Key Points
A CRISPR screen for HbF repressors identified polycomb protein BMI1, which is found to function within the canonical PRC1 complex.
RNA binding proteins LIN28B, IGF2BP1, and IGF2BP3 are the critical links between PRC1 and HbF repression in adult-type erythroid cells.
Abstract
The switch from fetal hemoglobin (HbF) to adult hemoglobin (HbA) is a paradigm for developmental gene expression control with relevance to sickle cell disease and β-thalassemia. Polycomb repressive complex (PRC) proteins regulate this switch, and an inhibitor of PRC2 has entered a clinical trial for HbF activation. Yet, how PRC complexes function in this process, their target genes, and relevant subunit composition are unknown. Here, we identified the PRC1 subunit BMI1 as a novel HbF repressor. We uncovered the RNA binding proteins LIN28B, IGF2BP1, and IGF2BP3 genes as direct BMI1 targets, and demonstrate that they account for the entirety of BMI1’s effect on HbF regulation. BMI1 functions as part of the canonical PRC1 (cPRC1) subcomplex as revealed by the physical and functional dissection of BMI1 protein partners. Lastly, we demonstrate that BMI1/cPRC1 acts in concert with PRC2 to repress HbF through the same target genes. Our study illuminates how PRC silences HbF, highlighting an epigenetic mechanism involved in hemoglobin switching.
Introduction
Functionally related genes can be organized into clusters but may be expressed at distinct developmental stages. In human erythroid cells, the genes encoding β-type globin chains are arranged in a manner reflecting the developmental stage of their expression.1 The HBE1, HBG1/2, and HBB genes are expressed during the embryonic, fetal, and adult stages of erythroid development, respectively. All these genes require a distal upstream enhancer called the locus control region.2-4 The switch from fetal HBG1/2 to adult HBB gene expression, which occurs perinatally, has received the most attention because its reversal benefits patients with sickle cell disease and some types of β-thalassemia.1
Several transcription factors, including BCL11A, LRF, and NFIA/NFIX, bind directly near the HBG1/2 genes to enforce their silencing.1,5-7 Perturbation of any of these triggers the reactivation of the HBG1/2 genes in adult erythroid cells.5-13 Many of these regulatory factors, including their coregulators such as NuRD, are themselves under developmental transcriptional control.7,14-24 For example, BCL11A and NFIA/NFIX messenger RNA (mRNA) production are markedly elevated in adult erythroblasts when compared with their fetal counterparts.7,19 In addition, in the case of BCL11A, posttranscriptional mechanisms contribute to its adult stage–specific expression.15,16,24 Hence, complex regulatory networks modulate the switch from fetal hemoglobin (HbF) to adult hemoglobin (HbA) production.
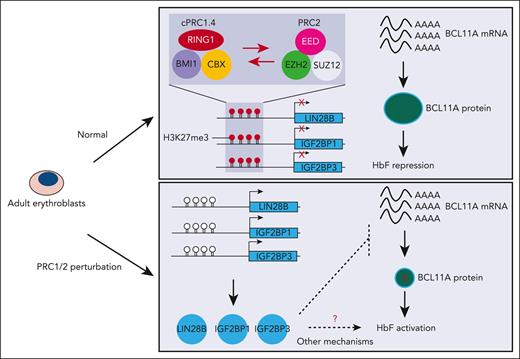
Polycomb group (PcG) proteins are broadly divided into 2 main classes, polycomb repressive complexes 1 and 2 (PRC1 and PRC2).25,26 PRC1 and PRC2 cooperate to establish and maintain a transcriptionally silent, facultative heterochromatic state. PRC1 catalyzes the monoubiquitylation of Lys119 of H2A (H2AK119ub1) via the E3 ligase RING1A/B,27,28 whereas PRC2 methylates lysine 27 of histone H3 (H3K27me3) through its enzymatic subunit EZH1/2.29-32
Prior studies uncovered that the core components of the PRC1 complex comprise a RING1A/B E3 ligase subunit and 1 of the 6 polycomb group RING finger (PCGF) proteins, which subdivide the complex into PRC1.1-PRC1.6.33 All of these subcomplexes are broadly classified as canonical (cPRC1) and noncanonical (ncPRC1).33,34 cPRC1 comprises RING1A/B, PCGF2/4 (PCGF4 is also called BMI1), 1 of the 3 polyhomeotic (PHC1-3) paralogues, and 1 of the 5 CBX proteins (CBX2/4/6/7/8) that load PRC1 onto H3K27me3-decorated chromatin.25,29,35 cPRC1 has low E3 ligase activity toward histone H2AK119. In contrast, ncPRC1 has strong E3 ligase activity because of the incorporation of RYBP/YAF2 polypeptides instead of CBX proteins, and accounts for the majority of cellular H2AK119ub1.33,34
The PRC2 complexes are likewise heterogeneous in their composition and mechanisms of recruitment. In their core, they share the catalytic subunits EZH1 or EZH2, EED, SUZ12, and RBBP subunits. PRC2.1 contains PCL proteins involved in targeting the complex to unmethylated CpG islands.36 JARID2 is an ancillary factor in the PRC2.2 complex that interacts with H2AK119ub1 to mediate cross talk between ncPRC1 and PRC2.37,38 Although evidence suggests that ncPRC1 and H2AK119ub1 can repress gene expression independently of PRC2,39 cPRC1 is recruited by H3K27me3 and maintains silencing of its targets in conjunction with PRC2.25,26
Polycomb proteins, including BMI1, are essential regulators of developmental transcriptional programs. In the hematopoietic system, BMI1 is involved in modulating cell division,40 lineage specification,41 mitochondrial function,42,43 and protein synthesis.44 Moreover, BMI1 has been implicated in the regulation of erythroid self-renewal and differentiation.45-47 Despite numerous studies on PRC complexes in development, surprisingly little is known about their roles in the regulation of fetal vs adult erythroid lineages and hemoglobin switching.25,26,48,49 One previous study reported that genetic perturbation of the EZH2 or EED subunit of PRC2 induces HbF expression in adult erythroid cells.50 Moreover, an EED inhibitor (FTX-6058) has been shown to elevate HbF production and has entered a clinical trial.51 Yet, it is unknown how, and in what composition, PRC1 and PRC2 complexes function during the transition from fetal- to adult-type erythroid cells, and through which target genes they regulate the switch in hemoglobin production.
Here, via a domain-focused CRISPR-Cas9 genetic screen, we identified BMI1/PCGF4 as a novel repressor of HbF. We found that 3 RNA binding proteins, LIN28B, IGF2BP1, and/or IGF2BP3, mediate BMI1’s HbF silencing activity. All 3 proteins have been reported to regulate BCL11A levels posttranscriptionally,15,16,24 suggesting that they may present the missing links between PcG and HbF. Mechanistic studies characterized the relevant PRC subcomplexes and their interplay, and defined their mechanism of recruitment.
Methods
Domain-focused CRISPR screening
Domain-focused CRISPR screening was performed as previously described.52 Oligonucleotide pools representing 2952 single guide RNAs (sgRNAs) (supplemental Table 1, available on the Blood website) obtained from Twist Bioscience were subcloned into pLRG2.1 (Addgene, #108098) by Gibson Assembly (NEB, #E2611). Lentivirus was produced in 293T cells as described in supplemental Methods. Human umbilical cord blood–derived erythroid progenitor 2 (HUDEP2)-Cas9 cells (3 × 106) were transduced at 0.3 to 0.5 multiplicity of infection with sgRNA library-carrying virus. Green fluorescent protein–positive (GFP+) cells were sorted and expanded for 7 days, followed by 7 days of erythroid differentiation induction. Differentiated HUDEP2 cells were stained with an allophycocyanin (APC) conjugated HbF antibody (Thermo Fisher Scientific, #MHFH05), and sorted on a FACSJazz instrument. Approximately 3 × 106 sorted cells (top and bottom 10% of antigen-presenting cell fluorescence intensity) were washed with phosphate-buffered saline and incubated with proteinase K at 65°C overnight, followed by isolation of genomic DNA using PureLink Genomic DNA Mini Kit (Invitrogen). sgRNA species were amplified by polymerase chain reaction and subjected to next-generation sequencing (2 × 75 base pairs) on the Illumina MiSeq platform. Reads were trimmed to contain only the sgRNA sequence and mapped to the reference sgRNA library without allowing any mismatches. Read counts were calculated for each sgRNA and then normalized to library size. sgRNA abundance in HbF-high and HbF-low populations were log2 transformed in R (version 4.1.0) and plotted as a scatter plot using ggplot2 (version 3.3.6).
More experimental details are described in supplemental Methods.
Results
Identification and validation of BMI1 as a novel HbF repressor
We previously reported the use of domain-focused CRISPR-Cas9 genetic screens to identify HbF regulators in HUDEP2 cells, which express low basal HbF.7,10,17,52-54 Here, we surveyed a sgRNA library targeting 453 human E3 ligase (RING and HECT) domains and others carrying enzymatic activity, with an average of 6 guides per gene. After the incorporation of the sgRNA library into Cas9-expressing HUDEP2 cells and induction of erythroid differentiation, we isolated HbF-high (top 10%) and HbF-low (bottom 10%) populations by fluorescence-activated cell sorting and determined the abundance of the integrated sgRNAs by next-generation sequencing (supplemental Table 1). Notably, all 6 sgRNAs targeting BMI1/PCGF4, a subunit of the PRC1 complex, were enriched in the HbF-high population (Figure 1A), implicating BMI1 as a HbF repressor.
A protein domain–focused CRISPR screen identified BMI1 as a novel HbF repressor. (A) Scatter plot of sgRNA abundance in HbF-high and HbF-low populations from the domain-focused CRISPR screen. Each dot represents 1 sgRNA. Control sgRNAs (n = 50) and sgRNAs targeting BMI1 (n = 6) are labeled in blue and red, respectively. (B-D) Representative immunoblots of BMI1, GATA1, and γ-globin protein, HBG:(HBG+HBB) mRNA level, and HbF+ cell fraction in control (sgNeg: nontargeting sgRNA; sgBCL11A+58: positive control) and BMI1-depleted HUDEP2 cells. n = 2. β-Actin was used as the loading control in immunoblot experiment. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control for RT-qPCR. Values are presented as mean ± SEM. SEM, standard error of the mean.
A protein domain–focused CRISPR screen identified BMI1 as a novel HbF repressor. (A) Scatter plot of sgRNA abundance in HbF-high and HbF-low populations from the domain-focused CRISPR screen. Each dot represents 1 sgRNA. Control sgRNAs (n = 50) and sgRNAs targeting BMI1 (n = 6) are labeled in blue and red, respectively. (B-D) Representative immunoblots of BMI1, GATA1, and γ-globin protein, HBG:(HBG+HBB) mRNA level, and HbF+ cell fraction in control (sgNeg: nontargeting sgRNA; sgBCL11A+58: positive control) and BMI1-depleted HUDEP2 cells. n = 2. β-Actin was used as the loading control in immunoblot experiment. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control for RT-qPCR. Values are presented as mean ± SEM. SEM, standard error of the mean.
To validate the screening results, Cas9-expressing HUDEP2 cells were transduced with lentivirus expressing either a negative control sgRNA, 2 independent sgRNAs targeting BMI1, or, as the positive control, a sgRNA disrupting the +58 erythroid enhancer of the BCL11A.23 BMI1-depleted HUDEP2 cells grew normally and displayed intact erythroid differentiation as suggested by normal expression of the erythroid cell–specific genes GATA1, HBB, and HBA (Figure 1B; supplemental Figure 1A-B). In accordance with our screening results, BMI1 depletion increased the number of HbF-producing cells by over 5-fold (Figure 1D), elevated the production of γ-globin protein (the product of the HBG genes, Figure 1B), and raised HBG1/2 mRNA levels in the absence of changes in expression of the adult β-globin gene HBB and the adult α-globin gene HBA (Figure 1C; supplemental Figure 1A-C). Thus, BMI1 depletion in HUDEP2 cells appears to specifically activate HBG1/2 transcription.
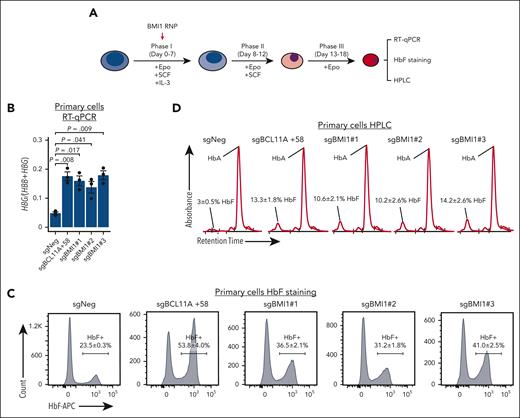
To test the effects of BMI1 loss in primary human adult erythroblasts, we electroporated immature proerythroblasts derived from healthy donor CD34+ hematopoietic stem and progenitor cells (HSPCs) with Cas9 ribonucleoprotein complexes targeting BMI1 (Figure 2A). All 3 BMI1 sgRNAs triggered robust HbF activation as assessed by reverse transcriptase quantitative PCR (RT-qPCR) (Figure 2B), HbF flow cytometry (Figure 2C), and hemoglobin high-performance liquid chromatography (HPLC, Figure 2D). There were also moderate increases of HBB and HBA mRNA in BMI1-depleted cells, likely a consequence of accelerated differentiation and slowed proliferation supplemental Figure 2A).55 However, the gains in HBG1/2 transcription were considerably more pronounced (supplemental Figure 2B). These results suggest that BMI1 represses HbF in adult erythroid cells.
BMI1 represses HbF in primary adult erythroblasts. (A) Schematic of the validation experiment in primary adult erythroid cells. CD34+ HSPCs isolated from peripheral blood of healthy donors were cultured with indicated cytokines and electroporated with Cas9 ribonucleoprotein at day 4 or 5 of the differentiation. (B-D) HBG:(HBB+HBG) mRNA levels (B) (n = 3 independent donors), HbF+ cell fraction (C) (n = 2), and hemoglobin HPLC profile (D) (n = 2) in control and BMI1-depleted primary adult erythroid cells. Values are presented as mean ± SEM. P values were calculated by unpaired 2-tailed Student t test. HPLC, high-performance liquid chromatography; SEM, standard error of the mean.
BMI1 represses HbF in primary adult erythroblasts. (A) Schematic of the validation experiment in primary adult erythroid cells. CD34+ HSPCs isolated from peripheral blood of healthy donors were cultured with indicated cytokines and electroporated with Cas9 ribonucleoprotein at day 4 or 5 of the differentiation. (B-D) HBG:(HBB+HBG) mRNA levels (B) (n = 3 independent donors), HbF+ cell fraction (C) (n = 2), and hemoglobin HPLC profile (D) (n = 2) in control and BMI1-depleted primary adult erythroid cells. Values are presented as mean ± SEM. P values were calculated by unpaired 2-tailed Student t test. HPLC, high-performance liquid chromatography; SEM, standard error of the mean.
IGF2BP1, IGF2BP3, and LIN28B mediate the HbF silencing activity of BMI1
To identify BMI1-regulated genes, we performed RNA sequencing (RNA-seq) in HUDEP2 cells after BMI1 depletion with 2 different Cas9 sgRNAs (n = 2 biological replicates). Differential gene expression analysis (fold change > 2; false discovery rate [FDR] < 0.1) identified 306 and 40 genes that were upregulated and downregulated, respectively, in both BMI1-depleted populations (Figure 3A-C; supplemental Tables 2 and 3). In line with our earlier findings (supplemental Figure 1), adult β-globin genes (HBB and HBD) remained unchanged in BMI1-depleted cells, whereas HBG1/2 genes were upregulated >16-fold (supplemental Tables 2 and 3). In addition, transcripts from the intergenic region between fetal and adult β-globin genes, BGLT3 and HBBP1, were all significantly upregulated, with BGLT3 preferentially affected (log2 fold change (BGLT3) of ∼4 vs log2 fold change (HBBP1) of ∼2; supplemental Tables 2 and 3). HBZ, encoding the embryonic form of α-globin, was upregulated ∼28-fold in BMI1-depleted HUDEP2 cells (supplemental Tables 2 and 3). The embryonic β−like globin gene HBE1 was upregulated ∼3-fold in BMI1-depleted cells in 1 replicate (supplemental Figure 3A; Rep1) and ∼90-fold in the second replicate (supplemental Figure 3A; Rep2), the latter being generated from cells with extended culture during the expansion phase after BMI depletion (see further discussion in “Results”). Indeed, HBG1/2, HBZ, and BGLT3 all exhibit more pronounced changes in replicate 2. Altogether, these data suggest that BMI1 represses expression of both fetal-type and embryonic-type globin genes in HUDEP2 cells.
IGF2BP1 and LIN28B mediate the HbF silencing activity of BMI1 in HUDEP2 cells. (A-B) Volcano plots of DEGs identified from RNA-seq in BMI1-depleted HUDEP2 cells (sgBMI1#1 or sgBMI1#2) by DESeq2. Cutoff threshold: fold change > 2; FDR < 0.1; n = 2. (C) Venn diagram of DEGs identified from RNA-seq in BMI1-depleted HUDEP2 cells by comparing control sample with sgBMI1#1 (A) and sgBMI1#2 (B) sample, respectively. (D-E) Enrichment plots of fetal enriched genes overrepresented in BMI1 depletion RNA-seq in HUDEP2 cells as determined by GSEA. (F-H) RT-qPCR analysis of HBG:(HBB+HBG) (F) (n = 2), representative immunoblots of BCL11A, IGF2BP1, LIN28B, BMI1, and γ-globin protein (G), and HbF+ fraction (H) in AsCas12a-expressing HUDEP2 cells that were transduced with lentivirus expressing indicated sgRNAs. Values are presented as mean ± SEM. RT-qPCR data were normalized to AHSP. GAPDH was used as loading control in immunoblot experiment. DEGs, differentially expressed genes; GSEA, gene set enrichment analysis; SEM, standard error of the mean.
IGF2BP1 and LIN28B mediate the HbF silencing activity of BMI1 in HUDEP2 cells. (A-B) Volcano plots of DEGs identified from RNA-seq in BMI1-depleted HUDEP2 cells (sgBMI1#1 or sgBMI1#2) by DESeq2. Cutoff threshold: fold change > 2; FDR < 0.1; n = 2. (C) Venn diagram of DEGs identified from RNA-seq in BMI1-depleted HUDEP2 cells by comparing control sample with sgBMI1#1 (A) and sgBMI1#2 (B) sample, respectively. (D-E) Enrichment plots of fetal enriched genes overrepresented in BMI1 depletion RNA-seq in HUDEP2 cells as determined by GSEA. (F-H) RT-qPCR analysis of HBG:(HBB+HBG) (F) (n = 2), representative immunoblots of BCL11A, IGF2BP1, LIN28B, BMI1, and γ-globin protein (G), and HbF+ fraction (H) in AsCas12a-expressing HUDEP2 cells that were transduced with lentivirus expressing indicated sgRNAs. Values are presented as mean ± SEM. RT-qPCR data were normalized to AHSP. GAPDH was used as loading control in immunoblot experiment. DEGs, differentially expressed genes; GSEA, gene set enrichment analysis; SEM, standard error of the mean.
Notably, BMI1 depletion activated genes encoding for 2 RNA-binding proteins, IGF2BP1 and LIN28B (Figure 3A-C; supplemental Figure 3A-B), that are normally expressed in fetal-type erythroid cells21,22 and could substantially raise HbF production when overexpressed in adult erythroid cells.15,16,24,56 In contrast, GCNT2, a known target of IGF2BP1 and LIN28B,16,22,24 was identified as downregulated in BMI1-depleted HUDEP2 cells (Figure 3A-C). Gene set enrichment analysis revealed that BMI1-depleted HUDEP2 cells upregulated fetal erythroid cell–specific transcripts (Figure 3D-E; supplemental Figure 3C-D), presumably because increases in IGF2BP1 and LIN28B promote a global fetal gene expression program.57
Prior studies have suggested that overexpression of IGF2BP1 or LIN28B in adult erythroid cells activates the HBG genes by interfering with BCL11A mRNA translation.15,16,24 BCL11A protein was only modestly reduced in the newly generated BMI1-depleted HUDEP2 cells (supplemental Figure 3B Rep1), however, BCL11A levels were substantially lower in BMI1-depleted HUDEP2 cells after an additional 5 days of culture (12 days total, supplemental Figure 3B Rep2). Accordingly, downstream targets of BCL11A58 were also further elevated in these cells (supplemental Figure 3A). Moreover, we detected further increases of LIN28B and IGF2BP1 at both mRNA and protein level (compare Rep1 and Rep2 in supplemental Figure 3A-B). We speculate that PcG silencing of LIN28B and IGF2BP1 might be epigenetically maintained even in the absence of BMI1, perhaps until repressive histone posttranslation modifications (PTMs) and their “readers” are diluted after several rounds of cell division.25 To test whether BCL11A loss is responsible for HbF activation in BMI1-depleted cells, we forced the expression of BCL11A using a lentiviral vector carrying BCL11A-HA cDNA along with GFP as a reporter to sort BCL11A-expressing cells (supplemental Figure 3E). As a control, we also infected BCL11A +58 erythroid enhancer–targeted HUDPE2 cells. Exogenous BCL11A expression largely restored HBG silencing in both BMI1-depleted (sgBMI1#1) and BCL11A-deficient cells (sgBCL11A+58) (supplemental Figure 3F-G), indicating that BCL11A loss is critical for HbF upregulation in BMI1-depleted cells.
To test whether the upregulation of IGF2BP1 and LIN28B accounted for the observed HbF activation in BMI1-depleted HUDEP2 cells, we employed an optimized CRISPR-AsCas12a system7,59 to combinatorially disrupt BMI1, IGF2BP1, and LIN28B genes (Figure 3F-H). As expected, codepletion of either IGF2BP1 or LIN28B with BMI1 partially attenuated the HbF activation, whereas triple disruption of IGF2BP1, LIN28B, and BMI1 genes lowered HbF production virtually to basal levels (Figure 3F-H).
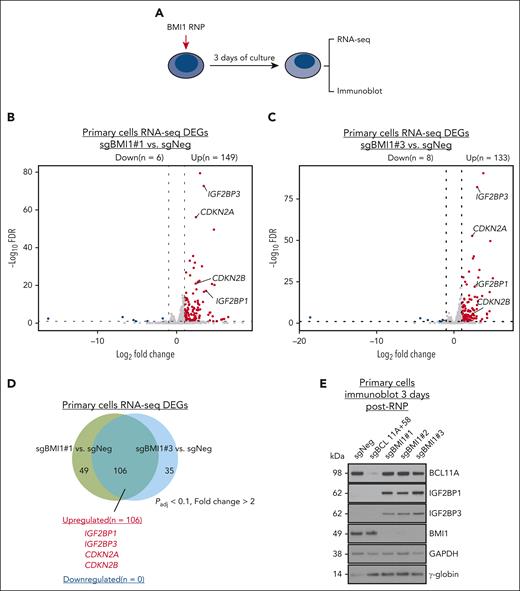
Next, we performed RNA-seq in erythroid progeny derived from CD34+ HSPCs in which BMI1 was targeted with sgRNA#1 and sgRNA#3 (Figure 4A). In total, 133 and 149 differentially expressed genes were identified, respectively, most of which were upregulated in BMI1 sgRNA targeted cells (Figure 4B-C; supplemental Tables 4 and 5); 106 genes were BMI1 repressed in both data sets (Figure 4D). Two genes that slow cell cycle progression, CDKN2A and CDKN2B, and are known BMI1 targets in other tissue contexts,40,60,61 displayed elevated expression in BMI1-depleted primary cells (Figure 4B-D). Notably, in HUDEP2 cells the expression of both genes was unchanged upon BMI1 depletion (supplemental Tables 2 and 3), consistent with our observation that BMI1-depleted HUDEP2 cells grew normally.
IGF2BP1 and IGF2BP3 mediate the HbF silencing activity of BMI1 in primary adult erythroblasts. (A) Schematic of BMI1 depletion experiments in primary adult erythroid cells. (B-C) Volcano plots of DEGs identified from RNA-seq in BMI1-depleted primary adult erythroblasts (sgBMI1#1 or sgBMI1#3) by DESeq2. Cutoff threshold: fold change > 2; FDR < 0.1; n = 2. (D) Venn diagram of overlapped DEGs identified from RNA-seq in BMI1-depleted primary adult erythroblasts by comparing control sample with sgBMI1#1 (B) and sgBMI1#3 (C) sample, respectively. (E) Representative immunoblots of BCL11A, IGF2BP1, IGF2BP3, BMI1, and γ-globin protein in control (sgNeg: nontargeting sgRNA; sgBCL11A+58, positive control) or BMI1-depleted primary adult erythroblasts. GAPDH was used as loading control. DEGs, differentially expressed genes.
IGF2BP1 and IGF2BP3 mediate the HbF silencing activity of BMI1 in primary adult erythroblasts. (A) Schematic of BMI1 depletion experiments in primary adult erythroid cells. (B-C) Volcano plots of DEGs identified from RNA-seq in BMI1-depleted primary adult erythroblasts (sgBMI1#1 or sgBMI1#3) by DESeq2. Cutoff threshold: fold change > 2; FDR < 0.1; n = 2. (D) Venn diagram of overlapped DEGs identified from RNA-seq in BMI1-depleted primary adult erythroblasts by comparing control sample with sgBMI1#1 (B) and sgBMI1#3 (C) sample, respectively. (E) Representative immunoblots of BCL11A, IGF2BP1, IGF2BP3, BMI1, and γ-globin protein in control (sgNeg: nontargeting sgRNA; sgBCL11A+58, positive control) or BMI1-depleted primary adult erythroblasts. GAPDH was used as loading control. DEGs, differentially expressed genes.
Surprisingly, the LIN28B gene was not on the list of misexpressed genes in BMI1-targeted primary cells, but IGF2BP1 and its functionally related homolog IGF2BP322,24 were both upregulated more than 10-fold (Figure 4E; supplemental Tables 4 and 5). To test the impact of gained IGF2BP1/3 expression, proerythroblasts derived from CD34+ HSPCs were transduced with a lentiviral vector expressing IGF2BP1/3-P2A-GFP at varying levels (based on GFP fluorescence intensity) and HbF production was measured. Forced expression of either IGF2BP1 or IGF2BP3 elevated HBG expression in primary adult erythroblasts in a dose-dependent manner (supplemental Figure 4A-C), as expected.24 This supports the idea that IGF2BP1/3 are drivers of HbF in BMI1-depleted primary adult erythroblasts.
BMI1 occupies proximal CpG islands at the IGF2BP1, IGF2BP3, and LIN28B genes
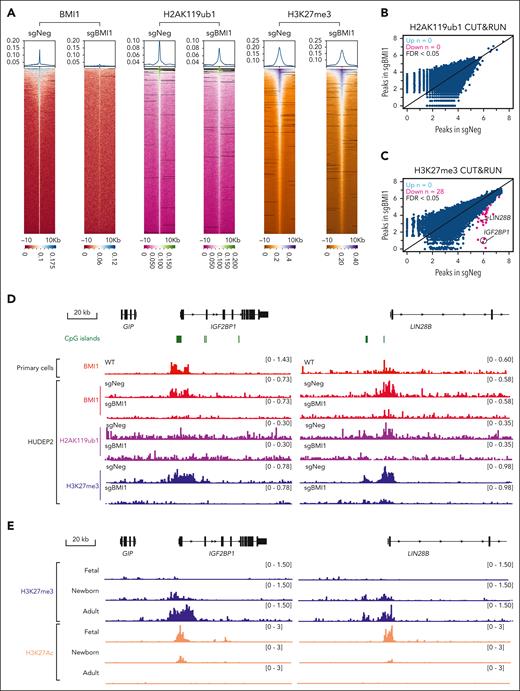
To gain insights into how BMI1 depletion led to derepression of LIN28B and IGF2BP1/3, we performed cleavage under targets and release using nuclease (CUT&RUN) of BMI1 in HUDEP2 cells and primary adult erythroblasts, and identified 5659 and 6766 peaks, respectively. In addition, we profiled 2 PcG-related chromatin marks, H2AK119ub1 and H3K27me3, in control and BMI1-depleted HUDEP2 cells. BMI1 depletion only modestly reduced the global signal intensities of either H2AK119ub1 or H3K27me3 in HUDEP2 cells (Figure 5A). Notably, at the β-globin gene locus, no enrichment of BMI1 proteins or H2AK119ub1/H3K27me3 was detected (supplemental Figure 5A), indicating that BMI1 regulates HbF indirectly.
BMI1 occupies proximal CpG islands at the IGF2BP1, IGF2BP3, and LIN28B genes. (A) Heatmaps of BMI1, H2AK119ub1, and H3K27me3 peaks identified from CUT&RUN experiment in control or BMI1-depleted HUDEP2 cells. (B-C) Scatter plots of H2AK119ub1 (B) and H3K27me3 (C) enrichments in control and BMI1-depleted HUDEP2 cells. Differential analysis was performed using DESeq2 and DiffBind packages. Differential peaks were identified with FDR < 0.05 and are labeled in red (n = 2 biological replicates). (D) Chromatin occupancy of BMI1 and enrichment of H2AK119ub1 and H3K27me3 at the IGF2BP1 and LIN28B genes in control and BMI1-depleted HUDEP2 cells. Chromatin occupancy of BMI1 in primary adult erythroblasts was used for comparison. CpG island track (hg38) was obtained from the University of California, Santa Cruz (UCSC) genome browser. (E) Enrichment of H3K27me3 and H3K27ac at the IGF2BP1 and LIN28B genes in primary erythroblasts derived from CD34+ HSPCs isolated from fetal liver (fetal), cord blood (newborn), and peripheral blood (adult); n = 2 healthy donors.
BMI1 occupies proximal CpG islands at the IGF2BP1, IGF2BP3, and LIN28B genes. (A) Heatmaps of BMI1, H2AK119ub1, and H3K27me3 peaks identified from CUT&RUN experiment in control or BMI1-depleted HUDEP2 cells. (B-C) Scatter plots of H2AK119ub1 (B) and H3K27me3 (C) enrichments in control and BMI1-depleted HUDEP2 cells. Differential analysis was performed using DESeq2 and DiffBind packages. Differential peaks were identified with FDR < 0.05 and are labeled in red (n = 2 biological replicates). (D) Chromatin occupancy of BMI1 and enrichment of H2AK119ub1 and H3K27me3 at the IGF2BP1 and LIN28B genes in control and BMI1-depleted HUDEP2 cells. Chromatin occupancy of BMI1 in primary adult erythroblasts was used for comparison. CpG island track (hg38) was obtained from the University of California, Santa Cruz (UCSC) genome browser. (E) Enrichment of H3K27me3 and H3K27ac at the IGF2BP1 and LIN28B genes in primary erythroblasts derived from CD34+ HSPCs isolated from fetal liver (fetal), cord blood (newborn), and peripheral blood (adult); n = 2 healthy donors.
Next, we performed differential peak analysis using DiffBind62 to examine histone PTM changes. H2AK119ub1 peaks identified from control and BMI1-depleted cells were merged to generate a reference data set comprising 36 361 H2AK119ub1 peaks (as called by MACS2). After normalization to library size and differential analysis (FDR < 0.05), surprisingly, no differential peaks were called between control and BMI1-depleted cells (Figure 5B; supplemental Table 6). However, with a more relaxed P value cutoff (P < .05), 771 differential peaks were called. Moreover, of the 36 921 H3K27me3 peaks, only 28 were identified as being reduced in BMI1-depleted cells (FDR < 0.05). These mild global effects are consistent with minimal changes of histone modification in bulk chromatin (see further sections for immunoblots of H3K27me3 and H2AK119ub1). However, H3K27me3 mark was dramatically reduced at the IGF2BP1 and LIN28B gene promoters (Figure 5C-D; supplemental Table 7).
IGF2BP1 and LIN28B harbor nearby CpG islands (Figure 5D), which are typical PRC1 and PRC2 binding locations.25,26 Indeed, H3K27me3 signals were highly enriched at CpG islands of both IGF2BP1 and LIN28B gene promoters in HUDEP2 cells (Figure 5D). In contrast, H2AK119ub1 was only modestly enriched at the LIN28B gene promoter (Figure 5D), overlapping with both BMI1 and H3K27me3 at these sites in HUDEP2 cells (Figure 5D). Depletion of BMI1 reduced the already low H2AK119ub1 signals at the LIN28B gene promoter (Figure 5D), suggesting that the presence of weak PRC1 enzymatic activity at this location. In addition, we detected strong enrichment of BMI1 at the IGF2BP3 gene promoter in primary adult erythroid cells but not in HUDEP2 cells (supplemental Figure 5B), which agrees with our observation that IGF2BP3 is a BMI1 target in primary adult erythroid cells but not in HUDEP2 cells (Figures 3C and 4C). Thus, BMI1 maintains a silent chromatin state at the IGF2BP1 and LIN28B genes in HUDEP2, whereas in primary adult erythroid cells, IGF2BP3 emerges as an additional target of BMI1.
LIN28B, IGF2BP1, and IGF2BP3 expression is extinguished during the fetal-to-adult developmental transition along with corresponding changes in histone PTM.21,22,63 We investigated H3K27me3 profiles in primary erythroblasts derived from fetal liver, cord blood, and peripheral blood, representing fetal, newborn, and adult stages, respectively (Figure 5E; supplemental Figure 5C). The CpG islands at the LIN28B, IGF2BP1, and IGF2BP3 gene promoters are devoid of H3K27me3 in fetal cells but display intermediate H3K27me3 levels in newborn erythroblasts with much higher levels in adult cells (Figure 5E; supplemental Figure 5C). H3K27ac, a chromatin mark associated with active genes, shows an inverse pattern (Figure 5E; supplemental Figure 5C). In addition, the dynamics of H3K27me3 and H3K27ac across the 3 developmental stages differ somewhat between the IGF2BP3, LIN28B, and IGF2BP1 genes. The major changes in chromatin status at the IGF2BP3 gene occur during the newborn-to-adult stage transition (supplemental Figure 5C), as opposed to the gradual changes of H3K27me3 and H3K27ac levels at the other 2 loci (Figure 5E). These observations, together with our genetic BMI1 depletion experiments, suggest that LIN28B, IGF2BP1, and IGF2BP3 are silenced by PRC complexes during the fetal-to-adult transition.
BMI1 associates with CBX proteins to repress HbF in adult erythroid cells
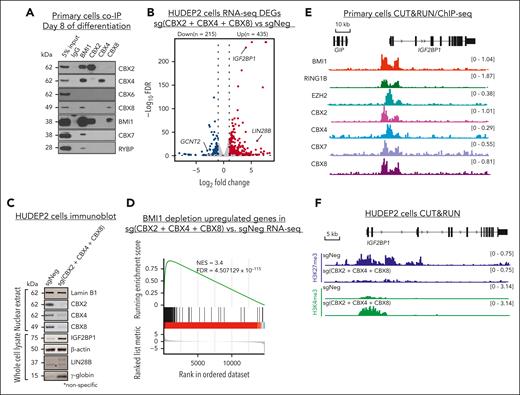
BMI1/PCGF4 can form 2 functionally distinct PRC1 subcomplexes, cPRC1 and ncPRC1.33,34 cPRC1 is defined by the presence of 1 of the 5 CBX (CBX2, 4, and 6-8) proteins, whereas RYBP/YAF2 are subunits specific to the ncPRC1 complex (supplemental Figure 6A). Coimmunoprecipitation experiments detected interactions of BMI1 with both CBX and RYBP in nuclear extracts isolated from primary adult erythroblasts (Figure 6A). Anti-CBX 2/4/8 immunoprecipitation failed to coprecipitate RYBP or other CBX proteins, suggesting that they form mutual-exclusive complexes with BMI1 (Figure 6A).
BMI1 associates with CBX proteins to repress HbF in adult erythroid cells. (A) Representative immunoblots of BMI1 and different ancillary subunits (CBX2, 4, 6-8, and RYBP) in primary adult erythroblasts coimmunoprecipitation (co-IP) experiments. (B) Volcano plots of DEGs identified from CBX2, 4, and 8 triple depletion RNA-seq experiment in HUDEP2 cells by DESeq2. Cutoff threshold: fold change > 2; FDR < 0.1; n = 2. (C) Representative immunoblots of CBX2, 4, and 8, and IGF2BP1, LIN28B, and γ-globin protein in nuclear extract or whole-cell lysate of control and CBX2, 4, and 8 triple-depleted HUDEP2 cells. Lamin B1 and GAPDH were used as loading controls for nuclear extract and whole cell lysate respectively. (D) Enrichment plots of BMI1-repressed genes overrepresented in CBX2, 4, and 8 triple depletion RNA-seq in HUDEP2 cells as determined by GSEA. (E) Chromatin occupancies of BMI1, RING1B, EZH2, CBX2, 4, 7, and 8 in primary adult erythroblasts at the IGF2BP1 gene region. (F) Chromatin enrichment of H3K27me3 and H3K4me3 at IGF2BP1 gene in control or CBX2, 4, and 8 triple-depleted HUDEP2 cells. DEGs, differentially expressed genes; GSEA, gene set enrichment analysis.
BMI1 associates with CBX proteins to repress HbF in adult erythroid cells. (A) Representative immunoblots of BMI1 and different ancillary subunits (CBX2, 4, 6-8, and RYBP) in primary adult erythroblasts coimmunoprecipitation (co-IP) experiments. (B) Volcano plots of DEGs identified from CBX2, 4, and 8 triple depletion RNA-seq experiment in HUDEP2 cells by DESeq2. Cutoff threshold: fold change > 2; FDR < 0.1; n = 2. (C) Representative immunoblots of CBX2, 4, and 8, and IGF2BP1, LIN28B, and γ-globin protein in nuclear extract or whole-cell lysate of control and CBX2, 4, and 8 triple-depleted HUDEP2 cells. Lamin B1 and GAPDH were used as loading controls for nuclear extract and whole cell lysate respectively. (D) Enrichment plots of BMI1-repressed genes overrepresented in CBX2, 4, and 8 triple depletion RNA-seq in HUDEP2 cells as determined by GSEA. (E) Chromatin occupancies of BMI1, RING1B, EZH2, CBX2, 4, 7, and 8 in primary adult erythroblasts at the IGF2BP1 gene region. (F) Chromatin enrichment of H3K27me3 and H3K4me3 at IGF2BP1 gene in control or CBX2, 4, and 8 triple-depleted HUDEP2 cells. DEGs, differentially expressed genes; GSEA, gene set enrichment analysis.
Next, we sought to determine which BMI1-associated subcomplexes are critical for HbF repression (supplemental Figure 6A). Notably, a previous chromatin reader domain–focused CRISPR-Cas9 screen failed to identify any of these BMI1 interaction partners as HbF repressors in HUDEP2 cells,53 which we suspected was because of redundancy among gene paralogues. Therefore, we used the CRISPR-AsCas12a system,7,59 which, because of its ability to multiplex sgRNAs, allows combinatorial targeting of different CBX or RYBP/YAF2 gene combinations. Various combinations of sgRNAs targeting CBX2, 4, 6, 7, and 8 as well as RYBP and YAF2 were examined for HbF production in HUDEP2 cells. Codepletion of any 2 of CBX2, 4, and 8 could reactivate the HBG genes (supplemental Figure 6B), whereas depletion of all 3 produced the strongest effect (supplemental Figure 6B), indicating that these proteins function redundantly during HbF repression. In contrast, disrupting RYBP plus YAF2, or some combinatorial depletions involving CBX6 or CBX7, did not significantly induce HbF expression (supplemental Figure 6B). Importantly, the combined depletion of CBX2, 4, and 8 activated IGF2BP1 and LIN28B expression (Figure 6B-C), and activated a fetal-specific gene expression signature (supplemental Figure 6C; supplemental Table 8), similar to what we observed in BMI1-depleted HUDEP2 cells. Moreover, the BMI1-repressed genes are significantly enriched in the CBX2, 4, and 8 triple-depleted cells as per gene set enrichment analysis (normalized enrichment score of 3.4; Figure 6D) suggesting that BMI1 and CBX2/4/8 are repressing similar genes. However, the CBX2/4/8- and BMI1-regulated genes do not completely overlap (supplemental Figure 6D), indicating that CBX2/4/8 may convey additional functions independent of PRC1.64
The CBX components in cPRC1 associate with H3K27me3-decorated chromatin.35 Hence, our findings support a model in which BMI1-containing cPRC1 is loaded onto select chromatin regions via H3K27me3 binding but in which PRC1 is also required for the maintenance of PRC2 function.25,26 To investigate the cooccupancy of BMI1 and other factors of the cPRC1 and PRC2 complexes, we determined their genomic localization by CUT&RUN (BMI1, RING1B, EZH2, and CBX2, 7, and 8) and chromatin immunoprecipitation followed by sequencing (ChIP-seq, CBX4). BMI1-bound regions displayed 30.8% overlap with RING1B-bound regions, 13.4%-19.9% overlap with CBX-bound regions (CBX2, 4, 7, and 8), and 11.7% overlap with EZH2-bound regions, respectively, in primary adult erythroblasts (supplemental Figure 6E-F). Notably, at the IGF2BP1 gene promoter both cPRC1 and PRC2 components were detected (Figure 6E). Although CBX7 was only weakly, if at all, represented in the cPRC1 complex in the immunoprecipitation experiments (Figure 6A), it displayed a binding pattern similar to that of CBX2, 4, and 8 (Figure 6E) at the IGF2BP1 gene promoter region. However, CBX7 was dispensable for HbF repression (supplemental Figure 6B), possibly because of differences in structure or expression level from other CBXs.65,66 Moreover, combined depletion of CBX2, 4, and 8 in HUDEP2 cells dramatically reduced H3K27me3 at the IGF2BP1 promoter with a concurrent gain of transcriptional activity, as reflected by increased H3K4me3 (Figure 6F). These observations, together with our prior chromatin mark analysis in BMI1-depleted HUDEP2 cells (Figure 5), suggest that cPRC1, once recruited by H3K27me3, is required for maintaining optimal PRC2 binding or activity.
cPRC1 and PRC2 cooperate to maintain the repression of IGF2BP1/3 and LIN28B in adult erythroid cells
PRC2 is required for cPRC1 recruitment because it lays down the H3K27me3 mark. To test whether PRC2 is involved in the silencing of IGF2BP1 and LIN28B genes in adult erythroid cells, we targeted the EZH2 gene via CRISPR-AsCas12a in HUDEP2 cells and examined the resulting gene expression changes by RNA-seq. HBG genes were significantly upregulated upon EZH2 depletion, accompanied by a modest upregulation of both IGF2BP1 and LIN28B genes (Figure 7A) as well as the enrichment of a fetal-specific gene signature (supplemental Figure 7A). These changes resemble those observed upon BMI1 or CBX2, 4, and 8 triple depletions. However, EZH2 depletion caused a considerable cell growth defect and more broad gene expression changes when compared with BMI1 depletion (supplemental Figure 7B); for example, several genes involved in apoptosis or cell proliferation including CDKN1A (p21), BID, and BIRC3 were dysregulated in EZH2-depleted HUDEP2 cells (supplemental Table 9). Next, we tested whether IGF2BP1 and LIN28B mediated the effect of EZH2 on HBG gene expression by AsCas12a multigene targeting. Similar to BMI1 depletion, codepletion of IGF2BP1 and LIN28B largely reversed EZH2 depletion–induced HbF activation (Figure 7B-C), suggesting that IGF2BP1 and LIN28B are the critical links between EZH2/PRC2 and HbF regulation in HUDEP2 cells.
cPRC1 and PRC2 cooperate to maintain the repression of IGF2BP1/3 and LIN28B in adult erythroid cells. (A) Volcano plots of DEGs identified from EZH2 depletion RNA-seq in HUDEP2 cells by DESeq2. Cutoff threshold: fold change > 2; FDR < 0.1; n = 2. (B-C) RT-qPCR analysis of the ratio of HBG:(HBB+HBG) mRNA levels (B), representative immunoblots (C) of EZH2, IGF2BP1, LIN28B, BMI1, H2AK119ub1, H3K27me3, and γ-globin protein in AsCas12a-expressing HUDEP2 cells that were transduced with lentivirus expressing indicated sgRNAs. Values are presented as mean ± SEM. RT-qPCR data were normalized to AHSP (n = 2). GAPDH was used as loading control in immunoblot experiment. (D) Schematic of EZH2 inhibition experiment in primary erythroid cells. (E) Representative immunoblots of IGF2BP1, IGF2BP3, H3K27me3, histone H3, and γ-globin in drug-treated primary adult erythroblasts. GAPDH were used as the loading control. (F) Relative IGF2BP1, IGF2BP3, and HBG mRNA level in primary adult erythroblasts (at day 10 or 12 of differentiation [equal to 3 or 5 days after drug treatment]) as detected by RT-qPCR. Values are presented as mean ± SEM. Data were normalized to AHSP; n = 4. (G) Summary of PRC1/2 perturbation experiments. In HUDEP2 cells, genetic perturbation of BMI1, CBX, or EZH2 led to upregulation of LIN28B and IGF2BP1, and thus, HbF activation. In primary adult erythroblasts, BMI1 depletion and EZH2 inhibition both resulted in derepression of IGF2BP1/3, and elevation of HbF. However, the molecular link between the 3 RNA binding proteins and HbF activation remains to be defined. Our observations support a major role of BCL11A in mediating the function of these RNA-binding proteins, especially in HUDEP2 cells, however we cannot rule out additional, yet to be identified targets of 3 RNA binding proteins involved in HbF activation.
cPRC1 and PRC2 cooperate to maintain the repression of IGF2BP1/3 and LIN28B in adult erythroid cells. (A) Volcano plots of DEGs identified from EZH2 depletion RNA-seq in HUDEP2 cells by DESeq2. Cutoff threshold: fold change > 2; FDR < 0.1; n = 2. (B-C) RT-qPCR analysis of the ratio of HBG:(HBB+HBG) mRNA levels (B), representative immunoblots (C) of EZH2, IGF2BP1, LIN28B, BMI1, H2AK119ub1, H3K27me3, and γ-globin protein in AsCas12a-expressing HUDEP2 cells that were transduced with lentivirus expressing indicated sgRNAs. Values are presented as mean ± SEM. RT-qPCR data were normalized to AHSP (n = 2). GAPDH was used as loading control in immunoblot experiment. (D) Schematic of EZH2 inhibition experiment in primary erythroid cells. (E) Representative immunoblots of IGF2BP1, IGF2BP3, H3K27me3, histone H3, and γ-globin in drug-treated primary adult erythroblasts. GAPDH were used as the loading control. (F) Relative IGF2BP1, IGF2BP3, and HBG mRNA level in primary adult erythroblasts (at day 10 or 12 of differentiation [equal to 3 or 5 days after drug treatment]) as detected by RT-qPCR. Values are presented as mean ± SEM. Data were normalized to AHSP; n = 4. (G) Summary of PRC1/2 perturbation experiments. In HUDEP2 cells, genetic perturbation of BMI1, CBX, or EZH2 led to upregulation of LIN28B and IGF2BP1, and thus, HbF activation. In primary adult erythroblasts, BMI1 depletion and EZH2 inhibition both resulted in derepression of IGF2BP1/3, and elevation of HbF. However, the molecular link between the 3 RNA binding proteins and HbF activation remains to be defined. Our observations support a major role of BCL11A in mediating the function of these RNA-binding proteins, especially in HUDEP2 cells, however we cannot rule out additional, yet to be identified targets of 3 RNA binding proteins involved in HbF activation.
To test the impact of PRC2 perturbation in primary adult erythroid cells, we treated CD34+ HSPCs-derived proerythroblasts (day 7 of differentiation) with the EZH2 inhibitor EPZ-6438 and determined the expression of IGF2BP1/3 and globin genes by RT-qPCR and immunoblot throughout the rest of differentiation protocol (day 10 and 12; Figure 7D).67 Compared with the vehicle control (dimethyl sulfoxide), 3 days of 1 μM or 5 μM EPZ-6438 treatment both reduced total H3K27me3 levels by >80% (Figure 7E) and activated IGF2BP1 and IGF2BP3 mRNA and protein production (Figure 7F). An additional 2-day treatment depleted bulk H3K27me3 levels more completely but did not further augment IGF2BP1/3 levels (Figure 7E). Consistent with prior reports on the role of PRC2 in regulating GATA1 target genes and erythroid cell maturation,68 all globin genes were upregulated in a dose-dependent manner after 5 days of EPZ-6438 treatment, however, as shown by HBG:(HBB+HBG), the gain of HBG expression was greater than that of the HBB gene (Figure 7F; supplemental Figure 7C). The induction of IGF2BP1/3 preceded that of the HBG genes, supporting the hypothesis that IGF2BP1 and IGF2BP3 are downstream effectors of PcG in primary adult erythroblasts. Together, these results suggest that both cPRC1 and PRC2 repress HbF through the developmental silencing of LIN28B, IGF2BP1, and/or IGF2BP3.
Polycomb complexes are critical epigenetic regulators with known mechanisms for propagation of repressive marks through cell division.69 Therefore, we asked whether transient interference with PcG activity might result in lasting effects on HbF induction. To this end, we treated HUDEP2 cells with EPZ-6438 (1.5 μM) for 3 days, followed by induction of erythroid differentiation in the absence of the drug (regimen 1, supplemental Figure 7D). Alternatively, cells were grown for an additional 5 days in the absence of the drug followed by differentiation induction (regimen 2). For comparison, we included the experimental HbF inducer pomalidomide70 following the same 2 regimens. Notably, the effects on HbF induction triggered by EPZ-6438 were sustained even in the regimen 2–treated cells (supplemental Figure 7D). This was not the case for pomalidomide, whose effects required its continued presence. This suggests that an epigenetic mechanism may propagate a transcriptionally active state after PcG inhibition. This observation has ramifications when considering PcG inhibition as a therapeutic approach.
Discussion
Here, via a domain-focused CRISPR genetic screen, we uncovered the cPRC1 component BMI1 as a novel HbF repressor. We further uncovered the fetal stage–specific RNA binding proteins LIN28B, IGF2BP1, and/or IGF2BP3 as critical mediators of the HbF silencing function of PRC complexes. First, all 3 genes are PRC-occupied predominantly in adult erythroid cells. Second, loss of BMI1 diminishes H3K27me3 and/or H2A119ub1 at these genes, accompanied by transcriptional derepression. Third, codepletion of these genes in combination with BMI1 restores HBG silencing.
Our dissection of the molecular composition of BMI1 interaction partners suggests that in this context, BMI1 predominantly functions as part of cPRC1. We further discovered that cPRC1 acts in conjunction with PRC2 to regulate LIN28B, IGF2BP1, and/or IGF2BP3 (Figure 7G). This is supported by our observation that EZH2 inhibition triggers upregulation of these genes, and conversely, that BMI1 depletion reduces the PRC2-associated histone mark H3K27me3. In support of our model, a prior study reported that in murine HSPCs, Ezh2 depletion led to upregulation of Lin28b and Igf2bp genes, accompanied by loss of H3K27me3 at the gene promoters.71
EZH2 was identified as a putative interaction partner of BCL11A,50 raising the possibility that BCL11A might recruit PRC2 and silence HBG1/2 genes directly via H3K27me3. However, we failed to detect EZH2 or H3K27me3 enrichment anywhere at the β-globin locus in HUDEP2 cells or primary adult erythroblasts. Because codepletion of IGF2BP1 and LIN28B almost completely rescued HBG1/2 repression in EZH2-deficient HUDPE2 cells as well as in BMI1-depleted cells, this further supports an indirect mechanism by which PRC complexes influence the fetal-to-adult hemoglobin switch.
HUDEP2 cells provide a powerful screening platform but, in select cases, they can behave differently from primary adult erythroblasts.54 Chromatin occupancies of BMI1 are distinct between these cellular models (only ∼16% overlap), and transcriptome analysis of BMI1-regulated genes revealed only 20 shared dysregulated genes (supplemental Tables 2-5). Because HUDEP2 cells are thought to resemble basophilic erythroblasts,8 this lack of concordance may in part be attributable to different differentiation stages of the cells. Moreover, HUDEP2 cells might carry some features of neonatal erythropoiesis72 because of their cord blood origin.73 These considerations, among others, might account for our observations that LIN28B repression is BMI1 dependent in HUDEP2 cells but not in primary cells, and IGF2BP3 is targeted by BMI1 only in primary adult erythroid cells. Regardless, BMI1 depletion reproducibly induced HbF expression in both experimental systems, enabling us to carry out our molecular dissection of the underlying mechanisms.
How the upregulation of LIN28B and IGF2BPs activates HbF in adult erythroid cells remains to be defined (Figure 7G). RNA binding protein LIN28B inhibits the biogenesis of let-7 family microRNAs (miRNAs)74 and can also affect the translation of select mRNAs. Gained expression of LIN28B in adult erythroid cells may suppress BCL11A mRNA translation through direct binding,15 and/or indirectly via let-7 miRNAs, which is the most abundant miRNA species in adult erythroid cells with a known role in supporting BCL11A expression.16 Although forced IGF2BP1 expression in adult erythroid cells has been shown to downregulate BCL11A through a posttranscriptional mechanism,24 how IGF2BP1 regulates BCL11A mRNA translation requires further investigation. In addition, we cannot rule out additional, yet to be identified targets of these RNA binding proteins involved in HbF regulation.
BMI1 deficiency in HUDPE2 cells led to loss of H3K27me3 at only a few genomic loci, most of which are in CpG islands, including those at the IGF2BP1 and LIN28B gene promoters. Future studies will be needed to determine what factors or mechanisms account for this selectivity of cPRC1. Given that targeting PRC2 would erase the H3K27me3 mark globally, our results suggest that targeting selective components of PRC1/2 might achieve a similar desirable outcome but with less potentially detrimental consequences. This study thus provides a platform for further dissection of cPRC1 complexes, including the fine mapping of the molecular structure of HbF-relevant cPRC1 components, which may pave the way for the development of novel PcG-targeted therapies with improved therapeutic indexes for the treatment of sickle cell disease and β-thalassemia.
Acknowledgments
The authors thank the CHOP Flow Cytometry Core for assistance in cell sorting, members of the Blobel laboratory for discussions, and Scott A. Peslak (University of Pennsylvania) for critical comments on the manuscript. HUDEP2 cells were a gift from R. Kurita and Y. Nakamura (RIKEN BioResource Center). The authors thank Emily C. Dykhuizen (Purdue University), Stephen V. Frye (The University of North Carolina), Hannele Ruohola-Baker (University of Washington), and Roberto Bonasio (University of Pennsylvania) for sharing reagents. The authors thank the DiGaetano family for their generous support.
The Fred Hutchinson Cancer Research Center Cooperative Center of Excellence in Hematology was supported by National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant DK106829. This work was supported by a grant from the NIH National Heart, Lung, and Blood Institute (NHLBI) (R01HL119479) and research funding from Fulcrum Therapeutics (G.A.B.); NIH, NIDDK grant R24DK106766 (G.A.B., R.C.H.); NIH, NHLBI T32 training grant HL007150-42; American Society of Hematology Research Training Award for Fellows (E.K.); Lingang Laboratory, China grant LG-QS-202204-05 (X.L.); and the St. Jude Children’s Research Hospital Collaborative Research Consortium on Novel Gene Therapies for Sickle Cell Disease.
Authorship
Contribution: X.L., J.S., and G.A.B. conceived the study; K.Q. and X.L. designed experiments and analyzed the data; X.L. and J.S. performed the CRISPR screen; X.L. performed BMI1 HUDEP2 and primary cell validation; K.Q. performed primary cell validation, AsCas12a CRISPR experiments, CUT&RUN, and chromatin immunoprecipitation with the help of M.S.S.; P.H. performed IGF2BP1/3 overexpression and drug treatment studies; E.K. and J.S. generated critical reagents for this study; O.A. performed Hb high-performance liquid chromatography; C.A.K. prepared the RNA-seq libraries and carried out all the next-generation sequencing runs; K.Q., C.A.K., B.G., and R.C.H. analyzed RNA-seq, CUT&RUN, and ChIP-seq data; R.C.H. and G.A.B. acquired funding and organized the study; and K.Q. and G.A.B. wrote the manuscript with input from all authors.
Conflict-of-interest disclosure: G.A.B. received funding from Pfizer and Fulcrum Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Gerd A. Blobel, 316H Abramson Research Center, 3615 Civic Center Blvd, Philadelphia, PA 19104; e-mail: blobel@chop.edu; and Kunhua Qin, 315H Abramson Research Center, 3615 Civic Center Blvd, Philadelphia, PA 19104; e-mail: qink@chop.edu.
References
Author notes
∗K.Q., X.L., and P.H. contributed equally to this study.
All RNA-seq, CUT&RUN, and ChIP-seq data generated in this study have been deposited in the NIH Gene Expression Omnibus (GEO) under accession number GSE218233.
Data are available on request from the corresponding authors, Gerd A. Blobel (blobel@chop.edu) or Kunhua Qin (qink@chop.edu).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.



![cPRC1 and PRC2 cooperate to maintain the repression of IGF2BP1/3 and LIN28B in adult erythroid cells. (A) Volcano plots of DEGs identified from EZH2 depletion RNA-seq in HUDEP2 cells by DESeq2. Cutoff threshold: fold change > 2; FDR < 0.1; n = 2. (B-C) RT-qPCR analysis of the ratio of HBG:(HBB+HBG) mRNA levels (B), representative immunoblots (C) of EZH2, IGF2BP1, LIN28B, BMI1, H2AK119ub1, H3K27me3, and γ-globin protein in AsCas12a-expressing HUDEP2 cells that were transduced with lentivirus expressing indicated sgRNAs. Values are presented as mean ± SEM. RT-qPCR data were normalized to AHSP (n = 2). GAPDH was used as loading control in immunoblot experiment. (D) Schematic of EZH2 inhibition experiment in primary erythroid cells. (E) Representative immunoblots of IGF2BP1, IGF2BP3, H3K27me3, histone H3, and γ-globin in drug-treated primary adult erythroblasts. GAPDH were used as the loading control. (F) Relative IGF2BP1, IGF2BP3, and HBG mRNA level in primary adult erythroblasts (at day 10 or 12 of differentiation [equal to 3 or 5 days after drug treatment]) as detected by RT-qPCR. Values are presented as mean ± SEM. Data were normalized to AHSP; n = 4. (G) Summary of PRC1/2 perturbation experiments. In HUDEP2 cells, genetic perturbation of BMI1, CBX, or EZH2 led to upregulation of LIN28B and IGF2BP1, and thus, HbF activation. In primary adult erythroblasts, BMI1 depletion and EZH2 inhibition both resulted in derepression of IGF2BP1/3, and elevation of HbF. However, the molecular link between the 3 RNA binding proteins and HbF activation remains to be defined. Our observations support a major role of BCL11A in mediating the function of these RNA-binding proteins, especially in HUDEP2 cells, however we cannot rule out additional, yet to be identified targets of 3 RNA binding proteins involved in HbF activation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/22/10.1182_blood.2022019578/2/m_blood_bld-2022-019578-gr7.jpeg?Expires=1767715523&Signature=V0FZ1N17O3~tcYf1gHGZrCsez3GmuBirvDoyjjKXPRTfP2mgT5hGw4nnQfXqkJf-HsE9bBzopgfeUWAGHAGk1iN-U9T5obBt0raelc44LtLJbEvbzpT7WjCJ3RXz5S0PmmaumSTlAbvpbijETstWqQrjHkY2X5OMcmiXACfE-PKQUeG5SC1f7VLKRoGMhhHZ7ZGahmYi1aDYGg8gZO5WqFfu25vQuFbtF8uerLnmadKRmssQ6K4sAku4BXmxTBtkFayJkczHaEVNddy2NtxzX2KHKa3-KXCnA0tu1nveXw1UyhqiBG91RvTQ5qOp9un0f9VCJwVjApD5CMtLxf7CXQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal