TO THE EDITOR:
Acute lymphoblastic leukemia (ALL) is an aggressive hematological malignancy with especially dismal outcomes in adults and at the recurrence of the disease. Somatic mutations in the tumor suppressor gene TP53 occur in 6% to 19% of ALL cases at diagnosis and are enriched in low hypodiploid-near triploid ALL and at reoccurrence of the disease. TP53 mutations in ALL play an important role in the evolution of treatment-resistant clones, associated with early relapses and poor overall survival.1-6 Furthermore, TP53 mutations have been reported as an early leukemogenic event in patients with acute myeloid leukemia, representing acute myeloid leukemia-initiating mutations and mediators of resistant disease, even at subclonal levels.7,8 However, for ALL, very limited data are available. Here, we retrospectively analyzed the kinetics of TP53 mutations in the bone marrow and peripheral blood during follow-up in a group of 43 adult patients with TP53-mutated ALL and compared the TP53 mutation burden with minimal residual disease (MRD) levels assessed by EuroMRD-standardized real-time quantitative polymerase chain reaction (PCR) of clonal immune gene rearrangements (IG/TR MRD).9,10
Analyses were performed in a cohort of 43 adult patients with ALL with known TP53 mutations at diagnosis or at relapse (supplemental Data, available on the Blood website),6 with available longitudinal MRD measurements9,10 and leftover DNAs of the respective bone marrow and/or blood samples in the Hematology Lab Kiel. All patients received treatment according to GMALL protocols and provided informed consent for the use of left-over samples for scientific purposes (supplemental Data). For each patient, the leukemic TP53 mutation profile was determined in at least 1 tumor-positive diagnostic and/or tumor-positive follow-up (54 samples) sample using a UMI-based amplicon next-generation sequencing approach (supplemental Table 1). The same method was applied to the 40 follow-up samples obtained from 30 patients who achieved molecular remission. All the samples were obtained before allogeneic hematopoietic stem cell transplantation. In patients with TP53 mutation positivity in an MRD-negative follow-up sample and availability of archived left-over viable cells in the respective samples, distinct precursor and mature hematopoietic populations were isolated using the FACS Aria sorter and analyzed for the presence of TP53 mutations using mutation-specific digital droplet PCR.
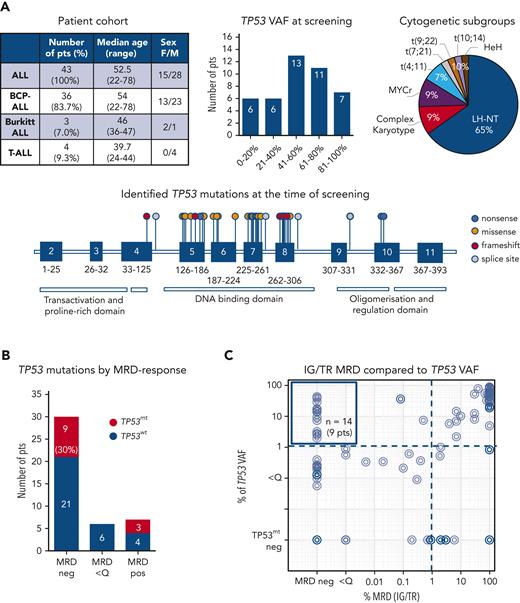
We analyzed the diagnostic and follow-up samples of 43 patients with ALL (36 B-cell-precursor ALL, 3 Burkitt leukemia, and 4 T-ALL) with known TP53 mutations at diagnosis or relapse. As expected, most of these patients (65%, 28/43) had ALL cases with a low hypodiploid-near triploid karyotype,5,6 all with the loss of 1 TP53 copy in a diploid or 2 copies in a tetraploid chromosome set. In total, 48 TP53 mutations (71% missense, 6% nonsense, 8% splice site, and 15% frameshift mutations) were identified as subclonal or clonal alterations, primarily in the DNA-binding domain (Figure 1A; supplemental Table 1). A total of 94 samples from these patients assessed for IG/TR MRD were retrospectively analyzed for TP53 mutation load at the time of diagnosis and during follow-up at the time of MRD-positivity (n = 54 samples) or in molecular remission (n = 40 samples), if applicable (Figure 1B). The IG/TR MRD persisted in 13 patients (30%) at quantifiable (MRD-positive, 7 patients, 16%) or nonquantifiable levels (MRD <Q, 6 patients, 14%). Thirty patients achieved molecular remission at some point (MRD-negative, 70%). Discrepancies in MRD kinetics between IG/TR MRD and TP53 mutant-allele burden were detected in 9 of the 30 MRD-negative patients (30%, Figure 1B). In 14 samples of these 9 patients, TP53 mutations were detected at levels between 2.0% and 42.9% (median 14.1%, upper left Figure 1C) despite IG/TR MRD-negativity. These data suggest that TP53 mutations occur as an early event in the preleukemic compartment, which can generate differentiated progeny and persist in nonleukemic cells during remission. Similarly, Salmoiraghi et al11 recently reported that TP53 alterations were correlated with very early rearrangements of the immunoglobulin receptor in ALL, also pointing to the occurrence of TP53 mutations as an early leukemogenic event and potentially their preleukemic origin. We selected 5 out of these 9 IG/TR MRD-negative and TP53-mutated patients (all B-ALL, Figure 2A) and analyzed the TP53 mutation load using mutation-specific digital droplet PCR (supplemental Table 2) in all available follow-up samples. The results were compared with the MRD kinetics over time (Figure 2A, top). In all 5 cases, TP53 mutations persisted in the follow-up samples despite MRD-negativity. To specify the origin of TP53 mutations, we sorted distinct mature and progenitor hematopoietic populations based on their characteristic immunophenotypes (supplemental Figure 1) in molecular remission at the indicated time point. Interestingly, early hematopoietic cells, including multipotent progenitors, common myeloid/lymphoid progenitors, myeloid progenitors, and B-cell precursors, as well as differentiated cells such as granulocytes and monocytes, contained TP53 mutations at levels ranging between 1.1% and 54.3% (Figure 2A, bottom), indicating the preleukemic origin of TP53 mutations (graphically shown in Figure 2B). Along this line, patients with preleukemic mutation patterns generally harbored a high TP53 mutation load (variant allele frequency >30%, supplemental Table 1) at the time of diagnosis, suggesting the presence of TP53 mutations in leukemia-originating cells and subsequently in all leukemic cells. TP53 mutations were generally very low/absent in mature T cells, except for patients 19 and 32 with late follow-up samples used in sorting experiments at 31 and 24 months after diagnosis, respectively, reflecting distinct developmental route and delayed recovery kinetics of T cells under/following ALL treatment.12,13
TP53 mutation profiles in a cohort of patients with ALL. (A) Description of a cohort of 43 patients with ALL with known somatic TP53 mutations at the time of diagnosis or relapse (top left), regarded as the screening timepoint. Distribution of TP53 mutant-allele burden (top middle) and cytogenetic subgroups (top right). Forty-two distinct TP53 mutations are shown on lollipop chart and color-coded as nonsense, missense, frameshift and splice site mutations (bottom). Some TP53 mutations occurred more than once, in total 48 TP53 mutations were detected. TP53 mutation load compared with IG/TR MRD in diagnostic and follow-up samples. (B) Patients are stratified based on the best molecular response (MRD-negative, MRD <Q, or MRD-positive) achieved before allogeneic stem cell transplantation. The proportion of patients with persisting TP53 mutations (red bars) in the respective follow-up samples is shown (left). IG/TR MRD (x-axis) is compared with the mutated TP53 variant allele frequency (VAF, y-axis) in all diagnostic and follow-up samples (94 samples). The dotted lines represent the sensitivity threshold for TP53 mutations (right). TP53mt, mutated TP53; TP53wt, wild-type TP53.
TP53 mutation profiles in a cohort of patients with ALL. (A) Description of a cohort of 43 patients with ALL with known somatic TP53 mutations at the time of diagnosis or relapse (top left), regarded as the screening timepoint. Distribution of TP53 mutant-allele burden (top middle) and cytogenetic subgroups (top right). Forty-two distinct TP53 mutations are shown on lollipop chart and color-coded as nonsense, missense, frameshift and splice site mutations (bottom). Some TP53 mutations occurred more than once, in total 48 TP53 mutations were detected. TP53 mutation load compared with IG/TR MRD in diagnostic and follow-up samples. (B) Patients are stratified based on the best molecular response (MRD-negative, MRD <Q, or MRD-positive) achieved before allogeneic stem cell transplantation. The proportion of patients with persisting TP53 mutations (red bars) in the respective follow-up samples is shown (left). IG/TR MRD (x-axis) is compared with the mutated TP53 variant allele frequency (VAF, y-axis) in all diagnostic and follow-up samples (94 samples). The dotted lines represent the sensitivity threshold for TP53 mutations (right). TP53mt, mutated TP53; TP53wt, wild-type TP53.
Discrepant dynamics of TP53 mutations during follow-up compared with IG/TR MRD. (A) Longitudinal DNA samples of 5 patients with ALL selected based on TP53 mutation positivity in an MRD-negative follow-up sample and availability of archived left-over viable cells were analyzed for the presence of specific TP53 mutations using mutation-specific digital droplet PCR. In all 5 cases, the respective TP53 mutations (red lines) were sustained despite MRD-negativity (blue lines) during remission (top panel). Pat32 relapsed with a clone harboring an entirely unrelated IG/TR (dotted blue line) rearrangement and gained a second TP53 mutation (dotted red line) while sustaining the diagnostic TP53 mutation. In patients 14, 19, and 29, false MRD negativity owing to potential clonal evolution of the MRD marker was excluded by IGH amplicon next-generation sequencing analysis that neither showed the diagnostic IGH clonotype nor a related IGH or any other unrelated highly abundant IGH clonotype in the remission sample (data not shown). Distinct hematopoietic cell populations (early hematopoietic cells; hematopoietic stem cells/multipotent progenitors [HSCs/MPPs]), common myeloid/lymphoid progenitors (L/M prog), myeloid progenitors (M prog), and B-cell precursors (B-cell prec), as well as the mature immune cell populations such as T cells, monocytes, and granulocytes of an MRD-negative follow-up sample, were isolated using the FACS Aria sorter. Sorted subpopulations were analyzed for the presence of specific TP53 mutations by mutation-specific digital droplet PCR. Values in percentages indicate the TP53 mutant-allele burden in the isolated populations. (B) TP53 mutations in ALL originate in a preleukemic compartment that can self-renew and cause clonal expansions in hematopoietic compartments; thus, they persist in tumor-free cells during remission and increase at the (re)occurrence of the disease. IGH, immunoglobulin heavy locus; TP53mt, mutated TP53; TP53wt, wild-type TP53. Panel B was created with BioRender.
Discrepant dynamics of TP53 mutations during follow-up compared with IG/TR MRD. (A) Longitudinal DNA samples of 5 patients with ALL selected based on TP53 mutation positivity in an MRD-negative follow-up sample and availability of archived left-over viable cells were analyzed for the presence of specific TP53 mutations using mutation-specific digital droplet PCR. In all 5 cases, the respective TP53 mutations (red lines) were sustained despite MRD-negativity (blue lines) during remission (top panel). Pat32 relapsed with a clone harboring an entirely unrelated IG/TR (dotted blue line) rearrangement and gained a second TP53 mutation (dotted red line) while sustaining the diagnostic TP53 mutation. In patients 14, 19, and 29, false MRD negativity owing to potential clonal evolution of the MRD marker was excluded by IGH amplicon next-generation sequencing analysis that neither showed the diagnostic IGH clonotype nor a related IGH or any other unrelated highly abundant IGH clonotype in the remission sample (data not shown). Distinct hematopoietic cell populations (early hematopoietic cells; hematopoietic stem cells/multipotent progenitors [HSCs/MPPs]), common myeloid/lymphoid progenitors (L/M prog), myeloid progenitors (M prog), and B-cell precursors (B-cell prec), as well as the mature immune cell populations such as T cells, monocytes, and granulocytes of an MRD-negative follow-up sample, were isolated using the FACS Aria sorter. Sorted subpopulations were analyzed for the presence of specific TP53 mutations by mutation-specific digital droplet PCR. Values in percentages indicate the TP53 mutant-allele burden in the isolated populations. (B) TP53 mutations in ALL originate in a preleukemic compartment that can self-renew and cause clonal expansions in hematopoietic compartments; thus, they persist in tumor-free cells during remission and increase at the (re)occurrence of the disease. IGH, immunoglobulin heavy locus; TP53mt, mutated TP53; TP53wt, wild-type TP53. Panel B was created with BioRender.
Half of the relapses in ALL evolve from an ancestral subclone present at diagnosis.14,15TP53 mutations are often enriched in relapse and contribute to resistance development in ALL.15,16 To that end, we observed that 1 patient with ALL (Figure 2A, Pat32), who relapsed after 2 years with a leukemic clone showing a completely different karyotype and IG/TR rearrangement fully unrelated to the initial diagnostic clone, gained a second TP53 mutation while sustaining the diagnostic one. This suggests underlying clonal heterogeneity and involvement of preleukemic TP53 mutations in ALL progression. To address the role of preleukemic TP53 and tumor-associated mutations in ALL progression, we compared the mutation profiles of 141 genes involved in leukemia pathogenesis in available paired samples of 9 patients obtained at diagnosis and/or disease progression (supplemental Figure 2; supplemental Data). Three patients had characteristic preleukemic TP53 mutation patterns and available remission samples. The initial TP53 mutations persisted in all patients with disease recurrence. Two patients gained (additional) TP53 mutations (Pat15; Pat32) at the time of relapse. The mutational profiles differed between the initial and paired relapse samples; however, no specific, reccurrent mutation patterns were identified in this relatively small patient group. In addition to TP53, candidate preleukemic genes (supplemental Figure 2) were identified as they persisted during remission in patients with ALL, requiring further studies to confirm their preleukemic origin.
In conclusion, we showed that a considerable number of adult patients with ALL harbor somatic TP53 mutations in a preleukemic compartment, are able to self-renew and clonally expand in different hematopoietic cells, and persist during remission. Therefore, the consideration of TP53 mutations as MRD markers requires careful interpretation. The primary selection criteria in this retrospective cohort were the presence of TP53 mutations in ALL and the availability of longitudinal MRD kinetics. Thus, valid statements regarding the frequency or clinical relevance of the preleukemic TP53 mutation pattern in ALL are not possible. Understanding the role of preleukemic TP53 mutations in ALL evolution and disease progression has broad clinical implications for leukemia treatment, MRD monitoring, treatment selection, and the prediction of treatment outcomes. Further studies in more homogeneous and prospective settings are needed to elucidate the clinical implications of preleukemic TP53 mutations in ALL.
Acknowledgments
The authors thank Henrik Knecht, Dietrich Hermann, and Martin Schwarz for their expert help. They also thank Petra Chall and Sandra Ussat for providing technical assistance.
This work was supported in part by the Deutsche Forschungsgemeinschaft (German Research Foundation) (project number 444949889) (KFO 5010/1 Clinical Research Unit “CATCH-ALL” to G.C., L.B., C.D.B., and M.B), and through the “Clinician Scientist Program in Evolutionary Medicine” (project number 413490537 to G.C. and A-S.S.).
Authorship
Contribution: G.C., C.J-K., J.B., K.P., and W.W. performed the experiments; G.C., J.B., W.W., H.T., and L.B. analyzed the results; B.K., H-H.O., M. Kotrova, F.D., M. Kelm, A-S.S., and N.D. helped with the experiments and data analysis; A.S., C.B., M.M., and C.H. provided the relevant patient information for this study; G.C., M.B., and C.H. designed the research; G.C. drafted the first version of the manuscript; and all authors discussed the results and contributed to the final manuscript.
Conflict-of-interest disclosure: M.B. is contracted to carry out research for Affimed, Amgen, Regeneron, and is a member of the advisory boards of Amgen and Incyte and the speaker bureaus of Amgen, Janssen, Pfizer, and Roche. C.D.B. is contracted to carry out research for Novartis and is a member of the advisory board of Amgen. C.H. declares partial ownership of the Munich Leukemia Laboratory. A.S., C.B., and M.M. are employed by Munich Leukemia Laboratory. P.M.A. is a consultant for CRISPR Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Monika Brüggemann, Medical Department II, Hematology and Oncology, Hematology Lab Kiel, University Hospital Schleswig-Holstein, Langer Segen 8-10, 24105 Kiel, Germany; e-mail: m.brueggemann@med2.uni-kiel.de.
References
Author notes
Mutation profiles of each patient may be found in a data supplement available with the online version of this article.
Data are available on reasonable request from the corresponding author, Monika Brüggemann (m.brueggemann@med2.uni-kiel.de)
The online version of this article contains a data supplement.

![Discrepant dynamics of TP53 mutations during follow-up compared with IG/TR MRD. (A) Longitudinal DNA samples of 5 patients with ALL selected based on TP53 mutation positivity in an MRD-negative follow-up sample and availability of archived left-over viable cells were analyzed for the presence of specific TP53 mutations using mutation-specific digital droplet PCR. In all 5 cases, the respective TP53 mutations (red lines) were sustained despite MRD-negativity (blue lines) during remission (top panel). Pat32 relapsed with a clone harboring an entirely unrelated IG/TR (dotted blue line) rearrangement and gained a second TP53 mutation (dotted red line) while sustaining the diagnostic TP53 mutation. In patients 14, 19, and 29, false MRD negativity owing to potential clonal evolution of the MRD marker was excluded by IGH amplicon next-generation sequencing analysis that neither showed the diagnostic IGH clonotype nor a related IGH or any other unrelated highly abundant IGH clonotype in the remission sample (data not shown). Distinct hematopoietic cell populations (early hematopoietic cells; hematopoietic stem cells/multipotent progenitors [HSCs/MPPs]), common myeloid/lymphoid progenitors (L/M prog), myeloid progenitors (M prog), and B-cell precursors (B-cell prec), as well as the mature immune cell populations such as T cells, monocytes, and granulocytes of an MRD-negative follow-up sample, were isolated using the FACS Aria sorter. Sorted subpopulations were analyzed for the presence of specific TP53 mutations by mutation-specific digital droplet PCR. Values in percentages indicate the TP53 mutant-allele burden in the isolated populations. (B) TP53 mutations in ALL originate in a preleukemic compartment that can self-renew and cause clonal expansions in hematopoietic compartments; thus, they persist in tumor-free cells during remission and increase at the (re)occurrence of the disease. IGH, immunoglobulin heavy locus; TP53mt, mutated TP53; TP53wt, wild-type TP53. Panel B was created with BioRender.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/141/13/10.1182_blood.2022017249/7/m_blood_bld-2022-017249-gr2.jpeg?Expires=1770140872&Signature=BlZU~a4~8LybYriJcOeJnOW3BvNMeTnpdmhzRhazIn63g0rqnsl-HECzZBN~1Kzze7BWhrnaW8u1PWzea1SRFu09GNCWCVTyJ7pBjmYZZhtma-2xV~2denDrztRcoCfRSPFZzvsBbTQnTghMewVQxRh-7UYHmhhUPLQMN6BYy2-uO8zdpzpsraS-SVI96f~KlBV4m9texHE5ru-2EZqAfei2X3YBOekLjBLHmEWqPNer8UqVUbBsOoRjTp7tMJk1luvDiA1imw~PUKxTq5frd7ArZRzTPGEOM09TMRWSVeO4SBfOfLivc3CAZXiAyzRDxoIrU9NZ8a2EkjWdjvODuA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal