Key Points
A high-resolution single-cell map of diagnostic CML bone marrow to annotate features of TKI resistance.
Gene expression signatures within the LSCs and NK cells are indicative of TKI response.
Abstract
Primary resistance to tyrosine kinase inhibitors (TKIs) is a significant barrier to optimal outcomes in chronic myeloid leukemia (CML), but factors contributing to response heterogeneity remain unclear. Using single-cell RNA (scRNA) sequencing, we identified 8 statistically significant features in pretreatment bone marrow, which correlated with either sensitivity (major molecular response or MMR) or extreme resistance to imatinib (eventual blast crisis [BC] transformation). Employing machine-learning, we identified leukemic stem cell (LSC) and natural killer (NK) cell gene expression profiles predicting imatinib response with >80% accuracy, including no false positives for predicting BC. A canonical erythroid-specifying (TAL1/KLF1/GATA1) regulon was a hallmark of LSCs from patients with MMR and was associated with erythroid progenitor [ERP] expansion in vivo (P < .05), and a 2- to 10-fold (6.3-fold in group A vs 1.09-fold in group C) erythroid over myeloid bias in vitro. Notably, ERPs demonstrated exquisite TKI sensitivity compared with myeloid progenitors (P < .001). These LSC features were lost with progressive resistance, and MYC- and IRF1-driven inflammatory regulons were evident in patients who progressed to transformation. Patients with MMR also exhibited a 56-fold expansion (P < .01) of a normally rare subset of hyperfunctional adaptive-like NK cells, which diminished with progressive resistance, whereas patients destined for BC accumulated inhibitory NKG2A+ NK cells favoring NK cell tolerance. Finally, we developed antibody panels to validate our scRNA-seq findings. These panels may be useful for prospective studies of primary resistance, and in assessing the contribution of predetermined vs acquired factors in TKI response heterogeneity.
Introduction
Chronic myeloid leukemia (CML) is both defined and driven by the BCR::ABL1 oncogenic kinase.1 Although targeting the BCR::ABL1 kinase with tyrosine kinase inhibitors (TKIs) has proved immensely successful, clinical responses remain highly heterogeneous with ∼40% of the patients having major molecular responses (MMRs) within 12 months, and ∼10% to 15% of patients progressing to blast crisis (BC).2,3 Intriguingly, recent work has demonstrated that rapid declines in BCR::ABL1 transcript levels within the first 3 months of TKI therapy predict clinical outcomes, including MMRs and treatment-free remissions.4-6 These striking observations suggest that factors contributing to primary TKI resistance (failure to respond to first-line TKIs) are already present at diagnosis. Importantly, the discovery of such factors will allow the development of biomarkers to predict TKI response and facilitate the generation of novel hypotheses aimed at understanding and overcoming primary resistance.
Prior work has suggested that both the leukemic and immune cell compartments contribute significantly to response heterogeneity,7 but few studies have comprehensively surveyed both compartments simultaneously, or have exploited the potential for biomarker discovery using high-dimensional approaches. Within the leukemic compartment, 2 independent studies have shown that bulk transcriptomes from CD34+ chronic phase (CP) cells obtained at presentation predict both TKI resistance and early BC transformation.8,9 Recent work from our group has suggested that epigenetic reprogramming via both DNA methylation and aberrant polycomb repressor complex activity contribute to a common set of gene expression (GE) changes observable in both data sets.10 Together, these studies suggest the existence of a subset of leukemic stem and progenitor cells within bulk CD34+ populations harboring prognostic GE signatures, but to date, their identity and the molecular features underpinning their differential TKI sensitivity are unknown. In addition, and within the immune compartment, several studies have described changes expected to impair antileukemic responses, while also enhancing suppressor activity.11,12 These include both numerical and functional changes among natural killer (NK) cells, cytotoxic T cells, dendritic cells, B cells, and myeloid-derived suppressor cells.13
To uncover cell types associated with differential TKI responses, we surveyed the entire bone marrow (BM) compartment of patients with CML with different clinical outcomes using single cell RNA sequencing (scRNA-seq). Applying machine learning (ML), we found in an unbiased data-driven manner that specific GE signatures within leukemic stem cells (LSCs) and NK cells best predicted TKI response at time of diagnosis. Using a combination of computational and functional approaches, we develop novel hypotheses to explain how early LSC fate–based decisions, as well as NK cell–hematopoietic stem and progenitor cell (HSPC) interactions might influence TKI sensitivity. Finally, we designed a custom antibody set for “cytometry by time of flight” (CyTOF) and conventional flow cytometry as a step toward converting cellular abundance and GE patterns into clinically informative biomarkers. We anticipate that our candidate biomarkers and the increased understanding that emerge from our study will not only predict a patient’s response to therapy at diagnosis (Figure 7), particularly those destined to transform to BC,9 but also suggest new therapeutic strategies.2
Generation and validation of a CML BM atlas at the single-cell resolution. (A) Diagnostic BM aspirates from normal controls and a cohort of patients with CP CML with differential imatinib responses were subjected to multimodal single-cell analysis using scRNA-seq. The patients with CP CML were classified, broadly based in line with European LeukemiaNet recommendations (supplemental Methods), as optimal responders to imatinib (group A), suboptimal responders to imatinib (group B), or as treatment failures if they progressed to BC (group C) (refer to supplemental Table 1 for patient details). The resulting single-cell atlas was subjected to data analysis pipelines to uncover features with predictive potential at the time of diagnosis. Mass cytometry, flow cytometry, and functional analyses were performed to validate the key predictive features. (B, left) The CD34+ and CD34− fractions were subjected to scRNA-seq analysis and analyzed as described in the supplemental Methods. The 4 major cell types from the scRNA-seq data set, namely B cells, myeloid cells, T and NK cells, and HSPCs were first identified. (B, right) The 4 major cell types were subclustered and cluster markers/GE signatures were used to identify 10 HSPC and 22 differentiated populations (8 myeloid; 7 B cell, and 7 T/NK metaclusters or cell types). (C) UMAP of single-cell transcriptomes, colored by control or CML cells. (D) HSPC subpopulations are plotted as a proportion of CD34+ cells. Each dot represents 1 patient. (E) T, NK, B, and myeloid cell types are plotted as a proportion of CD34− cells. (F) The 10 HSPC metaclusters were aggregated into a single pseudobulked population (CD34+ HSPC). GSEA comparisons were performed between groups A and C using gene sets curated in supplemental Table 12. Enrichment plots for differentially enriched gene sets are shown (false discovery rate q-value ≤0.1). (G) DNTT (TDT) expression in pseudobulked CD34+ cells of group C samples that transformed into lymphoid BC was contrasted against all remaining CML samples. A 2-tailed t test was used to test the statistical significance of data in Figure 1C-D,G. Supplemental Figures 1-3 and supplemental Tables 1-13 are linked with data shown in Figure 1. C Mono, classical monocytes; cDC2, classical dendritic cells; CD4+ TCM, CD4+ T central memory; CD8+ TEMRA, CD8+ T effector memory with CD45RA; EOBM, eosinophil-basophil mast cell progenitor; iMKP, immature megakaryocyte progenitor; I Mono, inflammatory monocytes; Lyp, lymphoid progenitor; Mono-T, monocyte-T; MKP, megakaryocyte progenitor; γδ T cells, T cells.
Generation and validation of a CML BM atlas at the single-cell resolution. (A) Diagnostic BM aspirates from normal controls and a cohort of patients with CP CML with differential imatinib responses were subjected to multimodal single-cell analysis using scRNA-seq. The patients with CP CML were classified, broadly based in line with European LeukemiaNet recommendations (supplemental Methods), as optimal responders to imatinib (group A), suboptimal responders to imatinib (group B), or as treatment failures if they progressed to BC (group C) (refer to supplemental Table 1 for patient details). The resulting single-cell atlas was subjected to data analysis pipelines to uncover features with predictive potential at the time of diagnosis. Mass cytometry, flow cytometry, and functional analyses were performed to validate the key predictive features. (B, left) The CD34+ and CD34− fractions were subjected to scRNA-seq analysis and analyzed as described in the supplemental Methods. The 4 major cell types from the scRNA-seq data set, namely B cells, myeloid cells, T and NK cells, and HSPCs were first identified. (B, right) The 4 major cell types were subclustered and cluster markers/GE signatures were used to identify 10 HSPC and 22 differentiated populations (8 myeloid; 7 B cell, and 7 T/NK metaclusters or cell types). (C) UMAP of single-cell transcriptomes, colored by control or CML cells. (D) HSPC subpopulations are plotted as a proportion of CD34+ cells. Each dot represents 1 patient. (E) T, NK, B, and myeloid cell types are plotted as a proportion of CD34− cells. (F) The 10 HSPC metaclusters were aggregated into a single pseudobulked population (CD34+ HSPC). GSEA comparisons were performed between groups A and C using gene sets curated in supplemental Table 12. Enrichment plots for differentially enriched gene sets are shown (false discovery rate q-value ≤0.1). (G) DNTT (TDT) expression in pseudobulked CD34+ cells of group C samples that transformed into lymphoid BC was contrasted against all remaining CML samples. A 2-tailed t test was used to test the statistical significance of data in Figure 1C-D,G. Supplemental Figures 1-3 and supplemental Tables 1-13 are linked with data shown in Figure 1. C Mono, classical monocytes; cDC2, classical dendritic cells; CD4+ TCM, CD4+ T central memory; CD8+ TEMRA, CD8+ T effector memory with CD45RA; EOBM, eosinophil-basophil mast cell progenitor; iMKP, immature megakaryocyte progenitor; I Mono, inflammatory monocytes; Lyp, lymphoid progenitor; Mono-T, monocyte-T; MKP, megakaryocyte progenitor; γδ T cells, T cells.
Identification of prognostic pretreatment cell types by ML. (A) Bootstrapped logistic regression pipeline to predict imatinib treatment outcome based on CML prognostic group. Briefly, given the transcriptome of a single cell, the output identifies GE patterns that predict to which group that cell belongs to. Specifically, the output builds distinct GE-based signatures that classify each cell type into a prognostic group. (B) Top table: accuracy scores (ACC) of (i) multiclass (A vs B vs C) and (ii) binary (C vs not C [AB]) logistic-regression classifiers. The ACC scores for the top 3 models of both multiclass and binary classifiers are highlighted in red. ACC scores are the ratio of true positives and true negatives to all positive and negative observations. Bottom table: left, confusion matrices displaying cell counts for the top 3 cell types in the multiclass classifiers, and right, for the top 3 binary classifiers. In a random model, the ACC = 0.33 for a multiclass classifier and 0.5 for a binary classifier. (C) A leave-one-patient-out classifier to identify marker genes in pseudobulked transcriptomes that can serve as prognostic markers. Only markers from the classifier in panel A are considered as the candidate features in the model. Confusion matrices for a patient-specific (D) multiclass HSC, (E) binary HSC, and NK cell classifier. Precision (Pre) is the ratio between true positives and all positives. Recall (Rec) is a measure of the model’s ability to identify true positives (true positive/true positive and false negative). (F) Top nonzero regression coefficients of the HSCs multiclass patient-specific classifier. (G) Top 20 nonzero regression coefficients identified by the patient-specific binary HSC classifier. (H) Top 20 nonzero regression coefficients identified by the patient-specific binary NK cell classifier. Statistical tests for the ML pipelines are described in the supplemental Methods. Supplemental Figures 4 and 5 and supplemental Tables 14-16 are linked with data shown in Figure 2.
Identification of prognostic pretreatment cell types by ML. (A) Bootstrapped logistic regression pipeline to predict imatinib treatment outcome based on CML prognostic group. Briefly, given the transcriptome of a single cell, the output identifies GE patterns that predict to which group that cell belongs to. Specifically, the output builds distinct GE-based signatures that classify each cell type into a prognostic group. (B) Top table: accuracy scores (ACC) of (i) multiclass (A vs B vs C) and (ii) binary (C vs not C [AB]) logistic-regression classifiers. The ACC scores for the top 3 models of both multiclass and binary classifiers are highlighted in red. ACC scores are the ratio of true positives and true negatives to all positive and negative observations. Bottom table: left, confusion matrices displaying cell counts for the top 3 cell types in the multiclass classifiers, and right, for the top 3 binary classifiers. In a random model, the ACC = 0.33 for a multiclass classifier and 0.5 for a binary classifier. (C) A leave-one-patient-out classifier to identify marker genes in pseudobulked transcriptomes that can serve as prognostic markers. Only markers from the classifier in panel A are considered as the candidate features in the model. Confusion matrices for a patient-specific (D) multiclass HSC, (E) binary HSC, and NK cell classifier. Precision (Pre) is the ratio between true positives and all positives. Recall (Rec) is a measure of the model’s ability to identify true positives (true positive/true positive and false negative). (F) Top nonzero regression coefficients of the HSCs multiclass patient-specific classifier. (G) Top 20 nonzero regression coefficients identified by the patient-specific binary HSC classifier. (H) Top 20 nonzero regression coefficients identified by the patient-specific binary NK cell classifier. Statistical tests for the ML pipelines are described in the supplemental Methods. Supplemental Figures 4 and 5 and supplemental Tables 14-16 are linked with data shown in Figure 2.
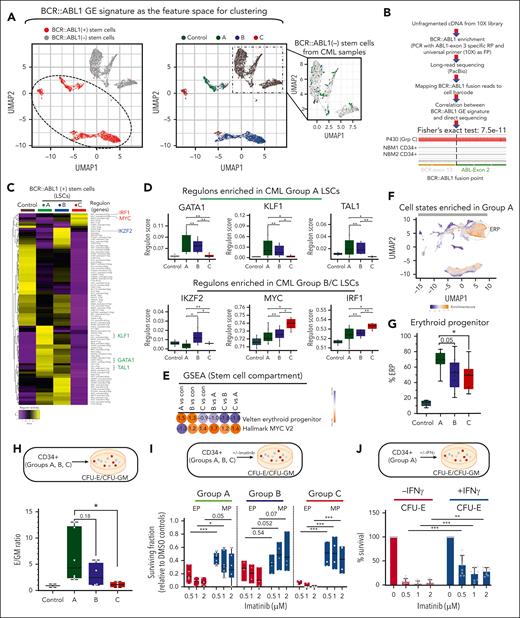
Identification and functional validation of LSC transcriptional programs. (A) HSCs were subclustered in a BCR::ABL1 GE feature space, as described in supplemental Figure 6A and in the supplemental Methods. (A, left) UMAP feature plots of HSCs colored by the presence or absence of the BCR::ABL1 GE signature. The dotted line demarcates the area that was identified as positive for the BCR::ABL1 GE signature. These cells are referred to as the BCR::ABL1+ stem cells or LSCs. (A, right) UMAP feature plots of HSCs colored by group label. The zoomed-in inset shows stem cells from CML samples clustering with normal stem cells from controls. These cells are referred to as BCR::ABL1− stem cells. (B) BCR::ABL1 fusion reads were amplified from 10× barcoded full-length complementary DNA using the universal primer from the 10× Genomics 5′ GE kit as the forward primer (FP) and a primer spanning exon 3 of ABL1 gene as the reverse primer (RP) from 4 CML and 2 NBM CD34+ HSPC fractions, and products were subjected to long-read sequencing (supplemental Methods). GE signature-based assignment of BCR::ABL1 status was correlated with the direct detection of BCR::ABL1 junction-spanning reads. Fisher exact test, 7.5e−11. (C) HSCs from controls and LSCs from patients with CML were analyzed using SCENIC. Scores for regulons that showed significant differential activity (P value < .05) between any pair of comparisons are shown. Up to 2 regulons are identified for each transcription factor (TF). (D) Patient-specific pseudobulk regulon scores for selected TFs are plotted. Each dot represents 1 patient (E) GSEA comparisons between LSCs from groups A and C for the selected gene sets. (For a complete analysis, refer to supplemental Tables 19 and 20; supplemental Figure 7.) (F) scRNA-seq UMAP, colored by fold enrichment of group A cells among 300 neighboring cells in transcriptome space. Enriched areas demarcated by the dotted line correspond to ERPs in the CML single-cell atlas. (G) scRNA-seq; ERP abundance is plotted as a proportion of total HSPCs in each patient. Each dot represents 1 patient. (H) CFU-E/CFU-GM ratio: CD34+ HSPCs were purified from all 3 patient groups and subjected to colony-forming assay (CFA). Colonies were scored after 14 days as erythroid (CFU-E) or myeloid (CFU-GM). E/GM ratio is plotted for each patient (n = 6 group A; n = 4 group B; and n = 4 group C). (I) CD34+ cells were purified from all groups, and subjected to CFAs in the presence or absence of imatinib. The surviving fraction with respect to dimethyl sulfoxide controls is plotted. (J) Purified CD34+ cells were plated in methylcellulose-based media in the presence/absence of imatinib/IFN-γ. Colony numbers for CFU-E were plotted relative to vehicle (dimethyl sulfoxide) control. The data for total colony numbers and for CFU-GM are shown in supplemental Figure 7E. Statistical tests: Figure 3D,F-I; 2-tailed t test. Supplemental Figures 6 and 7 and supplemental Tables 17-20 are linked with data shown in Figure 3. CFU-E, colony forming unit-erythroid; CFU-GM, colony forming unit-granulocyte-macrophage.
Identification and functional validation of LSC transcriptional programs. (A) HSCs were subclustered in a BCR::ABL1 GE feature space, as described in supplemental Figure 6A and in the supplemental Methods. (A, left) UMAP feature plots of HSCs colored by the presence or absence of the BCR::ABL1 GE signature. The dotted line demarcates the area that was identified as positive for the BCR::ABL1 GE signature. These cells are referred to as the BCR::ABL1+ stem cells or LSCs. (A, right) UMAP feature plots of HSCs colored by group label. The zoomed-in inset shows stem cells from CML samples clustering with normal stem cells from controls. These cells are referred to as BCR::ABL1− stem cells. (B) BCR::ABL1 fusion reads were amplified from 10× barcoded full-length complementary DNA using the universal primer from the 10× Genomics 5′ GE kit as the forward primer (FP) and a primer spanning exon 3 of ABL1 gene as the reverse primer (RP) from 4 CML and 2 NBM CD34+ HSPC fractions, and products were subjected to long-read sequencing (supplemental Methods). GE signature-based assignment of BCR::ABL1 status was correlated with the direct detection of BCR::ABL1 junction-spanning reads. Fisher exact test, 7.5e−11. (C) HSCs from controls and LSCs from patients with CML were analyzed using SCENIC. Scores for regulons that showed significant differential activity (P value < .05) between any pair of comparisons are shown. Up to 2 regulons are identified for each transcription factor (TF). (D) Patient-specific pseudobulk regulon scores for selected TFs are plotted. Each dot represents 1 patient (E) GSEA comparisons between LSCs from groups A and C for the selected gene sets. (For a complete analysis, refer to supplemental Tables 19 and 20; supplemental Figure 7.) (F) scRNA-seq UMAP, colored by fold enrichment of group A cells among 300 neighboring cells in transcriptome space. Enriched areas demarcated by the dotted line correspond to ERPs in the CML single-cell atlas. (G) scRNA-seq; ERP abundance is plotted as a proportion of total HSPCs in each patient. Each dot represents 1 patient. (H) CFU-E/CFU-GM ratio: CD34+ HSPCs were purified from all 3 patient groups and subjected to colony-forming assay (CFA). Colonies were scored after 14 days as erythroid (CFU-E) or myeloid (CFU-GM). E/GM ratio is plotted for each patient (n = 6 group A; n = 4 group B; and n = 4 group C). (I) CD34+ cells were purified from all groups, and subjected to CFAs in the presence or absence of imatinib. The surviving fraction with respect to dimethyl sulfoxide controls is plotted. (J) Purified CD34+ cells were plated in methylcellulose-based media in the presence/absence of imatinib/IFN-γ. Colony numbers for CFU-E were plotted relative to vehicle (dimethyl sulfoxide) control. The data for total colony numbers and for CFU-GM are shown in supplemental Figure 7E. Statistical tests: Figure 3D,F-I; 2-tailed t test. Supplemental Figures 6 and 7 and supplemental Tables 17-20 are linked with data shown in Figure 3. CFU-E, colony forming unit-erythroid; CFU-GM, colony forming unit-granulocyte-macrophage.
Association of NK cell states and markers with TKI response. (A) Expression heatmap of the top 10 marker genes for each NK cell subpopulation. The expression of literature-derived NK cell marker genes (CD56 [NCAM1], FCGR3A [CD16], CD57 [B3GAT1], NKG2C [KLRC2], and ZBTB16]) are plotted for comparison. Annotations were aided by Module Score computations for the 7 NK cell gene signatures compiled from literature (supplemental Table 21A; supplemental Figure 8C; “Methods”). Module scores were calculated for each NK cell type; note that adaptive NK cell signatures 1 to 3 are gene sets that are upregulated, whereas signature 4 is a gene set that is downregulated in the adaptive NK population. (B) Left: UMAP showing the 6 NK cell subpopulations identified through NK cell subclustering. Right: UMAP colored by control or CML labels. (C) NK subpopulations are plotted as a proportion of total NK in control and patients with CML. (D) The proportion of adaptive-like NK cells across control and prognostic groups is shown. Each dot represents 1 patient. (E) Expression heatmap of the top 15 genes upregulated and downregulated in adaptive-like NK cells, relative to other NK subpopulations. (F) Cell-to-cell communication analysis: NATMI was performed to identify potential interactions between ligands on HSPCs and receptors on NK cells. Ligand-receptor pairs predicted to mediate differential HSPC-NK interactions between control and group A (left), or groups C and A (right). x-axis: log2-transformed fold change of edge expression weight inferred by NATMI. The interactions marked by red asterisks represent NK cell–inhibitory interactions. (G) Expression of (left) HLA-E in HSPCs, and (right) NKG2A (KLRC1) in NK cells using pseudobulked populations. Each dot represents 1 patient. (H) Schematic summarizing prognostic NK cell features across groups. Statistical tests: Figure 4D,G; 2-tailed t test. Supplemental Figures 8 and 9 and supplemental Table 21 are linked with the data shown in Figure 4.
Association of NK cell states and markers with TKI response. (A) Expression heatmap of the top 10 marker genes for each NK cell subpopulation. The expression of literature-derived NK cell marker genes (CD56 [NCAM1], FCGR3A [CD16], CD57 [B3GAT1], NKG2C [KLRC2], and ZBTB16]) are plotted for comparison. Annotations were aided by Module Score computations for the 7 NK cell gene signatures compiled from literature (supplemental Table 21A; supplemental Figure 8C; “Methods”). Module scores were calculated for each NK cell type; note that adaptive NK cell signatures 1 to 3 are gene sets that are upregulated, whereas signature 4 is a gene set that is downregulated in the adaptive NK population. (B) Left: UMAP showing the 6 NK cell subpopulations identified through NK cell subclustering. Right: UMAP colored by control or CML labels. (C) NK subpopulations are plotted as a proportion of total NK in control and patients with CML. (D) The proportion of adaptive-like NK cells across control and prognostic groups is shown. Each dot represents 1 patient. (E) Expression heatmap of the top 15 genes upregulated and downregulated in adaptive-like NK cells, relative to other NK subpopulations. (F) Cell-to-cell communication analysis: NATMI was performed to identify potential interactions between ligands on HSPCs and receptors on NK cells. Ligand-receptor pairs predicted to mediate differential HSPC-NK interactions between control and group A (left), or groups C and A (right). x-axis: log2-transformed fold change of edge expression weight inferred by NATMI. The interactions marked by red asterisks represent NK cell–inhibitory interactions. (G) Expression of (left) HLA-E in HSPCs, and (right) NKG2A (KLRC1) in NK cells using pseudobulked populations. Each dot represents 1 patient. (H) Schematic summarizing prognostic NK cell features across groups. Statistical tests: Figure 4D,G; 2-tailed t test. Supplemental Figures 8 and 9 and supplemental Table 21 are linked with the data shown in Figure 4.
Genetic associations with progressive TKI resistance. (A) Cell type–specific clusters, in which >50% of cells are contributed by the CD34+ HSPCs of any 1 sample, were identified. Cell type clusters (eg, ERP-11, HSC-2, and LyP-1) are as described in supplemental Tables 9 and 10A,B. (B) CML samples with patient-specific clusters in panel A are shown in the UMAP space. Letters in parenthesis refer to the prognostic group (A, B, or C) each sample belongs to. (C) Genes that are frequently mutated in CML were compiled from the literature (supplemental Table 23), and their mutational status was assessed by WES. Oncoplot of somatic mutations in selected genes, colored by the type of mutation. The frequency of each mutation (%) is shown to the right of the plot. A total of 40 such variants were identified, as shown in supplemental Table 23. (D) To cross-reference the 40 SNVs identified in panel C, to our scRNA-seq data set, cb_sniffer was used (“Methods”). The asterisk indicates that the variant could be successfully detected within the scRNA reads of the same sample. (E) Cross-referencing variants identified in panel C to individual cell types. For any given variant, only the most recurrent cell type has been plotted. The complete data set is shown as supplemental Table 24B. Variants that can be detected in T/NK clusters have been highlighted with an orange rectangle. It should be noted that given the design of the 10× Genomics single cell technology, the absence of variant detection does not imply the absence of expression. Supplemental Figure 10 and supplemental Tables 22-24 are linked with the data in Figure 5.
Genetic associations with progressive TKI resistance. (A) Cell type–specific clusters, in which >50% of cells are contributed by the CD34+ HSPCs of any 1 sample, were identified. Cell type clusters (eg, ERP-11, HSC-2, and LyP-1) are as described in supplemental Tables 9 and 10A,B. (B) CML samples with patient-specific clusters in panel A are shown in the UMAP space. Letters in parenthesis refer to the prognostic group (A, B, or C) each sample belongs to. (C) Genes that are frequently mutated in CML were compiled from the literature (supplemental Table 23), and their mutational status was assessed by WES. Oncoplot of somatic mutations in selected genes, colored by the type of mutation. The frequency of each mutation (%) is shown to the right of the plot. A total of 40 such variants were identified, as shown in supplemental Table 23. (D) To cross-reference the 40 SNVs identified in panel C, to our scRNA-seq data set, cb_sniffer was used (“Methods”). The asterisk indicates that the variant could be successfully detected within the scRNA reads of the same sample. (E) Cross-referencing variants identified in panel C to individual cell types. For any given variant, only the most recurrent cell type has been plotted. The complete data set is shown as supplemental Table 24B. Variants that can be detected in T/NK clusters have been highlighted with an orange rectangle. It should be noted that given the design of the 10× Genomics single cell technology, the absence of variant detection does not imply the absence of expression. Supplemental Figure 10 and supplemental Tables 22-24 are linked with the data in Figure 5.
Toward development of antibody-based panels for predicting TKI responses. (A) A custom 38-antibody panel was designed for mass cytometry to detect the cell abundance gradients and GE profiles uncovered by our single-cell map. (B) The mean expression of indicated markers was assessed within the Lin−CD34+CD38− population of control and CML samples. (C) CD34+ HSPCs from the CyTOF data set was subjected to standard gating schemes to identify 6 HSPC populations: HSC, MPP, LMPP, CMP, megakaryocyte-erythroid progenitor (MEP), and GMP. MEP proportions were compared across CML groups A through C. (D) A flow cytometry panel was optimized to measure the frequency of ERPs across control and CML Groups A through C. An independent cohort of 6 patients of group A was used for this analysis, as well as the same set of group B and group C samples. (E) CD34− cells from the CyTOF data set were subjected to standard biaxial gating to identify NK cells (far right). The total abundance of NK cells with the CD34− population of samples and the mean expression of HLA-DR and CD7 within the NK cell population are shown. (F) A flow cytometry panel was optimized (supplemental Figure 13A) to measure the frequency of adaptive-like NK cells identified as CD57+NKG2C(KLRC2)+ cells within CD56dim CD16bright NK cell population. (G) A flow cytometry panel was optimized (supplemental Figure 13B) to measure the frequency of the NKG2A(KLRC1)+ NK subset within the CD56dim CD16bright NK cell population. Statistical tests for all data are shown in Figure 6; 2-tailed t test. Supplemental Figures 11-13 and supplemental Tables 25 and 26 are linked with the data in Figure 6.
Toward development of antibody-based panels for predicting TKI responses. (A) A custom 38-antibody panel was designed for mass cytometry to detect the cell abundance gradients and GE profiles uncovered by our single-cell map. (B) The mean expression of indicated markers was assessed within the Lin−CD34+CD38− population of control and CML samples. (C) CD34+ HSPCs from the CyTOF data set was subjected to standard gating schemes to identify 6 HSPC populations: HSC, MPP, LMPP, CMP, megakaryocyte-erythroid progenitor (MEP), and GMP. MEP proportions were compared across CML groups A through C. (D) A flow cytometry panel was optimized to measure the frequency of ERPs across control and CML Groups A through C. An independent cohort of 6 patients of group A was used for this analysis, as well as the same set of group B and group C samples. (E) CD34− cells from the CyTOF data set were subjected to standard biaxial gating to identify NK cells (far right). The total abundance of NK cells with the CD34− population of samples and the mean expression of HLA-DR and CD7 within the NK cell population are shown. (F) A flow cytometry panel was optimized (supplemental Figure 13A) to measure the frequency of adaptive-like NK cells identified as CD57+NKG2C(KLRC2)+ cells within CD56dim CD16bright NK cell population. (G) A flow cytometry panel was optimized (supplemental Figure 13B) to measure the frequency of the NKG2A(KLRC1)+ NK subset within the CD56dim CD16bright NK cell population. Statistical tests for all data are shown in Figure 6; 2-tailed t test. Supplemental Figures 11-13 and supplemental Tables 25 and 26 are linked with the data in Figure 6.
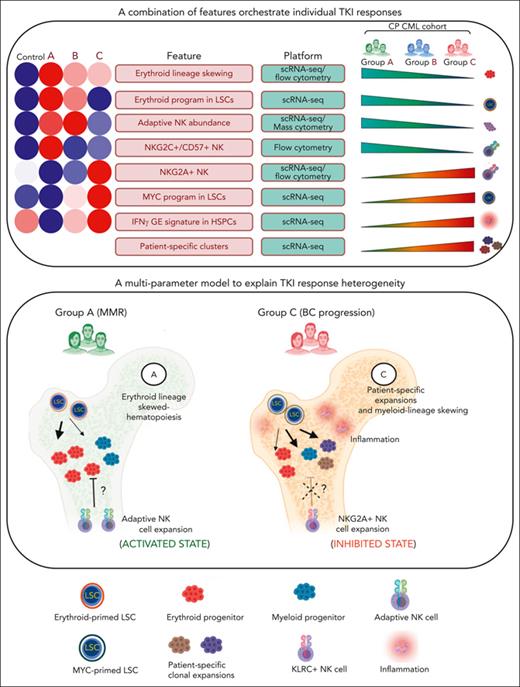
A multiparameter model of primary TKI resistance. (A) Eight pretreatment features associated with differential group-specific TKI responses are summarized here and in supplemental Table 27. For each feature, we provide a heatmap of the average degree of enrichment in each of the control and prognostic groups (supplemental Table 27), a technology platform for assessing each feature (mass or flow cytometry, or scRNA-seq), and whether the feature increases or decreases across groups. (B) Schematic summarizing the biological features contributing to heterogeneous TKI responses, including LSC/HSPC-, NK cell–, and tumor microenvironment–dependent factors.
A multiparameter model of primary TKI resistance. (A) Eight pretreatment features associated with differential group-specific TKI responses are summarized here and in supplemental Table 27. For each feature, we provide a heatmap of the average degree of enrichment in each of the control and prognostic groups (supplemental Table 27), a technology platform for assessing each feature (mass or flow cytometry, or scRNA-seq), and whether the feature increases or decreases across groups. (B) Schematic summarizing the biological features contributing to heterogeneous TKI responses, including LSC/HSPC-, NK cell–, and tumor microenvironment–dependent factors.
Methods
Prognostic groups and sample preparation for scRNA-seq
Samples from patients with primary CML were obtained from the Singapore General Hospital, in accordance with institutional review board protocols and after informed consent by the patients. Patients were broadly classified into prognostic categories based on European LeukemiaNet recommendations (supplemental Tables 1-3, available on the Blood website). We studied 3 groups of patients with CML with increasing TKI resistance: optimal response (MMR) to first-line imatinib (group A, n = 9), suboptimal responses to imatinib but favorable responses to second- or third-line TKIs (group B, n = 9); and pan-TKI resistance with eventual BC progression (group C, n = 6). Among patients in group C, 3 patients (P605, BM157, and P430) transformed into lymphoid BC at 76, 187, and 497 days, respectively, whereas 3 patients (LEU9, P309, and P147), transformed into myeloid BC at 519, 1518, and 2771 days, respectively. The 8 normal BM (NBM) mononuclear cells (MNCs) used in this study were harvested from healthy donors (aged, 36-55 years) and obtained from commercial sources (StemCell Technologies). Frozen BM-MNCs were thawed at 37°C, and CD34+ and CD34− fractions were isolated using the MACS technology (Miltenyi Biotec). scRNA-seq was done using the 10× 5′ V3 kit (10× Genomics).
scRNA-seq analysis pipelines
We devised ML pipelines to identify genes with prognostic value using the code base of the logistic regression classifier contained in the TEPIC framework.14 To identify the transcriptional networks operational in the LSCs, we applied the SCENIC algorithm.15 To identify candidate receptor-ligand pairs mediating cell-to-cell communication, we used the NATMI pipeline.16 Pseudobulked single-cell transcriptomes were analyzed using gene set enrichment analysis (GSEA, version 4.1.0) and module scores were computed using Seurat AddModuleScore implementation.
Mass cytometry and flow cytometry
CML BM-MNCs were stained with a 39-antibody panel (supplemental Table 4), and acquired on a CyTOF-II mass cytometer (Fluidigm). Data were analyzed using the FlowJo software (version 10.2; FlowJo) or subjected to FlowSOM clustering (Cytobank Inc, Beckman Coulter). For flow cytometry, cells were stained using the antibody panels described in supplemental Table 5.
Clonogenic assays
CD34+ cells were plated in methylcellulose (H4230; StemCell Technologies), supplemented with cytokines (supplemental Methods), and colonies were enumerated after 14 days in a blinded manner.
WES
Whole-exome sequencing (WES) libraries were sequenced using Novaseq 6000 (Illumina) and downstream analysis was performed as described in the supplemental Methods.
Results
Generation and validation of a single-cell CML BM atlas
To identify cell types contributing to differential TKI responses, we conducted a single-cell analysis of pretreatment CP BM samples. We studied the groups of patients with CML with increasing TKI resistance, groups A (n = 9), B (n = 9), and C (n = 6), as described in “Methods” (Figure 1A; supplemental Tables 1-3). Using scRNA-seq (10× Genomics Chromium), we interrogated 87 815 CD34+ as well as 75 331 CD34− BM MNC–derived single cells and detected an average of 11 935 unique molecular identifiers representing 2627 genes per cell (supplemental Figure 1A-C). After reference component analysis, we identified 4 major cell types (supplemental Figure 1D), which were subjected to major cell type–specific quality control and de novo clustering. Overall, we identified 64 transcriptional states (supplemental Tables 9 and 10A,D), which were merged into 32 cell populations (Figure 1B-C). Of these, 10 and 22 cell populations belonged to the CD34+ HSPC and the CD34− differentiated compartments (T/NK, B, and myeloid cells), respectively (Figure 1B). Within the HSPC compartment, subpopulations were assigned labels based on cluster-specific expression of known marker genes (supplemental Tables 6-9).17-19 Hematopoietic stem cells (HSCs) were identified as clusters that were positive for literature-derived HSC signature genes, HLF, CRHBP, and AVP, and lacking lineage markers. The identity of differentiated cell populations was ascertained through canonical marker GE (supplemental Figure 1E-G).17,18,20
Next, we used our atlas to assess the cellular hierarchy of CP HSPCs. Inferences from colony-forming assay-based studies (which provide readout for lineage potential and not actual in vivo cell fate) predict a relative reduction in primitive progenitors and stem cells together with significant expansion of erythroid, myeloid, and megakaryocytic progenitors compared with NBM.21 We confirmed that the proportions of major cell states within NBM controls were consistent with literature (Figure 1D-E), and that CP BM recapitulated the major cell types (HSPCs, and myeloid, T/NK, and B cells) found in NBM (supplemental Figure 2A-C; supplemental Table 10F,I, for cell type proportions in controls and patient groups).22 As expected, we observed an increase in the fraction of myeloid and neutrophil HSPC subpopulations (immature megakaryocyte progenitor, megakaryocyte progenitor, erythroid progenitor [ERP], immature neutrophil progenitor [iNeP], and NeP) in the total MNC compartment of patients with CML compared with controls (supplemental Table 10G; supplemental Figure 2D). Within the CD34+ fraction subjected to scRNA-seq, we observed a 7-fold relative decrease in the proportion of HSCs in patients with CML (CML vs control; 4.5% ± 11.1% vs 32.8% ± 7.2%; P < .0001) together with an expansion of differentiated progenitors that skewed toward erythropoietic (CML vs control; 56.6% ± 19.1% vs 13.4% ± 3.7%; P < .0001) and megakaryocytic (CML vs control; 4.9% ± 4.1% vs 1.2% ± 0.74%; P < .0001) lineages, and a decrease in iNePs and lymphoid progenitors (Figure 1D; supplemental Table 10G). However, 2 of the 3 patients with CP in group C (P605 and BM175) who eventually underwent lymphoid BC transformation, exhibited an increase in lymphoid progenitors and pro-B cells, respectively (for the complete data set on CD34+ HSPC cell type proportions across controls and prognostic groups, please refer to supplemental Figure 10E-F). In the CD34− compartment, only erythroblasts and mature B cells exhibited differential proportions between normal and CML samples (Figure 1E, complete data set on CD34− cell type proportions in supplemental Figure 10H-I). Comparing GE changes between control and CML samples, both known (increased GAS2 expression in CML HSCs) and novel GE alterations were uncovered across cell types (supplemental Table 11).
Pretreatment CD34+ cells from patients with CP who develop TKI resistance or early BC transformation share GE signatures reminiscent of BC GE changes.8-10 We tested the hypothesis that GE signatures found in BC HSPCs are detectable in group C HSPCs. Using GSEA, we found that multiple hallmark BC features were enriched in group C compared with Group A HSPCs (Figure 1F; supplemental Tables 12 and 13; supplemental Figures 2D and 3). These signatures included a BC signature, stemness, and inflammatory signaling (interferon gamma [IFN-γ], IFN-α, interleukin-2 [IL-2]-STAT5, and IL-6-JAK-STAT3 signaling), relative quiescence, and enrichment of polycomb repressor complex 1 components. Consistent with their intermediate status, group B samples exhibited an GE phenotype between that of groups A and C (supplemental Figure 2E, complete data set on GSEA analysis in supplemental Figure 3). As a second independent test, and because increased DNTT or TDT expression has been shown to predict early vs late lymphoid BC transformation at diagnosis,23 we confirmed that this was also the case in pseudobulked group C CD34+ HSPCs (Figure 1G). Taken together, our atlas represents an accurate map, at unprecedented resolution, of both cell-state perturbations in CML and prognostic GE information (http://scdbm.ddnetbio.com/).21,24 Next, we used ML to identify cell types and GE programs that could classify both cells and patients to prognostic groups in an unbiased manner.
ML identifies prognostic cell types
Here, we first collapsed all cells of the same cell type from each patient into a single “pseudobulked” transcriptome. Following pseudobulking, and through comparisons across the prognostic groups, we identified several differentially expressed genes (DEGs). For the complete list of DEGs at the pseudobulk level for every cell type (supplemental Table 14), with details on comparison methods and their known functions, please refer to supplemental Figure 4 and supplemental Table 15. Of these DEGs, HSPA8, PSME1, and TXN have been previously associated with drug resistance or progression in CML and/or acute myeloid leukemia.25-29 However, although valuable, simple DEG analysis does not exploit the predictive power of gene combinations, nor would such analyses directly assess predictive accuracy in assigning cell types or patients to our prognostic groups.
To overcome these limitations, we designed an ML pipeline based on logistic regression with elastic net regularization to identify combinations of cell type–specific genes that could determine which prognostic group a particular cell type was from (Figure 2A). The classifier was either trained as a multiclass (A vs B vs C) or binary (A/B vs C) classifier. Accuracy (ACC) scores were computed for every cell type, based on “predicted” and “true” label measurements. As a multiclass classifier, inflammatory monocytes (Inf-Mono) were the most accurate, followed by plasmacytoid dendritic cells (pDCs), and HSCs; whereas for the binary classifier, HSCs and pDCs performed the best, closely followed by NK cells (Figure 2B). We note that NK cells and pDCs have been described as being prognostically important in TKI responses,13 including in the setting of treatment-free remissions.30-33 Next, to complement our marker-based analysis, we used an independent approach, Milo,34 to identify cell states enriched or depleted in group C, relative to the union of groups A and B. Analyzing HSPC, lymphoid, and myeloid compartments separately, we found that the abundance of specific HSC and NK cell subpopulations was again associated with TKI response (supplemental Figure 5A-C).
Because 2 independent approaches highlighted the prognostic power of HSCs and NK cells, we asked if we could classify patients using either of these 2 cell types. Using a leave-1-sample-out logistic regression pipeline (Figure 2C), we developed both multiclass (Figure 2D,F; supplemental Figure 5D) and binary patient classifiers for HSCs (Figure 2E,G; supplemental Figure 5E), and a binary classifier for NK cells (Figure 2E,H; supplemental Figure 5E). All 3 classifiers showed an accuracy of >80%, with no false positives in predicting group C. However, sensitivity for group C was lower (50%-67%) (Figure 2D-E), likely because of the interpatient heterogeneity in this group. Interestingly, >80% of the genes identified by the patient classifiers have been previously reported in the context of CML or imatinib resistance (supplemental Table 16), including GAS2 for LSCs, and KLRC1 in NK cells.35 Given that HSCs and NK cells emerged as important prognostic cell types, we next focused on understanding the biological consequences of the differential GE programs in LSCs and NKs.
Identification and functional validation of LSC transcriptional programs
Next, and because primitive stem cells within CML samples comprise both BCR::ABL1+ and BCR::ABL1− cells,36 we used a combination of GE- and direct sequencing–based approaches to identify and segregate these 2 populations. First, we compiled a list of DEGs by comparing populations (normal stem cells, BCR::ABL1+ stem cells, and BCR::ABL1− stem cells) within CD34+CD38− CML and NBM samples from Giustacchini et al36 (supplemental Table 2B-D of ref Giustacchini et al36). Using these DEGs as the feature space (described in the supplemental Methods), we reclustered normal and CML stem cells into 9 subclusters (supplemental Figure 6). As expected, most cells from the CML samples clustered within the BCR::ABL1 GE signature–containing clusters (Figure 3A, dotted oval in left-hand panel), whereas cells from the control samples clustered separately (compare left- and right-hand panels in Figure 3A). In addition, and consistent with CML BM containing a minority of BCR::ABL1− cells, we were also able to visualize a small number of cells (98 of 3810) from the CML samples that clustered with control stem cells (Figure 3A, note BCR::ABL1− stem cells plotted in the inset), suggesting that our GE-based analysis was able to separate BCR::ABL1+ and BCR::ABL1− primitive cells. To confirm this, we used 2 independent sequencing–based approaches to determine the presence or absence of BCR::ABL1 fusion transcripts within the GE-defined BCR::ABL1+ and BCR::ABL1− cells. In the first approach, we designed polymerase chain reaction primers to amplify BCR::ABL1-containing transcripts encompassing cell-specific barcodes that could then be mapped back to our single-cell data set (Figure 3B; supplemental Methods). We found that 23 BCR::ABL1-specific fusion reads mapped to single cells that we had assigned as BCR::ABL1+, and that none of the BCR::ABL1-specific fusion reads mapped to single cells assigned as BCR::ABL1− (Fisher exact test, 7.5e−11). In control experiments, as expected, the BCR::ABL1 fusion only mapped to CML samples and none to controls (66 and 0 reads per sample, respectively), whereas the housekeeping gene, B2M, mapped back to equivalent numbers of CML and controls (1827 and 2250 reads per sample, respectively). In the second approach, we computationally retrieved any short reads from the raw FASTQ files that happened to cross the BCR::ABL1 fusion junction that were previously discarded (because they are not part of the reference genomes). We found 6 BCR::ABL1 fusion reads among the stem cells we had designated as BCR::ABL1+ based on their GE signature, and 0 fusion reads within the BCR::ABL1− cells (Fisher exact test, 0.067). Based on these experiments, we conclude that our assignment of BCR::ABL1 status is highly accurate.
Using these assignments, we identified DEGs across BCR::ABL1+ LSC groups (supplemental Table 17), and then also explored the functional consequences of LSC transcriptional heterogeneity. Specifically, we determined whether altered LSC fates or hierarchical frameworks affect TKI responses, as recently suggested for other myeloid malignancies.37-40 We used SCENIC15 to identify differential regulon activity. Regulons or gene regulatory networks comprise a small number of transcription factors and their target genes that determine lineage commitment. We first calculated a single pseudobulk regulon score for each patient’s LSC and observed distinctive regulon activity across prognostic groups (supplemental Table 18). Group A LSCs showed increased activity of a canonical core erythroid-like network (CEN), comprising TAL1, GATA1, and KLF1,41 whereas group C LSCs showed increased activity of a MYC regulon, and regulons related to inflammatory responses (IRF1 and CEBPD) (Figure 3C-D). Group B LSCs demonstrated intermediate levels of erythroid regulon activity, but showed notable levels of IKZF2 activation, a transcription factor implicated in acute myeloid leukemia LSC self-renewal, and like Group A LSCs, a lack of MYC activity (Figure 3C-D).42
Given the physiologic role of the CEN in specifying erythroid commitment,41 we determined whether group A HSPCs would show functional evidence of CEN-like activity compared with controls and groups B/C. First, using GSEA, we found that Group A LSCs exhibited enrichment for functional erythroid GE programs (Figure 3E; supplemental Figure 7A-B; supplemental Tables 19 and 20). Second, and assessing the cellular hierarchy, we observed a fourfold expansion of ERPs from normal to group A HSPCs, followed by a progressive decrease across groups B and C (Figure 3F-G) and enrichment for erythroid GE programs in the HSPC compartment of patients in group A (supplemental Figure 7C-D). Third, using colony-forming assay-based readouts, we found that HSPCs from all group A samples had an intrinsic erythroid bias, notably a 2- to 12-fold increase in ERP/MP ratio, whereas group C samples consistently showed an increase of <2-fold (average of 6.3-fold in group A vs 1.09-fold in group C), and group B samples had intermediate ratios (Figure 3H). Importantly, we also found that ERPs retained high levels of TKI-sensitivity across all groups, and that MPs are intrinsically less TKI-sensitive than ERPs (Figure 3I).
Lastly, because HSPCs in groups B and C had evidence of increased inflammatory signaling compared with group A, including an IFN-γ signaling signature (Figure 1F), we determined the impact of exogenous IFN-γ on TKI sensitivity. Using Group A HSPCs, we found that IFN-γ induced TKI resistance (Figure 3J; supplemental Figure 7E). Together, our single-cell atlas describes differential regulon usage in CML LSCs, most strikingly a CEN-like regulon in group A and a MYC regulon in C. Functional assays suggest a model in which CEN-bearing Group A LSCs induce a bias toward erythroid fates, resulting in expansion of TKI-sensitive ERPs and an ERP-skewed hierarchical structure,40 whereas in groups B and C, there is an increased accumulation of non-ERPs within an inflamed environment, together with an absence of ERP skewing.
Association of NK cell states and markers with TKI response
Encouraged that pretreatment NK cell GE patterns were prognostically discriminating (Figure 2), we set out to identify NK cell states that gave rise to these signatures, and within specific NK subtypes, DEGs that could be clinically exploited using more robust antibody-based platforms. In addition, because the antileukemic activity of NK cells is dependent on the net balance of activating and inactivating ligand-receptor interactions between NK and target cells, we also sought to identify group-specific interactions that might contribute to differential TKI responses.
As an initial step, we performed de novo clustering of 8617 NK cells to identify 6 major subtypes: adaptive, active, CD56-bright, mature, NKT-like, and cytokine-induced memory-like NK cells (Figure 4A-B; supplemental Figure 8; supplemental Table 21A, refer to the supplemental Methods “subclustering of NK cells” for inference of annotations). Strikingly, NK cells from controls and patients with CML had minimal overlap in the UMAP (uniform manifold approximation and projection) space, suggesting distinct transcriptional states (Figure 4B). Indeed, NK cells from the NBM controls were mainly composed of mature and CD56-bright NK cells, with a minor proportion of adaptive NK cells (mainly from 1 patient), in close agreement with recent literature (Figure 4C).43,44 In contrast, NK cells from CML BM displayed higher transcriptional diversity, with adaptive and active NK cells comprising the majority (Figure 4C). The adaptive NK cells from patients with CML are henceforth referred to as adaptive-like NK cells. Strikingly, adaptive-like NK cells accumulated significantly in groups A and B as compared with group C or control BM (Figure 4D; supplemental Table 21B). We identified several markers for the adaptive-like NK population, including 2 surface molecules, HLA-DR and CD7, which were upregulated and downregulated respectively, (Figure 4E).
Next, using the NATMI algorithm,16 we characterized signaling interactions between HSPC ligands and NK cell receptors. Remarkably, compared with the control, group A NK-HSPC interactions were notable for enrichment and depletion of multiple activating and inhibitory interactions, respectively (Figure 4F). In contrast, comparing group C vs A NK-HSPC interactions, we found a striking upregulation of a key inhibitory interaction between HLA-E and NKG2A, as well as downregulation of 2 activating interactions (ICAM4/TGM2 and ICAM4/ITGB1) (Figure 4F-G; supplemental Figure 9A for complete data set of NATMI-predicted HSPC-NK cell interactions). Notably, HLA-E-NKG2A interaction plays a critical role in target cell inhibition of NK-mediated cytolysis and can be targeted with specific antibodies.45,46
We conclude that the CML BM NK cell repertoire is skewed toward an activated state in imatinib-sensitive cohorts, and an HSPC-tolerant state in group C (Figure 4H). The discovery of these 2 contrasting NK cell states suggest that these cells may be exploited as biomarkers for prognostication as well as stratification of differential NK cell–based therapies.47-49
Genetic associations with progressive TKI resistance
Genetic alterations, including copy number variants (CNVs) and single-nucleotide variants (SNVs), are associated with the development of TKI resistance and BC transformation, but a causal role for these events as well as their utility as biomarkers is less clear.7,50 Accordingly, we determined the frequency of both types of events in our samples by using inferCNV, a method to predict CNVs from scRNA-seq data, and by WES to detect SNVs and small insertions/deletions.
Because genetic events that drive transformation are expected to result in clonal expansion of the affected cell, we first examined the relationship between cluster abundances and CNVs within our scRNA-seq data set. We observed that 11 clusters were dominated (≥50%) by cells from a single sample (Figure 5A-B). These clusters were attributable to 4 of 9 patients in group B and 4 of 6 patients in group C. Notably, none of the 9 group A samples contributed to sample-specific cell clusters in HSPCs (Fisher exact test, 0.0095). We asked whether the 11 patient-specific cell clusters could be attributed to the clonal expansion of CNVs in the corresponding samples. We found evidence of CNVs in 3 of 8 patients (BM157 [group C], P309 [group C], and P387 [group B]) in which BM157 has Pro-B, P309 has ERP, and P387 has ERP and iNeP cluster expansions (supplemental Figure 10; supplemental Table 22). Together, these findings suggest that patient-specific cell clusters, some of which may be attributable to CNV-driven clonal expansion, are more common in patients in groups B and C, and are thus predictive of imatinib resistance.
Regarding SNVs, and in line with the literature, we found that the most frequent mutations were ASXL1, SETD1B, and TET2 (23.8%,14%, and 7%, respectively),50 whereas ABL1 kinase domain mutations were absent, consistent with the notion that primary TKI resistance is a BCR::ABL1-independent phenomenon (Figure 5C; supplemental Table 23).2 We also found no relationship between the overall frequency of SNVs across groups (6 of 8, 4 of 7, and 4 of 6 individuals in groups A, B, and C, respectively) but that frameshift insertions/deletions were only found in groups B (1 of 7) and C (2 of 6). Mutations that were only found in group C included IKZF1 (1 frameshift insertion), KMT2D (1 nonframeshift deletion [nsSNV]), EP300 (1 nsSNV), SETD2 (1 nsSNV), L3MBTL3 (1 nsSNV), and SFMBT2 (1 nsSNV), whereas all other group C mutations could be found in groups A and B including ASXL1 (A and B), SETD1B (A and B), and KMT2E (A) (supplemental Table 24A).
Because prior work has identified the existence of preleukemic subclones harboring myeloid malignancy–related mutations in CP CML,51 we determined their frequency in our data set. First, we mapped the myeloid mutations found by WES to our scRNA-seq–derived cell clusters, and were able to do this for 66% of mutations (the inability to map all myeloid mutations to the single-cell data set is because of the low sequencing depth inherent in the 10× Genomics scRNA-seq platform) (Figure 5D; supplemental Table 24B). Next, and based on prior work confirming that the majority (65%-95%) of T and NK cells in CML samples arise from Philadelphia chromosome–negative clones,52,53 we inferred the existence of a preleukemic subclone if the specific myeloid mutation could be found in a T or NK cell cluster in addition to an HSPC cluster, which are predominantly (>90%) Philadelphia chromosome positive.54 Using this approach, we found that 28% (2 of 8 group A, 1 of 7 group B, and 3 of 6 group C) or 6 of 21 of CML samples had evidence suggestive of preleukemic subclones (Figure 5E), and 16% were reported by Kim et al using this approach.51 In summary, our genetic analysis suggests that CNV-associated clonal expansion may be a marker for imatinib resistance, and that whereas specific mutations were only found in group C (eg, IKZF1), the most common recurrent mutations in CML, including ASXL1, TET2, and SETD1B, are also found in individuals who achieve optimal TKI response (group A). Because certain mutations may also be found in preleukemic subclones (TET2, CBX4, KMT2E CBLB, CBX8, SETD2, ZCCHC7, and L3MBTL3), our findings, as well as that of others,51 indicate that larger cohorts will also need to be evaluated using single-cell approaches.
Toward the development of antibody-based panels for predicting TKI response
GE-based biomarkers are difficult to develop into clinical tests given the challenges of reliably measuring messenger RNA in clinical specimens.7 Accordingly, we developed a custom 38-antibody panel (Figure 6A; supplemental Figure 11A) to determine whether the GE- and cell-state abundance-based features we identified above could be detected by mass cytometry (CyTOF) and conventional flow cytometry.
In agreement with our scRNA-seq findings, we observed an expansion of CD34+ HSPC cells in CP CML compared with controls, which comprised a decrease in HSCs as well as an increase in common myeloid progenitors and megakaryocyte-erythroid progenitors (supplemental Figure 11B-D). In addition, we found that although LSC markers (CD25/IL1RAP/CD26)55 as well as lineage and immune signaling markers were increased in CML compared with controls (particularly IL1RAP in groups B and C) (Figure 6B; supplemental Table 25), stemness markers (c-kit, CD133, and CD62L) were decreased.56 Comparison across groups showed that megakaryocyte-erythroid progenitors were significantly expanded in group A compared with controls and that the degree of expansion diminished with increasing TKI resistance (Figure 6C). Interestingly, immunophenotypically defined granulocyte-monocyte progenitors (GMPs) demonstrated the opposite relationship with clinical resistance (supplemental Figure 11E). To develop a flow cytometry panel for the detection of ERPs, we first confirmed previous reports that CD34+ cells coexpressing CD71 and CD105 (endoglin) are reliable markers of ERPs (supplemental Figure 11F), and included these markers in our validation experiments.57 Using conventional flow cytometry on an independent cohort of patients characterized into group A, we confirmed that both erythroid lineage-primed (CD45+ lin−CD34+CD71+) (supplemental Figure 11G), as well as erythroid lineage-committed (CD45+ lin−CD34+CD71+CD105+) progenitors accumulated in patients in group A (Figure 6D).
Next, we conducted an unbiased assessment of the CD34− compartment using FlowSOM clustering (supplemental Figure 12A-C; supplemental Table 26). Comparing across groups, we found that cells with an NK immunophenotype were significantly expanded in groups A compared with group C in FlowSOM analysis (supplemental Figure 12D). We confirmed through biaxial gating of the FlowSOM data set that CD3−CD56+NK cells, specifically CD56dim CD16bright NK cells, were expanded in group A compared with controls, but not in patients in group C (Figure 6E; supplemental Figure 12E). We also confirmed the differential expression of scRNA-seq–derived NK markers, HLA-DR and CD7, on adaptive-like NK cells, and found that these markers were increased and decreased respectively, when comparing groups A and B NK cells to those of group C by CyTOF (Figure 6E).
Finally, using conventional flow cytometry, we examined the frequency of adaptive-like NK cells using NKG2C (KLRC2) and CD57 (B3GAT1) double positivity (supplemental Figure 13A).58 Concordant with our scRNA-seq findings, we observed an expansion of CD56dim CD16brightCD57+NKG2C+ NK cells specific to group A (Figure 6F). We also validated that NKG2A-marked group C NK cells (supplemental Figure 13B), and this subset was much lower in abundance in groups A and B (Figure 6G).
In summary, our cytometric analyses validated the findings of our single-cell atlas in an orthogonal manner and confirmed the existence of several cellular abundance gradients and distinct GE patterns associated with progressive TKI resistance. The flow cytometric panels we describe (Figure 6D-G) may represent important new tools to assess the likelihood of encountering primary TKI resistance but will need an independent evaluation in larger cohorts.
Discussion
We have generated a single-cell atlas from 6 healthy and 24 pretreatment CML BM samples, and obtained a transcriptional and cellular landscape of progressive TKI resistance, which allowed us to identify 8 prominent features associated with primary TKI resistance (Figures 1 and 7). Using ML, we determined that GE signatures of at least 2 cell types, LSCs and NK cells, are predictive of TKI responses.
The prognostic value of LSC GE signatures suggested an underlying LSC biology contributing to TKI response heterogeneity. By characterizing their differential regulon usage, we discovered a striking activation of the CEN in Group A LSCs compared with controls and group B and C LSCs, and conversely in group C LSCs, activation of MYC- and IRF1-driven regulons (Figure 3C-D).41 Our data suggest a model in which pretreatment LSCs with erythroid-biased fates are associated with optimal outcomes. Interestingly, the favorable effect of BM ERP expansion on patient survival was noted even in patients treated with interferon-α2b and hydroxyurea or busulfan,59 indicating that ERPs are intrinsically more drug sensitive than non-ERPs, and which our functional validation and that of others support.60 The expansion of exquisitely TKI-sensitive ERPs in patients with good prognoses may provide the biological explanation for the prognostic value of rapidly declining BCR::ABL1 transcripts.4 Future studies using multimodal analysis of serial samples will determine whether this is indeed the case.
To understand the biology underlying the prognostic value of NK cells, we performed a detailed transcriptional analysis (Figure 4A-B). We identified an HLA-DR+ adaptive-like NK population that underwent a fourfold increase in patients in groups A and B compared with controls but was unchanged in group C (Figures 4C-D and 6E). Adaptive NK cells are epigenetically reprogrammed long-lived populations with high antileukemic potential, unique properties that current clinical trials are exploiting with adaptive NK cell infusions.47,61-63 Based on our observations, such approaches may be of particular utility in group C.47,64 By exploring NK target cell interactions, we also discovered that NK cells and HSPCs from group C samples had increased expression of NKG2A and HLA-E, respectively (Figure 4F-G). Interestingly, dasatinib has been shown to downregulate KLRC1 and promote NK cell cytotoxicity in CML samples.35 Thus our studies not only uncover prognostic NK cell states but suggest specific NK cell–based therapies guided by our biomarkers.47
Because we were able to examine the entire BM compartment of each patient in a single experiment, we also gained an appreciation that multiple parameters, not all acting in the same direction, contribute to an individual patient’s ultimate TKI response. For example, although most (7 of 8) patients in group A demonstrated ERP expansion, P713 did not; instead, this patient exhibited high numbers of adaptive-like NK cells (supplemental Table 27). Conversely, although 1 patient in group C (P309) had elevated ERPs, this patient’s adaptive-like NK cell counts were among the lowest across all groups (supplemental Table 27). Indeed, because of the elevated ERPs at diagnosis, this individual may also represent patients who have initial rapid responses but who eventually progress because of other adverse features (supplemental Table 27). Despite the small numbers, our overall observations imply that comprehensive biomarkers will necessarily include readouts for erythroid fate, NK cell activity, and inflammation. Interestingly, the core features indicative of BC transformation could also be detected in the 2 patients in group C in whom BC transformation took the longest time (P309 and P147; time to transformation 1518 and 2771 days, respectively) (supplemental Table 27). Thus, a major conclusion of our work is that, in addition to TKI resistance that is acquired “on-therapy,”2 a comprehensive understanding of primary resistance and response heterogeneity will need to include additional features present “pretherapy,” as we have demonstrated. We anticipate that larger studies will discover additional factors, beyond the 8 that we have defined (Figure 7), that could predict primary TKI resistance, and overcome the limitations of smaller cohort studies. Future studies may also need to convert our GE-based findings into more robust protein-based assays that can be reliably performed on tissues collected in the course of routine clinical care.7 In addition, although we identified features segregating groups B vs A (Figure 3C), larger studies are needed to uncover additional discriminators. Although our work also suggests that the 3 prognostic groups are on a biologic continuum, we should highlight that group C has undergone additional molecular events that renders these patients completely TKI resistant. Consequently, some but not all the molecular features across groups A through C will demonstrate a unidirectional change (ie, lie on a continuum). We have also identified genetic features associated with differential TKI responses, including the presence of preleukemic clones harboring recurring mutations, but the predictive value of these findings remains controversial51,65 and awaits resolution by larger studies.
In summary, the creation of 29 high-resolution maps from normal and CML BM has enabled us to study pretreatment cell states at unprecedented resolution and to correlate our findings with progressive TKI resistance. We have identified candidate pretreatment biomarkers for future prospective validation in larger cohorts, as well as potential therapeutic targets to prevent or delay TKI resistance and BC transformation. Our computational and functional analyses also led us to propose novel hypotheses to account for heterogeneous TKI responses (Figure 7), including for LSCs, and NK-HSPC interactions, and suggest bespoke therapeutic interventions for each patient.
Acknowledgments
The authors acknowledge Joanna Sinnakannu for assistance in the initial stages of this work, Bobby Ranjan and Liew Jun Xian for software troubleshooting with CellRanger and Seurat, and Quy Xiao Xuan Lin for assistance with data analysis. The authors acknowledge Bei Jun Chen for extensive discussions, and testing of new computational algorithms. Figure 7 was created with BioRender.com.
This work was supported by grants from the National Medical Research Council Singapore (NMRC/CIRG/1468/2017, MOH-000602, MOH-000059, CIRG16nov032), A∗STAR (1727600056, H18/01/a0/020), Khoo Pilot Award (Collaborative) (KP(Coll)/2019/0014B).
Authorship
Contribution: V.K. contributed to CyTOF data acquisition, data analysis and interpretation, scRNA-seq data interpretation, in vitro validation experiments, and manuscript writing; F.S. performed scRNA-seq data analysis and interpretation, contributed to CyTOF data analysis, designed and executed pipelines to detect prognostic marker genes, advised V.K., Z.N., and M.M. in their data analysis tasks, and contributed to manuscript writing; Z.N. performed scRNA-seq experiments, library generations, data processing using CellRanger, scRNA-seq data analysis, Module Score analysis, cell-cell interaction through NATMI, and contributed to manuscript writing; X.R. conducted the bioinformatic analyses shown in Figures 3B and 5D-E at the time of manuscript revision; P.N.V. helped with scRNA-seq experiments and library preparations; Z.E.C. and M.Y. performed validation experiments; K.L.L. performed GSEA analysis of the scRNA-seq data sets; M.M. assisted with running NATMI and prognostic marker gene detection across patient-specific clusters; J.O. and O.R. created the shinycell interactive R package; T.Z.T. conducted the WES data analysis; N.A.R. and M.G.L.L. contributed towards scRNA-seq optimization experiments; A.M.S.C. contributed toward validation experiments involving colony-forming assays; S.B. and W.Y.K.H. contributed toward optimization experiments and advised on CyTOF-based HSPC analysis; H.T. contributed to the annotation of the clinical responses of patients; W.J.C. provided clinical samples that were used for the optimization of long-read sequencing; C.C. provided annotated clinical samples and participated in discussions; S.P. conceived and oversaw the project, while also providing financial support, data interpretation, and manuscript writing; and S.T.O. conceived and oversaw the project, while also providing financial support, data interpretation, and manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shyam Prabhakar, Genome Institute of Singapore, 60 Biopolis St, A∗STAR, 138672 Singapore; e-mail: prabhakars@gis.a-star.edu.sg; and S. Tiong Ong, Cancer and Stem Cell Biology Program, Duke-NUS Medical School, 8 College Rd, 169857 Singapore; e-mail: sintiong.ong@duke-nus.edu.sg.
References
Author notes
∗V.K., F.S., and Z.N. contributed equally to this study.
Fastq files are available online at the EGA under accession ID (EGAS00001005509).
Raw data and R codes are deposited at our Zenodo repository and will be available on request. The scRNA-seq data from this manuscript can be accessed at http://scdbm.ddnetbio.com.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


![Identification of prognostic pretreatment cell types by ML. (A) Bootstrapped logistic regression pipeline to predict imatinib treatment outcome based on CML prognostic group. Briefly, given the transcriptome of a single cell, the output identifies GE patterns that predict to which group that cell belongs to. Specifically, the output builds distinct GE-based signatures that classify each cell type into a prognostic group. (B) Top table: accuracy scores (ACC) of (i) multiclass (A vs B vs C) and (ii) binary (C vs not C [AB]) logistic-regression classifiers. The ACC scores for the top 3 models of both multiclass and binary classifiers are highlighted in red. ACC scores are the ratio of true positives and true negatives to all positive and negative observations. Bottom table: left, confusion matrices displaying cell counts for the top 3 cell types in the multiclass classifiers, and right, for the top 3 binary classifiers. In a random model, the ACC = 0.33 for a multiclass classifier and 0.5 for a binary classifier. (C) A leave-one-patient-out classifier to identify marker genes in pseudobulked transcriptomes that can serve as prognostic markers. Only markers from the classifier in panel A are considered as the candidate features in the model. Confusion matrices for a patient-specific (D) multiclass HSC, (E) binary HSC, and NK cell classifier. Precision (Pre) is the ratio between true positives and all positives. Recall (Rec) is a measure of the model’s ability to identify true positives (true positive/true positive and false negative). (F) Top nonzero regression coefficients of the HSCs multiclass patient-specific classifier. (G) Top 20 nonzero regression coefficients identified by the patient-specific binary HSC classifier. (H) Top 20 nonzero regression coefficients identified by the patient-specific binary NK cell classifier. Statistical tests for the ML pipelines are described in the supplemental Methods. Supplemental Figures 4 and 5 and supplemental Tables 14-16 are linked with data shown in Figure 2.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/22/10.1182_blood.2022017295/2/m_blood_bld-2022-017295-gr2ac.jpeg?Expires=1767706656&Signature=bvMx2eozTNtvtROUokpGwRBkZJdo~fFtTJPQz1DUD-OdmLPvxAcHTA8b7m3lbCh3iQcPfyeyxO5BrhLo2S6oHbEbWG4DwGXGu7Q0YxAj~buC8VHpbw128HVOJQdMxAKSGo4mH4nAY3eRlZIzhP-gIZ9U3NLafYYIQgUNx5BhVLUSgWpdnTH8qUCvsp6XWCyDTEED9mKv7hMR~wjQwrF9LImVPPDmlrpUnHB7SPFECnc6mAxf6mG-QgmKC0Gg9-9zeKP-Klbl6nX01poSZImNluCrTbxWysi7kAmZRFeue8EXlmkFlwWIpMSGWnX9126u5UnkbMe~H0JeUQQebcOU3w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Identification of prognostic pretreatment cell types by ML. (A) Bootstrapped logistic regression pipeline to predict imatinib treatment outcome based on CML prognostic group. Briefly, given the transcriptome of a single cell, the output identifies GE patterns that predict to which group that cell belongs to. Specifically, the output builds distinct GE-based signatures that classify each cell type into a prognostic group. (B) Top table: accuracy scores (ACC) of (i) multiclass (A vs B vs C) and (ii) binary (C vs not C [AB]) logistic-regression classifiers. The ACC scores for the top 3 models of both multiclass and binary classifiers are highlighted in red. ACC scores are the ratio of true positives and true negatives to all positive and negative observations. Bottom table: left, confusion matrices displaying cell counts for the top 3 cell types in the multiclass classifiers, and right, for the top 3 binary classifiers. In a random model, the ACC = 0.33 for a multiclass classifier and 0.5 for a binary classifier. (C) A leave-one-patient-out classifier to identify marker genes in pseudobulked transcriptomes that can serve as prognostic markers. Only markers from the classifier in panel A are considered as the candidate features in the model. Confusion matrices for a patient-specific (D) multiclass HSC, (E) binary HSC, and NK cell classifier. Precision (Pre) is the ratio between true positives and all positives. Recall (Rec) is a measure of the model’s ability to identify true positives (true positive/true positive and false negative). (F) Top nonzero regression coefficients of the HSCs multiclass patient-specific classifier. (G) Top 20 nonzero regression coefficients identified by the patient-specific binary HSC classifier. (H) Top 20 nonzero regression coefficients identified by the patient-specific binary NK cell classifier. Statistical tests for the ML pipelines are described in the supplemental Methods. Supplemental Figures 4 and 5 and supplemental Tables 14-16 are linked with data shown in Figure 2.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/22/10.1182_blood.2022017295/2/m_blood_bld-2022-017295-gr2dh.jpeg?Expires=1767706656&Signature=qrINTprYurJIuSAOoDwaHTlylApU5VjEL~-iUvIvJm0qtF7SS9tmF2T-hOs4T8BkHb-1UfkRPtQ06sTaDpGrcpygwPmOSv6nubGgNaC6UoGuISbhEl29sATtGW8N7UqY-grEuLp0o8VD5FSFQNABXUAsS~E9jL1Ozigcwv1sDAGPgfbHUHA5oKD8Ntdh1-uUYxxMhc5uQYWMvFtazcQaJUF5nAijSdlQtxYvWFkewUPV452pOtB6vAhJaYg8xEyoB9S~9SlCcJ5Tw04GvmJpoilAyDJrsIoOIbLSj4pOyEc6KH7vTsxFFRmwvZe3Xc~y3SP75fDqw8RXKMflcZZ1Jg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Association of NK cell states and markers with TKI response. (A) Expression heatmap of the top 10 marker genes for each NK cell subpopulation. The expression of literature-derived NK cell marker genes (CD56 [NCAM1], FCGR3A [CD16], CD57 [B3GAT1], NKG2C [KLRC2], and ZBTB16]) are plotted for comparison. Annotations were aided by Module Score computations for the 7 NK cell gene signatures compiled from literature (supplemental Table 21A; supplemental Figure 8C; “Methods”). Module scores were calculated for each NK cell type; note that adaptive NK cell signatures 1 to 3 are gene sets that are upregulated, whereas signature 4 is a gene set that is downregulated in the adaptive NK population. (B) Left: UMAP showing the 6 NK cell subpopulations identified through NK cell subclustering. Right: UMAP colored by control or CML labels. (C) NK subpopulations are plotted as a proportion of total NK in control and patients with CML. (D) The proportion of adaptive-like NK cells across control and prognostic groups is shown. Each dot represents 1 patient. (E) Expression heatmap of the top 15 genes upregulated and downregulated in adaptive-like NK cells, relative to other NK subpopulations. (F) Cell-to-cell communication analysis: NATMI was performed to identify potential interactions between ligands on HSPCs and receptors on NK cells. Ligand-receptor pairs predicted to mediate differential HSPC-NK interactions between control and group A (left), or groups C and A (right). x-axis: log2-transformed fold change of edge expression weight inferred by NATMI. The interactions marked by red asterisks represent NK cell–inhibitory interactions. (G) Expression of (left) HLA-E in HSPCs, and (right) NKG2A (KLRC1) in NK cells using pseudobulked populations. Each dot represents 1 patient. (H) Schematic summarizing prognostic NK cell features across groups. Statistical tests: Figure 4D,G; 2-tailed t test. Supplemental Figures 8 and 9 and supplemental Table 21 are linked with the data shown in Figure 4.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/22/10.1182_blood.2022017295/2/m_blood_bld-2022-017295-gr4ae.jpeg?Expires=1767706656&Signature=tF5egFoITgcfpbZZyJBHbIkT1LlPQWj9fQWi7cJLOIeqdhQEyJCFT-GhthowFqLuOjDUJvLAvzju4hD0tlVoo~P~ZdDCVmeKjTBJ-5koyv~rI6XeNADmMhmocxm1YGEtQtSQHTdKKQFJALRcH2S0k2iobaCjvqofAGj0yoZuXyarqc~zJMOmh4dbJr6-wwLC6dvvGaJmNgmeD1h4iADmNkQvQoWTR6Y7Nys9pea5EjFL5yogtnicmLUKHbdYSHayBBjrDwMpLjUgSCrLAon~pRDfSNqYYjiZS7Kb-J3bW1w~aNJPusFv3wGSJ8uSO7kfJzUivfRxdmT~CEhzAnpdAQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Association of NK cell states and markers with TKI response. (A) Expression heatmap of the top 10 marker genes for each NK cell subpopulation. The expression of literature-derived NK cell marker genes (CD56 [NCAM1], FCGR3A [CD16], CD57 [B3GAT1], NKG2C [KLRC2], and ZBTB16]) are plotted for comparison. Annotations were aided by Module Score computations for the 7 NK cell gene signatures compiled from literature (supplemental Table 21A; supplemental Figure 8C; “Methods”). Module scores were calculated for each NK cell type; note that adaptive NK cell signatures 1 to 3 are gene sets that are upregulated, whereas signature 4 is a gene set that is downregulated in the adaptive NK population. (B) Left: UMAP showing the 6 NK cell subpopulations identified through NK cell subclustering. Right: UMAP colored by control or CML labels. (C) NK subpopulations are plotted as a proportion of total NK in control and patients with CML. (D) The proportion of adaptive-like NK cells across control and prognostic groups is shown. Each dot represents 1 patient. (E) Expression heatmap of the top 15 genes upregulated and downregulated in adaptive-like NK cells, relative to other NK subpopulations. (F) Cell-to-cell communication analysis: NATMI was performed to identify potential interactions between ligands on HSPCs and receptors on NK cells. Ligand-receptor pairs predicted to mediate differential HSPC-NK interactions between control and group A (left), or groups C and A (right). x-axis: log2-transformed fold change of edge expression weight inferred by NATMI. The interactions marked by red asterisks represent NK cell–inhibitory interactions. (G) Expression of (left) HLA-E in HSPCs, and (right) NKG2A (KLRC1) in NK cells using pseudobulked populations. Each dot represents 1 patient. (H) Schematic summarizing prognostic NK cell features across groups. Statistical tests: Figure 4D,G; 2-tailed t test. Supplemental Figures 8 and 9 and supplemental Table 21 are linked with the data shown in Figure 4.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/22/10.1182_blood.2022017295/2/m_blood_bld-2022-017295-gr4fh.jpeg?Expires=1767706656&Signature=oBwPVFJGq-cihYveMK2HyNmNRaae3U6gG~NWz4ZEZt4TOIv6n-2UYc39f~9ak9DwLvor53QXNn79jkptKirWqierF9FtZZ4T3VvQ1co3I4mmsni3GspgdEC8nf1y7vG2fSq943avimGOuBob6XMuBnexubqFCzZqBF8YW1NEI5fqFF61ktpIqWfrWTLr9JaaKZ~TBlcc8mGtppNDQ3k0ev5HTolsiy3t~0odeB6JXp6TymTqJJmwkOe4OGq4-lFDhdEKWRpIOuNO9irey3lSVah3VjvxMO6QUpmAw7ynPHtowJhNbxrJfn5bFkgp5OzBKaWw6aP04y3cKpM~Q47VaQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
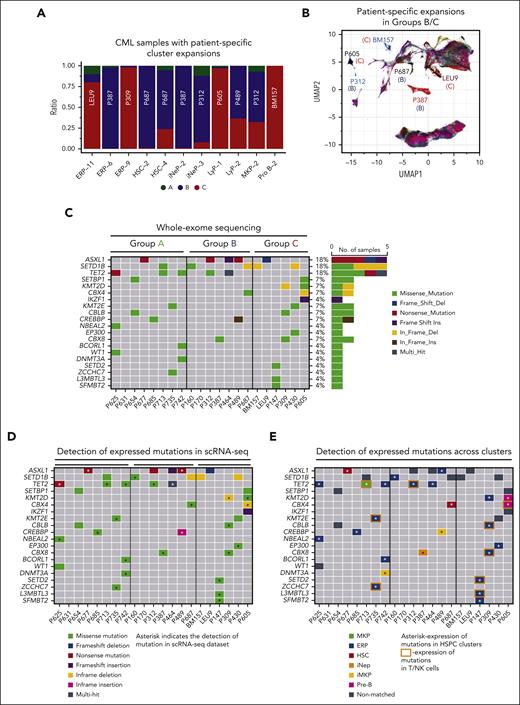

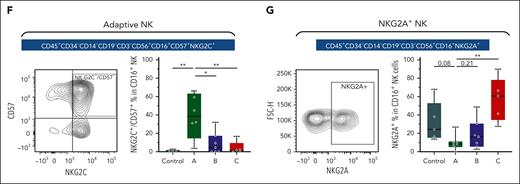
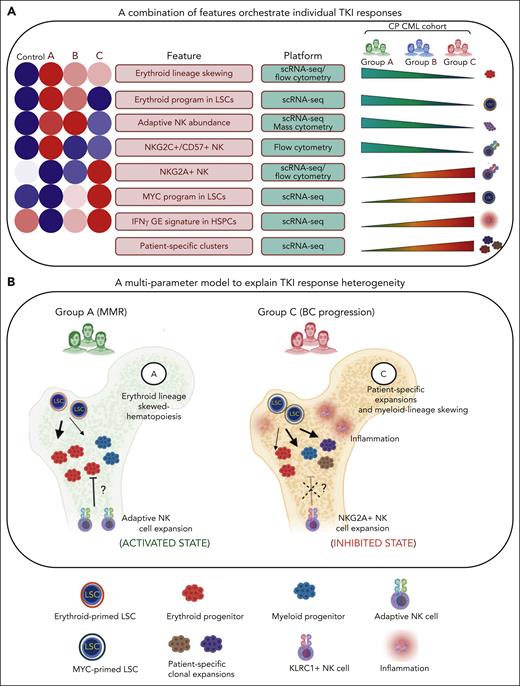
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal