Biorender.com.
The study was supported by a Max-Eder-Grant of German Cancer Aid (70114435; C.P.) and funding by the German Research Foundation (DFG) (PA 2815/2-1; C.P.). This project was cofunded by the European Union (European Research Council [ERC], epiNicheAML, 101044873). The authors acknowledge the data storage service SDS@hd supported by the Ministry of Science, Research and the Arts Baden-Württemberg (MWK) and the DFG through grant INST 35/1314-1 FUGG and INST 35/1503-1 FUGG.
Views and opinions expressed are those of the author(s) only and do not necessarily reflect those of the European Union or the ERC. Neither the European Union nor the granting authority can be held responsible for them.
Authorship
Contribution: A.M. performed experiments, performed clinical data analysis, performed computational analyses, generated most figures, codeveloped the shiny online application, and wrote the manuscript; X.W. collected samples and information from patients with AML, performed experiments, performed clinical data and sample analyses, helped with computational analyses, generated figures, and wrote the manuscript; D.S. generated CD33.CAR T-cells, performed CAR-T and ELISpot experiments, generated figures, and wrote the manuscript; R.H. collected patient information and assisted CAR-T experiments; A.K. developed the shiny online application; M.H. provided primary human samples and supervised the FACS staining; Y.L. generated the 3G.CD33.CAR construct and CRISPR/Cas9 engineered HL60 cells; S.V. and P.L. performed and analyzed GPR56 staining in patient samples; C.M.-T., P.D., and T.L. provided samples, clinical information, and edited the manuscript; T.S. and M.S. provided primary human PBMC for CAR-T production, expertise on CAR-T systems, and edited the manuscript; J.B.Z. supervised all computational analyses, wrote the manuscript, and co-supervised the project; and C.P. directed the project, performed clinical data and sample analyses, generated figures, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Caroline Pabst, Department of Medicine V, Hematology, Oncology and Rheumatology, University Hospital Heidelberg, Im Neuenheimer Feld 410, 69120 Heidelberg, Germany; email: caroline.pabst@med.uni-heidelberg.de; and Judith B. Zaugg, Structural and Computational Biology Unit, European Molecular Biology Laboratory, Meyerhofstr 1, 69117 Heidelberg, Germany; email: judith.zaugg@embl.de.
References
Author notes
A.M. and X.W. contributed equally to this work.
Single-cell sequencing data contained in this manuscript have been uploaded to the Gene Expression Omnibus database (accession number GSE216187).
The study data set can be browsed at https://apps.embl.de/allosct/.
Data are available upon reasonable request from the corresponding authors, Caroline Pabst (caroline.pabst@med.uni-heidelberg.de) and Judith B. Zaugg (judith.zaugg@embl.de).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




![CD8+ T cells in REL samples have lower cytotoxic potential. (A) Differential TF activity analysis using SCENIC. Heat map indicating log2 odds ratios; only significantly enriched TFs are colored (Fisher exact test false discovery rate < 0.05) (left). Characterization of target genes based on known gene sets (right). Assignment of target genes to known functions was performed using publicly available gene sets (supplemental Methods). The colored bars represent the fraction of target genes per TF, which belong to the different gene sets (IFN, IFN response; Activation, Immune cell activation; TNF, TNF signaling). (B) CD8+ T gene regulatory network (GRN) of exemplary differentially active TFs (TBX21, REL, and FOS) and their target DEGs. (C) UMAP highlighting the T-cell clusters that belong to the CD8+ effector memory (EM) cells and were used for the differential expression analysis. (D) Heat map depicting scaled expression (z-score) across all 6 samples of 235 DEGs (CR, 144 genes; REL, 91 genes) in CD8+ EM clusters. Analysis was performed using the MAST algorithm (log2FC > 0.5; adjusted P value [p.adj] < 0.05 after Bonferroni correction). (E) Gene ontology and hallmark enrichment analysis on the DEGs from D. Terms were selected from the top-enriched terms. Full list provided in supplemental Table 8. (F) UMAP indicating the normalized gene expression of ADGRG1/GPR56. (G) GPR56 and CD27 expression across pseudotime of CD8+ cells, split per condition. The bar plots below indicate the percentages of GPR56+ and CD27+ cells in REL and CR samples when considering all CD8+ clusters. (H) Percentage of GPR56 positive cells in the indicated fractions determined by flow cytometry in the same 6 samples used for scRNA-seq. (Top left cartoon) Gating strategy used to identify naive, central memory (TCM), EM (TEM), and CD45RA+ EM (TEMRA) cells using CCR7 and CD45RA. P values were calculated using Student t test. (I) Volcano plot illustrating the DEGs between GPR56+ (purple) and GPR56– (orange) CD8+ TEM cells of patients in CR. The y-axis represents P value after Bonferroni correction (p.adj), and points were colored according to absolute log2FC > 0.5 and P.adj < .05 (purple/orange). (J) Box plots illustrating the percentage of GZMB+ (top) and PRF1+ (bottom) T cells in the GPR56+ and GPR56− fractions assessed by intracellular flow cytometry. Connected points indicate fractions originating from the same sample. P value was calculated using paired Wilcoxon test, n = 10.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/13/10.1182_blood.2023021815/1/m_blood_bld-2023-021815-gr3hj.jpeg?Expires=1767698736&Signature=HfgtjvEC2QfxAdb4AyhMLloM9vCA2d5~vwxyGmV0~qIB-YnQi5CrPSBgL9aKSaxvireylbzIbyeA78EhhroDS8o4F4KYVnR1qTdJwURvsdFFGRhuOoC7ptU-VtkzqumBjdTULVxTf4LqtRqM9NvLJXvBq-Pth-rRr3Lv6ReD6lfZRRLKxLhfM64AbQRRe6un1mOxc96WZf8-F9-bt0PWhmafHu0vnPGMIgKTnwxe18iBkIjdpKBl4F0ljfzeepjnkZnD~Ck~5AdZOmFg2woMHcZhwNSRQBfMJRIBpZrQ5ydNk-X9XDhprpjYKK9ebM2jsvQpAxTHOtxIX1sSbC45QA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![GPR56 increases after non-self recognition by allo-reactive T cells after allo-HCT. (A) Percentage of CD8+ TEM in BM (left) and percentage of GPR56+ on CD8+ TEM (right). Numbers below plots show the median percentage. Numbers between groups of patients without (no-Allo), before (pre-Allo), and after (post-Allo) allo-HCT indicate the P values (unpaired Wilcoxon test). Note that the total fraction of CD8+ TEM in BM does not significantly differ between the 3 groups. Box plots showing medians and quartiles, each dot represents an individual sample. (B) Representative FACS plots showing CD27 and GPR56 expression on CD3+CD8+ cells in healthy BM (left), in a patient with low GPR56 upregulation (patient 1 [middle]), and a patient with a dominant GPR56+CD27– fraction (patient 2 [right]). (C) Time course of percentage of GPR56–CD27+ (blue), GPR56+CD27+ (purple), and GPR56+CD27– (pink) in the CD8+ compartment in patients with CR after allo-HCT. Numbers above the box plots indicate the median percentages. Box plots represent medians, quartiles, and outliers. (D) Proposed model of CD8+ T-cell phenotype switch after allo-HCT. (E) Percentage of GPR56+ on CD8+ TEMRA in recipients with CMV IgG negativity (left) and positivity (right). Numbers above box plots indicate the median percentages. Box plots represent medians, quartiles, and outliers. (F) Time course of the percentage of the indicated cell types in patient GXW165. As indicated, CMV status was positive for the recipient and negative for the donor (CMV R/D pos/neg); donor sex was male. The text below the x-axis provides clinical information on the course of the disease. Note the drop in chimerism, neutrophils, and GPR56 positivity accompanied by FACS MRD positivity around day +100. IS was reduced, and the patient developed skin GVHD requiring steroids, which was accompanied by a steady increase in GPR56+, re-establishment of full donor chimerism, and CR. (G) Schematic illustrating the experimental strategy for measuring IFN-γ production using ELISpot. GPR56+ and GPR56– CD8+ T cells were sorted using FACS from PBMCs of 6 patients with AML in CR after allo-HCT. Sorted populations were then cocultured for 24 hours with primary AML blasts from initial diagnosis of the same patients. ELISpot assay was performed to detect and quantify IFN-γ production by the T cells in response to AML blasts. (H) Representative wells from the ELISpot assay after coculturing primary AML blasts with GPR56– (orange) and GPR56+ (purple) CD8+ T cells for 24 hours. (I) ELISpot results showing the numbers of spots generated in the GPR56+ and GPR56– T-cell fractions of 6 patients with AML in remission within 24 hours of contact with matched AML blasts. P value was calculated using paired Wilcoxon test. Individual points indicate biological replicates (mean across technical replicates) and connected points indicate fractions originating from the same sample. Chim, donor chimerism; IgG, immunoglobulin G; IS, under immunosuppression; MRD, minimal residual disease.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/143/13/10.1182_blood.2023021815/1/m_blood_bld-2023-021815-gr5gi.jpeg?Expires=1767698736&Signature=n1cdeKLo2~D4km9XUbgqt6ftdjZT3xn6laEVLJ68w32JNaXTFTVUrqochNhRrEx8pGDtP3yJWP~JDZlvJJ9rLwm22tM9zCN54RQ89ml6KadDx4cPf4I~q30v4M8cTDhV9y9cMd61-WS~G9UGVu5b9q725z5c9X7lYmLNJGKQkd1Kcbu4ydWWczAxt3~YIF34ID6WncRKa7svYEsbJL7-rAkuvdbWoXL14zqyVA7108m10KnIpkBsiIYmwSfN4zKTrbncKOOAlF92adQm7sWsmTTNyORPjSLy-3CX2nmI0ffl2vCpheOF1uTbs-~R0H0ldqjncL7VD0dEEAvhwL1xog__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal