Key Points
Noncanonical interactions of β-catenin with FOXO3 transcription factor promote LIC activity in early T-cell precursor ALL.
β-Catenin– and FOXO3-dependent gene signature identifies LIC-enriched CD82+CD117+ cell subsets, highly selected in MRD.
Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is a T-cell malignancy characterized by cell subsets and enriched with leukemia-initiating cells (LICs). β-Catenin modulates LIC activity in T-ALL. However, its role in maintaining established leukemia stem cells remains largely unknown. To identify functionally relevant protein interactions of β-catenin in T-ALL, we performed coimmunoprecipitation followed by liquid chromatography–mass spectrometry. Here, we report that a noncanonical functional interaction of β-catenin with the Forkhead box O3 (FOXO3) transcription factor positively regulates LIC-related genes, including the cyclin-dependent kinase 4, which is a crucial modulator of cell cycle and tumor maintenance. We also confirm the relevance of these findings using stably integrated fluorescent reporters of β-catenin and FOXO3 activity in patient-derived xenografts, which identify minor subpopulations with enriched LIC activity. In addition, gene expression data at the single-cell level of leukemic cells of primary patients at the time of diagnosis and minimal residual disease (MRD) up to 30 days after the standard treatments reveal that the expression of β-catenin– and FOXO3-dependent genes is present in the CD82+CD117+ cell fraction, which is substantially enriched with LICs in MRD as well as in early T-cell precursor ALL. These findings highlight key functional roles for β-catenin and FOXO3 and suggest novel therapeutic strategies to eradicate aggressive cell subsets in T-ALL.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is a hematological malignancy that affects both children and adults. Although, with current chemotherapy regimens, cure is achieved in ∼80% of pediatric patients, adults fare more poorly, with only 40% 5-year overall survival.1,2 Restricted cellular subsets with asymmetrically enriched leukemia-initiating cell (LIC) activity have been reported in human and mouse models of T-cell leukemia,3-12 suggesting that more efficient targeting of LICs could lead to dramatic improvements in patient outcomes. Recent studies have demonstrated that LICs are functionally distinct from bulk cells and modulated by distinct molecular signaling pathways and epigenetic mechanisms.13 High expression levels of β-catenin have been observed in LICs of both human leukemias and mouse models of T-ALL, highlighting the crucial role of β-catenin signaling in T-ALL maintenance and progression.12,14-16
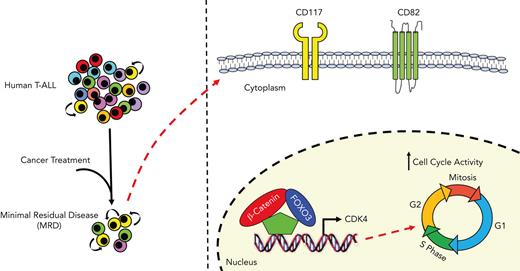
In the canonical Wnt/β-catenin pathway, Wnt ligands bind to the N-terminal extracellular cysteine-rich domain of Fzd receptors that interact with other coreceptors, including lipoprotein receptor related protein 5/6, tyrosine-protein kinase transmembrane receptor related 2, and receptor tyrosine kinase.17 The binding of Wnt ligands leads to the stabilization of β-catenin in the cytoplasm and its subsequent translocation into the nucleus to form an active transcriptional complex with the transcription factors of lymphoid enhancer factor (LEF) and T-cell factor (TCF) families.18,19 Wnt target genes such as c-Myc20 are master regulators of cancer progression and are directly involved in cell growth and tumor-initiating activity.21 Of interest, it has been reported that in the PTEN-null mouse model of T-cell leukemia, the LIC-enriched c-Kitmid CD3+ Lin− cell subsets express high levels of unphosphorylated β-catenin and c-Myc.12 Nonetheless, the mechanisms that regulate β-catenin activity in LICs remain largely unknown. Here, we report a novel noncanonical functional interaction between β-catenin and Forkhead box O3 (FOXO3), which promotes cyclin-dependent kinase 4 (CDK4) expression and LIC activity in cell subsets, mostly enriched in minimal residual disease (MRD), revealing novel potential therapeutic targets for T-ALL treatment.
Materials and methods
Cell culture
Primary mouse leukemia cells from bone marrow or spleen of moribund mice were cultured in RPMI 1640 medium supplemented with 20% fetal bovine serum (FBS), 1 mM sodium pyruvate, 2 mM L-glutamine, antibiotics (Invitrogen), and supplemental cytokines interleukin 2 (IL-2) and IL-7 (10 ng/mL each, Peprotech). Mouse primary leukemias were typically cultured in vitro for 4 to 7 days before viral transduction. Xenograft-expanded primary human T-ALLs were cultured on MS5-DL1 feeders in Iscove modified Dulbecco medium (Gibco) with 10 ng/mL IL-2, 10 ng/mL IL-7, and 0.75 μM SR1 (StemCell Technologies) as described previously.22 All established mouse and human T-ALL cell lines were cultured in RPMI 1640 medium supplemented with 10% FBS, 1 mM sodium pyruvate, 2 mM L-glutamine, and antibiotics.
Drugs
Small-molecule phosphatidylinositol 3-kinase (PI3K)/AKT pathway inhibitors, PI-103 (Calbiochem, Sigma) and LY294002 (Calbiochem, Sigma) were resuspended in dimethyl sulfoxide to a stock concentration of 10 mM and 50 mM, respectively, and diluted serially in Hanks balanced salt solution before addition to culture media.
Cell transfection and luciferase reporter assay
Human T-ALL cell lines were cultured in RPMI 1640 medium supplemented with 10% FBS, 1 mM sodium pyruvate, and 2 mM L-glutamine, without antibiotics, and transfected at 90% confluence with the plasmids indicated by electroporation using an Amaxa Nucleofector Device (Lonza, Germany), following the manufacturer’s instructions. Luciferase assays were performed after 48 hours of transfection using the Dual-Luciferase Reporter Assay System (Promega) and a luminometer (Glomax 20/20; Promega), according to the manufacturer's instructions. The level of firefly-luciferase activity was normalized to the activity of Renilla.
Viral transduction
Lentiviral particles were produced by transient cotransfection of 293T cells with packaging/envelope vectors and concentrated by ultracentrifugation as described.23 Viral transduction was performed by spinfection in the presence of Polybrene, as reported.3 Virally transduced cells were fluorescence-activated cell sorter (FACS) sorted as applicable.
Flow cytometry
We stained human leukemia cells with fluorochrome or biotin-conjugated antibodies against CD45, CD3, CD4, CD8, CD99, CD7, CD117, and CD82 (eBioscience, Thermo Fisher Scientific). We used anti–human CD271 (hCD271), anti-hCD8, and anti–mouse CD8 (eBioscience; Thermo Fisher Scientific) to detect the lentiviral truncated nerve growth factor receptor (NGFR), mouse CD8, and hCD8 markers. We performed intracellular staining with phycoerythrin-conjugated antibody against CDK4 (1:5 dilution; catalog #12790, Cell Signaling) after paraformaldehyde fixation and permeabilization with 90% ice-cold methanol as specified by the manufacturer. We measured cell proliferation by 5-bromo-2′-deoxyuridine incorporation, according to the manufacturer’s instructions (BrdU kit; BD Biosciences). We measured cell viability by propidium iodide exclusion. We performed FACS analysis and sorting on the FACS Calibur, Canto2, and Aria2 cytometers (BD Biosciences) and the MoFlo Astrios cell sorter (Beckman Coulter). We analyzed flow cytometry data using FlowJo software (BD Biosciences).
Coimmunoprecipitation
Cells were lysed with NETN lysis buffer (Tris-HCl, pH 7.5, 50 mM; NaCl, 150 mM; NP-40, 10 μL; EDTA, pH 8, 0.5 mM; proteinase inhibitor [phenylmethylsulfonyl fluoride], 1 mM; sodium orthovanadate, 100 μM). The lysate was spun for 15 minutes, 14 000 revolutions per minute at 4°C and subsequently incubated overnight at 4°C with 125 ng of indicated antibody against β-catenin (catalog #9582, Cell Signaling), nonphospho (active) β-catenin (catalog #8814, Cell Signaling), or FOXO3 (catalog #12829, Cell Signaling) together with Dynabeads protein G (catalog #10003D, Thermo Fisher Scientific), Pierce Anti-HA Magnetic Beads (catalog 88836, Thermo Fisher Scientific) or Pierce Anti-c-Myc Magnetic Beads (catalog #88842, Thermo Fisher Scientific), previously blocked with 0.5 mg/mL bovine serum albumin overnight at 4°C. The immunocomplexes were washed with NETN buffer and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
Statistics
GraphPad PRISM 8.4.3 software was used for the analyses of quantitative data, including the 2-way analysis of variance with Dunnett test and the Fisher exact test, corrected for multiple comparisons by false discovery rate using a 2-stage linear step-up procedure of Benjamini et al.24 LIC frequencies were calculated as previously described.25
Results
β-Catenin functionally interacts with FOXO3 transcription factor
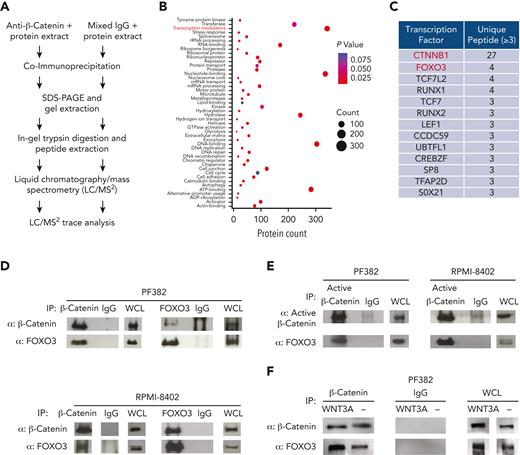
To address the role of β-catenin in T-ALL, we first assessed the expression of total and nonphospho (active) β-catenin proteins in human T-ALL cell lines by western blot. We noted that β-catenin protein levels varied among cell lines and that levels of active β-catenin were higher in PF382, RPMI-8402, HPB-ALL, JURKAT, and CCRF-CEM cell lines (supplemental Figure 1, available on the Blood website) and the highest in the nuclear fraction of PF382 cells (supplemental Figure 2). Using publicly available RNA sequencing (RNA-seq) data,26 we also performed the gene set variation analysis to assess variations of β-catenin pathway activity over the different T-ALL cell lines in an unsupervised manner. Consistently with the levels of active β-catenin protein, we observed that PF382, RPMI-8402, HPB-ALL, JURKAT, and CCRF-CEM cell lines were enriched in genes of Wnt/β-catenin signaling pathway (supplemental Figure 3). Interestingly, we did not observe a full correspondence between the protein expression of active β-catenin and the enrichment scores for Wnt/β-catenin signaling. This enforces the idea that different levels of regulation between transcripts and protein products,27 as well as potential mechanisms of β-catenin–independent regulation of Wnt target genes28 exist. Furthermore, to identify functionally relevant protein interactions of β-catenin in T-ALL, we performed coimmunoprecipitation followed by liquid chromatography–mass spectrometry using protein extracts of PF382 cell line (Figure 1A). To focus on highly confident interactions, the enrichment was independently repeated 3 times, and the limit of detection was fixed at a minimum of 2 unique peptides per protein. Of interest, we identified a total of 1495 proteins, including canonical β-catenin–interacting proteins such as TCF7 and LEF1. Gene ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses assigned functional annotations and likely molecular functions, highlighting the transcription modulators as the main category of enriched β-catenin–interacting proteins (Figure 1B-C). Within the subgroup of the transcription factors, we identified TCF7 and LEF1 as canonical β-catenin interactors. Of interest, we also found noncanonical interactions between β-catenin and transcription factors, such as FOXO3 and Runt-related transcription factors. To explore this further, we focused on FOXO3 as a putative novel interactor of β-catenin in T-ALL (Figure 1C). Of interest, functional interactions between β-catenin and FOXO have previously been reported in Caenorhabditis elegans under conditions of oxidative stress.29 Therefore, to validate our observations, we performed coimmunoprecipitations between FOXO3, β-catenin, and nonphospho (active) β-catenin in PF382, RPMI-8402, HPB-ALL, JURKAT, and CCRF-CEM cell lines (Figure 1D-E; supplemental Figures 4 and 30). As expected, FOXO3 effectively bound total and active β-catenin in all these lines. Of interest, the FOXO3 protein increased its binding to β-catenin in PF382, RPMI-8402, and NIH/3T3 cell lines following the addition of Wnt3a ligand to in vitro culture media (Figure 1F; supplemental Figure 4). Furthermore, deletion mutants into the transactivation domain of β-catenin or FOXO3 proteins negatively affect their binding (supplemental Figures 5-8 and 30), supporting the idea of noncanonical protein interactions between β-catenin and FOXO3.
β-Catenin binds FOXO3 transcription factor in human T-ALL cells. (A) Schematic representation of coimmunoprecipitation followed by liquid chromatography–mass spectrometry for the identification of interacting proteins of β-catenin. (B) Distribution of transcription regulators, subcellular component, molecular function, and biological processes of identified β-catenin interactors by liquid chromatography–mass spectrometry. Benjamini adjusted P values and protein counts were determined by the database for Annotation, Visualization and Integrated Discovery resource. Functional annotations were collected from DAVID’s “UP_Keywords,” “UP_SEQ_Feature,” and “COG_Ontology” categories. (C) List of the top 12 β-catenin-interacting transcription factors with >3 unique peptides identified by mass spectrometry and determined as transcription factors by Proteome Discoverer software. Coimmunoprecipitation between β-catenin and FOXO3 (D) and nonphospho (active) β-catenin and FOXO3 (E) in PF382 and RPMI-8402 T-ALL cell lines, followed by immunoblot analysis using the indicated antibodies. (F) Coimmunoprecipitation between β-catenin and FOXO3 in PF382 cell line after in vitro growth into L-Wnt3A–conditioned medium or control-conditioned medium for 48 hours. Protein lysate were immunoprecipitated with indicated antibodies followed by immunoblot analysis. ADP, adenosine diphosphate; IgG, immunoglobulin G; rRNA, ribosomal RNA; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; WCL, whole cell lysate.
β-Catenin binds FOXO3 transcription factor in human T-ALL cells. (A) Schematic representation of coimmunoprecipitation followed by liquid chromatography–mass spectrometry for the identification of interacting proteins of β-catenin. (B) Distribution of transcription regulators, subcellular component, molecular function, and biological processes of identified β-catenin interactors by liquid chromatography–mass spectrometry. Benjamini adjusted P values and protein counts were determined by the database for Annotation, Visualization and Integrated Discovery resource. Functional annotations were collected from DAVID’s “UP_Keywords,” “UP_SEQ_Feature,” and “COG_Ontology” categories. (C) List of the top 12 β-catenin-interacting transcription factors with >3 unique peptides identified by mass spectrometry and determined as transcription factors by Proteome Discoverer software. Coimmunoprecipitation between β-catenin and FOXO3 (D) and nonphospho (active) β-catenin and FOXO3 (E) in PF382 and RPMI-8402 T-ALL cell lines, followed by immunoblot analysis using the indicated antibodies. (F) Coimmunoprecipitation between β-catenin and FOXO3 in PF382 cell line after in vitro growth into L-Wnt3A–conditioned medium or control-conditioned medium for 48 hours. Protein lysate were immunoprecipitated with indicated antibodies followed by immunoblot analysis. ADP, adenosine diphosphate; IgG, immunoglobulin G; rRNA, ribosomal RNA; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; WCL, whole cell lysate.
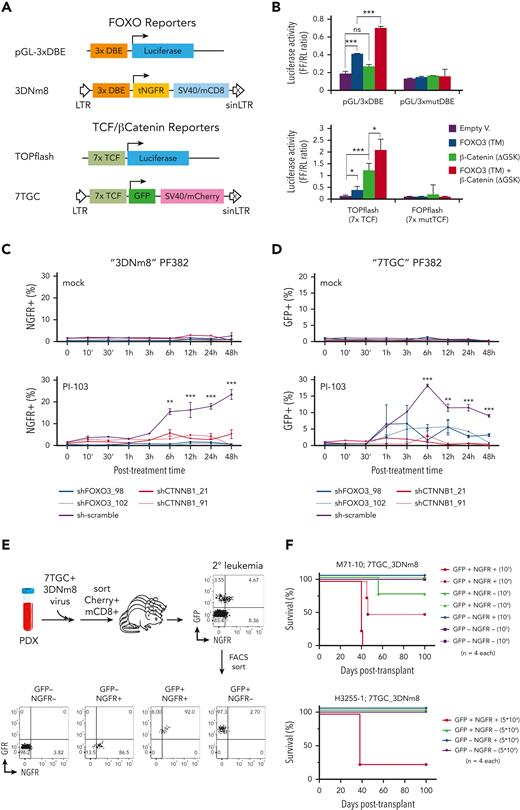
FOXO proteins promote the transcription of its target genes by binding as monomer to the DAF-16–binding element and the insulin-responsive element with an essential T(A/G)TTT motif.30,31 To determine whether β-catenin was able to modulate the transcriptional activity of FOXO3 in T-ALL, we transiently cotransfected the luciferase FOXO reporter construct, pGL-3xDAF-16–binding element, with an active FOXO3 triple mutant (TM)32 alone or in combination with a stable isoform of β-catenin (ΔGSK)33 into PF382 cells as well as MOLT-4, DND-41, and CUTLL134 T-ALL cell lines, which show a low expression level of active β-catenin (supplemental Figures 1 and 2). Here, the transcriptional activity of FOXO reporter was induced by the constitutive expression of active FOXO3 and unexpectedly increased after coexpression of β-catenin (Figure 2A-B; supplemental Figure 9). FOXO3 activity was also tested on a TCF/β-catenin signaling reporter consisting of 7 TCF/LEF consensus binding sites upstream of a minimal promoter driving expression of luciferase.33 In PF382 and PEER cells, the expression of both β-catenin (ΔGSK) and FOXO3 (TM) synergistically promoted the TCF/β-catenin reporter activity (Figure 2B; supplemental Figure 8). Moreover, deletion of mutant isoforms in the transactivation domain of both β-catenin and FOXO3 proteins negatively affects their synergistic activity (supplemental Figures 5 and 6), suggesting that β-catenin and FOXO3 can cooperate to modulate the transcription through noncanonical interactions.
β-Catenin and FOXO together modulate the transcriptional and LIC activity in human T-ALL. (A) Schematic map of the FOXO and TCF/β-catenin reporters. The FOXO lentiviral reporter (3DNm8) is composed of 3 DAF-16–binding elements (DBEs) upstream of a minimal promoter encoding a truncated NGFR, followed by a separate SV40 promoter expressing a truncated mouse CD8 marker. The fluorescent TCF/β-catenin lentiviral reporter (7TGC) is composed of 7 Tcf/Lef-binding sites upstream of a minimal promoter and green fluorescent protein (GFP), followed by a separate SV40-mCherry cassette. (B) The TCF/β-catenin and FOXO transcriptional activities were evaluated by the pGL-3xDBE and TOPFlash luciferase reporters, respectively. A construct containing 3 mutated DBE and the FOPFlash reporter with mutant TCF/LEF-binding sites were also included as control. PF382 cell lines were also cotransfected with an active FOXO3-TM alone or in combination with a stable isoform of β-catenin (ΔGSK) as indicated. Data were normalized to the Renilla luciferase reporter vector and given as mean and standard deviation. The graphs report the result of 3 independent experiments performed in triplicate. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 (Student t test). (C-D) Flow cytometric analysis of NGFR+ and GFP+ cell abundance in PF382 cell line transduced respectively with FOXO (3DNm8) or TCF/β-catenin (7TGC) reporters. Leukemia cells were doubly transduced with 3DNm8 and short hairpin RNA/GFP (C) or 7TGC and short hairpin RNA/NGFR (D) lentiviral constructs, FACS sorted and cultured in vitro with PI-103, a PI3K inhibitor, or mock control. NGFR+ or GFP+ alive cells were measured at the indicated time points by flow cytometry for DRAQ7 exclusion. The graphs report the result of 3 independent experiments performed in triplicate. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 (2-way analysis of variance with Dunnett test, comparing the sh-scramble control mean with the other values). (E) Schematic diagram of experimental approach. Leukemia cells of patient-derived xenografts were transduced with 7TGC and 3DNm8 lentiviruses, FACS sorted for Cherry and mouse CD8 expression and then transplanted into recipient mice, all of which subsequently developed leukemia. These secondary 7TGC_3DNm8-transduced leukemias were then analyzed by flow cytometry for GFP and NGFR expression. GFP+NGFR+, GFP+NGFR−, GFP−NGFR+, and GFP−NGFR− subsets were then FACS sorted and transplanted at limiting dilution into new recipients. (F) Survival of recipient NSG mice after transplantation with FACS-sorted β-catenin active (GFP+) and/or FOXO active (NGFR+) subsets from 7TGC_3DNm8-transduced xenograft-expanded human leukemias. The cell doses injected in each of 4 recipient animals are indicated in parentheses. Two separate experiments are depicted using independent patient-derived xenograft clones as indicated. The calculated LIC frequency in GFP+NGFR+ is 1 in 14 241 (95% CI, 1 in 3698-54 839), in GFP+NGFR− is 1 in 387 857 (95% CI, 1 in 54 949-2 737 697), and in mock-treated cells is >1 in 146 876. DBEmut, mutated DBE; Empty V, empty vector; NS, not significant; tNGFR, truncated NGFR.
β-Catenin and FOXO together modulate the transcriptional and LIC activity in human T-ALL. (A) Schematic map of the FOXO and TCF/β-catenin reporters. The FOXO lentiviral reporter (3DNm8) is composed of 3 DAF-16–binding elements (DBEs) upstream of a minimal promoter encoding a truncated NGFR, followed by a separate SV40 promoter expressing a truncated mouse CD8 marker. The fluorescent TCF/β-catenin lentiviral reporter (7TGC) is composed of 7 Tcf/Lef-binding sites upstream of a minimal promoter and green fluorescent protein (GFP), followed by a separate SV40-mCherry cassette. (B) The TCF/β-catenin and FOXO transcriptional activities were evaluated by the pGL-3xDBE and TOPFlash luciferase reporters, respectively. A construct containing 3 mutated DBE and the FOPFlash reporter with mutant TCF/LEF-binding sites were also included as control. PF382 cell lines were also cotransfected with an active FOXO3-TM alone or in combination with a stable isoform of β-catenin (ΔGSK) as indicated. Data were normalized to the Renilla luciferase reporter vector and given as mean and standard deviation. The graphs report the result of 3 independent experiments performed in triplicate. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 (Student t test). (C-D) Flow cytometric analysis of NGFR+ and GFP+ cell abundance in PF382 cell line transduced respectively with FOXO (3DNm8) or TCF/β-catenin (7TGC) reporters. Leukemia cells were doubly transduced with 3DNm8 and short hairpin RNA/GFP (C) or 7TGC and short hairpin RNA/NGFR (D) lentiviral constructs, FACS sorted and cultured in vitro with PI-103, a PI3K inhibitor, or mock control. NGFR+ or GFP+ alive cells were measured at the indicated time points by flow cytometry for DRAQ7 exclusion. The graphs report the result of 3 independent experiments performed in triplicate. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 (2-way analysis of variance with Dunnett test, comparing the sh-scramble control mean with the other values). (E) Schematic diagram of experimental approach. Leukemia cells of patient-derived xenografts were transduced with 7TGC and 3DNm8 lentiviruses, FACS sorted for Cherry and mouse CD8 expression and then transplanted into recipient mice, all of which subsequently developed leukemia. These secondary 7TGC_3DNm8-transduced leukemias were then analyzed by flow cytometry for GFP and NGFR expression. GFP+NGFR+, GFP+NGFR−, GFP−NGFR+, and GFP−NGFR− subsets were then FACS sorted and transplanted at limiting dilution into new recipients. (F) Survival of recipient NSG mice after transplantation with FACS-sorted β-catenin active (GFP+) and/or FOXO active (NGFR+) subsets from 7TGC_3DNm8-transduced xenograft-expanded human leukemias. The cell doses injected in each of 4 recipient animals are indicated in parentheses. Two separate experiments are depicted using independent patient-derived xenograft clones as indicated. The calculated LIC frequency in GFP+NGFR+ is 1 in 14 241 (95% CI, 1 in 3698-54 839), in GFP+NGFR− is 1 in 387 857 (95% CI, 1 in 54 949-2 737 697), and in mock-treated cells is >1 in 146 876. DBEmut, mutated DBE; Empty V, empty vector; NS, not significant; tNGFR, truncated NGFR.
FOXO factors have been proposed to be sensors/modulators of stress conditions rather than essential mediators of normal cell physiology.35 Of interest, FOXO transcription factors are negatively modulated by growth factors, including insulin-like growth factor 1 and insulin, through PI3K and protein kinase B (c-Akt).36 In fact, inhibition of PI3K/Akt pathway causes FOXO proteins to persist in the nucleus and subsequently promote the expression of its target genes.37 To assess whether β-catenin was able to modulate the transcriptional activity of FOXO3 induced by the inhibition of PI3K/Akt pathway, PF382, RPMI-8402, and HPB-ALL cell lines were transduced with 3DNm8 or 7TGC lentiviruses, reporters of FOXO and TCF/β-catenin signaling activity, respectively, (Figure 2A) as well as with shFOXO3 or shCTNNB1 constructs for knocking down FOXO3 or β-catenin, respectively, or scramble control (supplemental Figure 10). Interestingly, FOXO-dependent reporter activity of 3DNm8-transduced cells increased after exposure to PI3K inhibitors, PI-103 and LY294002, and significantly decreased after short hairpin RNA–mediated knockdown or CRISPR/Cas9-mediated deletion of FOXO3 or β-catenin (Figure 2C; supplemental Figures 11 and 12). In agreement with these results, the inhibition of the PI3K/Akt signaling pathway increased the β-catenin–dependent reporter activity (Figure 2D; supplemental Figures 11 and 12).
β-Catenin signaling plays relevant roles in the maintenance of normal hematopoietic stem cells and LICs.14,38,39 To explore the functional cross talk of FOXO3 and β-catenin signaling in established human T-ALLs, we selected 2 patient-derived xenografts (PDXs) with the highest transcriptional levels of both CTNNB1 and FOXO3 genes (supplemental Figure 13) and introduced the FOXO (3DNm8) and TCF/β-catenin (7TGC) fluorescent reporter constructs by lentiviral transduction. After FACS sorting purification, 3DNm8 and 7TGC doubly transduced cells were injected into secondary recipients, all of which subsequently developed clinically morbid disease. Interestingly, only a small fraction of the totality of leukemia cells exhibited green fluorescent protein (GFP) fluorescence indicative of endogenous β-catenin activity and NGFR expression for FOXO activity (Figure 2E). To assess the relative leukemogenic capacity in each of the cell subsets, GFP+NGFR+, GFP−NGFR+, GFP+NGFR−, and GFP−NGFR− fractions were FACS sorted and transplanted into new recipients at limiting dilution. Strikingly, LIC activity was enriched in the GFP+NGFR+ fraction (Figure 2F) and gave rise to leukemias comprising both positive and negative cells and similarly low percentages of reporter activity, resembling the original unsorted cells (supplemental Table 2). Similar observations were also generated using NOTCH1-induced mouse T-cell leukemias (supplemental Figure 14; supplemental Table 3).3,40,41 Of note, CRISPR/Cas9-mediated deletion of both FOXO3 and β-catenin in PDX samples of T-ALL resulted in a marked decrease in LIC frequency (supplemental Figure 15; supplemental Table 4). On the contrary, the constitutive expression of both β-catenin (ΔGSK) and FOXO3 (TM) in leukemia cells of 2 independent PDX clones increased the LIC frequency (supplemental Figure 16; supplemental Table 5). All these findings suggest that both FOXO and β-catenin signaling may reciprocally contribute to LIC activity in T-ALL.
β-Catenin and FOXO3 mutually induce the expression of LIC-related genes
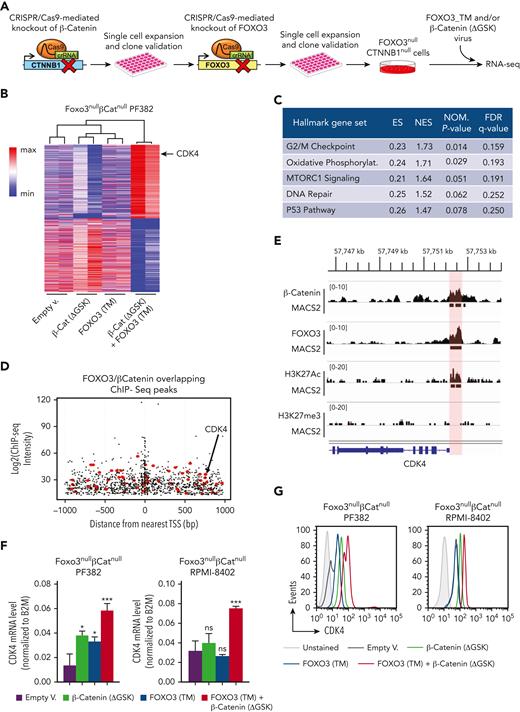
To define the gene expression profiling modulated by either β-catenin or FOXO3 in T-ALL, we generated double knockout FOXO3nullCTNNB1null cell lines using the CRISPR/Cas9 technology. Specifically, DNA mutations in CTNNB1 and FOXO3 genes were serially induced by the direct delivery of ribonucleoprotein Cas9 complex to block the expression of β-catenin and FOXO3 proteins. Independent clones of FOXO3nullCTNNB1null cells were able to grow after serial dilutions into a 96-well plate and were subsequently validated by DNA sequencing of target regions and western blot analysis to confirm the inactivation of CTNNB1 and FOXO3 genes (Figure 3A; supplemental Figure 17). Afterward, FOXO3nullCTNNB1null PF382 cells were cotransduced with β-catenin (ΔGSK) and FOXO3 (TM) lentiviruses or empty vector control to perform RNA-seq assay. We ranked genes according to their expression, identifying a total of 2114 genes with a log2 fold change ≤ −0.5 and a false discovery rate <0.05. Moreover, coexpression of FOXO3 and β-catenin in FOXO3null CTNNB1null PF382 cells defined a distinct gene expression profiling and partitioned the RNA-seq samples into a distinctive group based on the principal components 1 and 2 dimensions of the principal component analysis (Figure 3B; supplemental Figure 17). Of interest, the gene set enrichment analysis of RNA-seq data highlighted the “G2/M checkpoint” as the most enriched hallmark gene set in the gene signature, modulated by FOXO3 and β-catenin and CDK4 in the genes contributing the most to the “G2/M checkpoint” (Figure 3C; supplemental Figure 18 and supplemental Table 8).
β-Catenin and FOXO3 mutually promote the expression of LIC-related genes. (A) Schematic overview of experimental approach. Inactivating DNA mutations in CTNNB1 and FOXO3 genes were serially induced by the direct delivery of ribonucleoprotein Cas9 complex. Positive clones were selected after serial dilutions into a 96-well plate and validated by DNA sequencing of target regions and western blot analysis for protein expression. Afterward, the FOXO3null CTNNB1null cells were cotransduced with an active FOXO3-TM alone or in combination with a stable isoform of β-catenin (ΔGSK) for performing RNA-seq and functional assays. (B) Heat map and hierarchical clustering of gene expression RNA-seq data. FOXO3null CTNNB1null PF382 cells were transduced as reported in panel A and sorted by FACS for RNA isolation and sequencing. Differentially expressed genes, scaled with mean is 0 and standard deviation is 1, are represented (adjusted P ≤ .1 and log fold change ≤ −0.5) by Morpheus-Broad Institute software (https://software.broadinstitute.org/morpheus). (C) Table of the hallmark gene sets significantly enriched in FOXO3null CTNNB1null PF382 cells, doubly transduced with β-catenin (ΔGSK) and FOXO3 € with respect to other cell conditions by gene set enrichment analysis of RNA-seq data. (D) Location of predicted FOXO3 and β-catenin sites relative to transcription start site and FOXO3 and β-catenin chromatin immunoprecipitation (ChIP-seq) signal intensity (ChIP-seq score in log2). In the plot, each dot represents overlapping FOXO3 and β-catenin peaks within 1 kilobase around the transcription start site. The ChIP-seq peaks over 140 genes highly expressed in the FOXO3null CTNNB1null PF382 cells cotransduced with FOXO3-TM and β-catenin (ΔGSK) are highlighted in red. (E) ChIP-seq analysis. ChIP-seq was performed with the anti–β-catenin, anti-FOXO3, anti-H3K27me3, and anti-H3K27Ac antibodies in PF382 cell line. Peaks of aligned reads over the CDK4 locus are shown along with MACS2 peak calls (P ≤ .05). The active genomic region identified as overlapping between FOXO3 and β-catenin ChIP-seq peaks in the CDK4 locus is highlighted in red. (F) CDK4 messenger RNA expression level of human FOXO3null CTNNB1null PF382 and RPMI-8402 cell lines with engineered levels of the active FOXO3-TM mutant alone or in combination with the stable ΔGSK isoform of β-catenin. Transduced cells were sorted by FACS sorting and RNA was isolated to perform the TaqMan reverse transcription droplet digital polymerase chain reaction assay. (G) Protein expression level of intracellular CDK4 by flow cytometric analysis in the transduced cells as reported in panel F. crRNA, crispr RNA; ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score; NOM, nominal P value.
β-Catenin and FOXO3 mutually promote the expression of LIC-related genes. (A) Schematic overview of experimental approach. Inactivating DNA mutations in CTNNB1 and FOXO3 genes were serially induced by the direct delivery of ribonucleoprotein Cas9 complex. Positive clones were selected after serial dilutions into a 96-well plate and validated by DNA sequencing of target regions and western blot analysis for protein expression. Afterward, the FOXO3null CTNNB1null cells were cotransduced with an active FOXO3-TM alone or in combination with a stable isoform of β-catenin (ΔGSK) for performing RNA-seq and functional assays. (B) Heat map and hierarchical clustering of gene expression RNA-seq data. FOXO3null CTNNB1null PF382 cells were transduced as reported in panel A and sorted by FACS for RNA isolation and sequencing. Differentially expressed genes, scaled with mean is 0 and standard deviation is 1, are represented (adjusted P ≤ .1 and log fold change ≤ −0.5) by Morpheus-Broad Institute software (https://software.broadinstitute.org/morpheus). (C) Table of the hallmark gene sets significantly enriched in FOXO3null CTNNB1null PF382 cells, doubly transduced with β-catenin (ΔGSK) and FOXO3 € with respect to other cell conditions by gene set enrichment analysis of RNA-seq data. (D) Location of predicted FOXO3 and β-catenin sites relative to transcription start site and FOXO3 and β-catenin chromatin immunoprecipitation (ChIP-seq) signal intensity (ChIP-seq score in log2). In the plot, each dot represents overlapping FOXO3 and β-catenin peaks within 1 kilobase around the transcription start site. The ChIP-seq peaks over 140 genes highly expressed in the FOXO3null CTNNB1null PF382 cells cotransduced with FOXO3-TM and β-catenin (ΔGSK) are highlighted in red. (E) ChIP-seq analysis. ChIP-seq was performed with the anti–β-catenin, anti-FOXO3, anti-H3K27me3, and anti-H3K27Ac antibodies in PF382 cell line. Peaks of aligned reads over the CDK4 locus are shown along with MACS2 peak calls (P ≤ .05). The active genomic region identified as overlapping between FOXO3 and β-catenin ChIP-seq peaks in the CDK4 locus is highlighted in red. (F) CDK4 messenger RNA expression level of human FOXO3null CTNNB1null PF382 and RPMI-8402 cell lines with engineered levels of the active FOXO3-TM mutant alone or in combination with the stable ΔGSK isoform of β-catenin. Transduced cells were sorted by FACS sorting and RNA was isolated to perform the TaqMan reverse transcription droplet digital polymerase chain reaction assay. (G) Protein expression level of intracellular CDK4 by flow cytometric analysis in the transduced cells as reported in panel F. crRNA, crispr RNA; ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score; NOM, nominal P value.
To highlight putative common targets of β-catenin and FOXO3, we generated chromatin immunoprecipitation sequencing (ChIP-seq) data for β-catenin and FOXO3 in the PF382 cell line and analyzed them together with publicly available ChIP-seq data sets (β-catenin, Gene Expression Omnibus: GSE5392742 and FOXO3, Gene Expression Omnibus: GSE3548643) to determine the overlapping genomic peaks of both factors within 1 kilobase around transcription start sites (TSSs) (Figure 3D; supplemental Figure 19). We found 2739 DNA sequences associated with 1380 TSSs, which revealed shared consensus motifs for β-catenin and FOXO3, including canonical binding sites for TCF7 and FOXO transcription factors as well as novel predicted sites (supplemental Figure 20). We focused our attention on 50 top gene transcripts, highly expressed in the FOXO3nullCTNNB1null PF382 cells cotransduced with β-catenin (ΔGSK) and FOXO3 (TM) lentiviruses compared with other cell conditions (supplemental Figure 21). Of interest, we noted that the genomic region over the TSS of CDK4 gene was enriched in DNA binding sites for both β-catenin and FOXO3, and its expression was significantly induced by β-catenin (ΔGSK) and FOXO3 (TM) in FOXO3nullCTNNB1null PF382 cells (Figure 3D-E). We validated these findings in the unmanipulated PF382 cell line by local ChIP using a series of 5 primer sets spanning the CDK4 promoter and found a substantial enrichment of FOXO3 and β-catenin binding in an overlapping region over the TSS of human CDK4 gene (supplemental Figure 22). Subsequent ChIP-seq analyses also showed a high H3K27Ac density over the CDK4 promoter, suggesting that this region can be regarded as active in PF382 cell line (Figure 3E).
CDK4 is a major modulator of cell cycle during G1/S transition44 and promotes disease progression in T-ALL.45-47 We confirmed the specific upregulation of CDK4 by β-catenin (ΔGSK) and FOXO3 (TM) in FOXO3nullCTNNB1null PF382 and FOXO3nullCTNNB1null RPMI-8402 cell lines at the messenger RNA (mRNA) level by quantitative reverse transcription polymerase chain reaction (Figure 3F) and also at the protein level by intracellular flow cytometry analysis (Figure 3G). Moreover, to address whether FOXO3 and β-catenin were able to promote cell cycle in our system, we assessed the cell proliferation by 5-bromo-2′-deoxyuridine incorporation in FOXO3nullCTNNB1null PF382 and RPMI-8402 cell lines after lentiviral transduction of β-catenin (ΔGSK) alone or in combination with FOXO3 (TM). We found that β-catenin and FOXO3 together induce cycling of FOXO3nullCTNNB1null cell lines (supplemental Figure 23). Of note, a similar phenotype was also observed after constitutive expression of a stable isoform of CDK4 (R24L)48 (supplemental Figure 23). In contrast, short hairpin RNA–mediated knock down of CDK4 expression in FOXO3nullCTNNB1null cell lines, doubly transduced with β-catenin (ΔGSK) and FOXO3 (TM) (supplemental Figure 23), as well as in unmanipulated PF382 and RPMI-8402 cell lines (supplemental Figure 24), significantly inhibits in vitro cell proliferation and expansion. Taken together, these data support the conclusion that FOXO3 and β-catenin might drive cell cycling in T-ALL cell lines and that this effect is largely mediated through CDK4 induction. To assess the relevance of β-catenin, FOXO3, and CDK4 transcriptional axis in human T-ALL, we determined the expression level of CDK4 in PDX samples, doubly transduced with FOXO (3DNm8) and TCF/β-catenin (7TGC) lentiviral reporters, and expanded in vivo in immunocompromised (NSG) mice. CDK4 was highly expressed in TCF/β-catenin (GFP+) and FOXO (NGFR+) active subsets with respect to the other cell populations by intracellular flow cytometry analysis (supplemental Figure 25), suggesting that active β-catenin and FOXO signaling pathway together promote CDK4 expression.
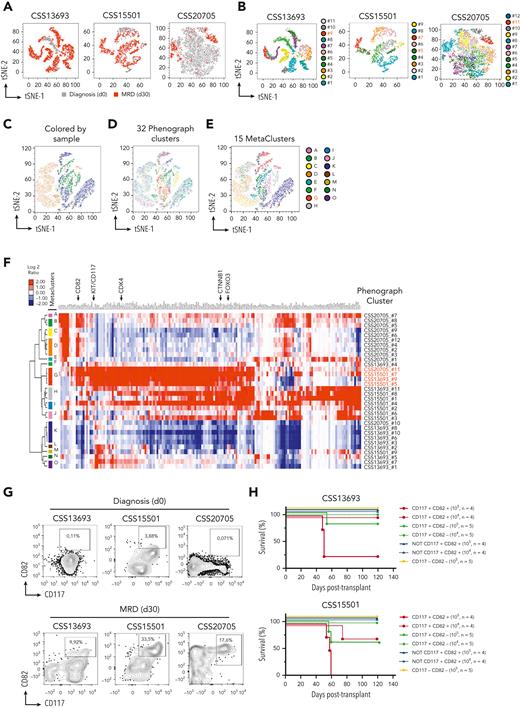
β-Catenin and FOXO3 genes are highly expressed in LIC-enriched CD117+CD82+ cell subsets
To define the phenotypic profiling of leukemia cell subsets with high levels of β-catenin and FOXO3, we performed single-cell RNA-seq (scRNA-seq) profiling, simultaneously with a panel of 42 oligoconjugated antibodies against cell-surface protein markers (AbSeq assay) (supplemental Table 9) in CSS13693, CSS15501, and CSS20705 primary samples of human T-ALLs at the time of diagnosis and MRD 30 days after the start of the therapy without any expansion into immunocompromised mice (supplemental Figure 26; supplemental Tables 6 and 10). To discriminate between cell subsets with related phenotypes from highly dimensional data, we used the Phenograph clustering algorithm, which identified in total 32 distinct population clusters from 3 scRNA-seq assays, each including a different fraction of leukemia cells at days 0 and 30 (Figure 4A-B). We used this distinction to determine which clusters showed the highest level of CTNNB1 and FOXO3 transcripts. To determine common phenotypes among different patient samples, we performed a meta-clustering analysis based on gap statistic49 and identified 15 meta-clusters (MCs) from the original 32 Phenograph clusters (Figure 4C-E). Of note, the MC-G group, which included the CSS13693_#9, CSS15501_#5, CSS15501_#7, and the CSS20705_#11 Phenograph clusters, was highly enriched in both CTNNB1 and FOXO3 transcripts (Figure 4F; supplemental Figure 27). Of interest, the highlighted cell subset was primarily constituted of MRD cells and interestingly showed a high transcriptional level of β-catenin– and FOXO3-dependent genes, including CDK4 (Figure 4F; supplemental Figure 27). To design a lower dimensional conventional flow cytometry assay and validate these findings, we performed a differential expression analysis between all Phenograph clusters and determined the level of transcripts associated with cell-surface markers, as well as the proteins recognized by the oligoconjugated antibodies in the AbSeq assay. Of interest, all CSS13693_#9, 15501_#5, CSS15501_#7, and CSS20705_#11 clusters of the MC-G group were characterized by high levels of CD117/KIT and CD82 cell-surface markers (Figure 4D; supplemental Figure 27D), showing similar gene and protein expression profiles (supplemental Figure 28). We validated these findings by conventional flow cytometry (Figure 4G) and applied the same gating strategy for isolation of CD117+CD82+ cells by FACS sorting using parallel vials of frozen MRD cells. Of interest, we also found that the expression levels of CD82 and CD117 cell-surface markers are not modulated by FOXO3 and β-catenin activity, suggesting that KIT and CD82 genes are not direct targets of FOXO3 and β-catenin (supplemental Figure 29). To assess the LIC frequency of CD117+CD82+ fraction compared with the other subsets, FACS-sorted cells were injected into NSG mice at a limiting dilution. Strikingly, we found that the LIC activity was enriched in the CD117+CD82+ fraction in 2 independent clones of primary T-ALLs (Figure 4H; supplemental Table 7). Specifically, the calculated LIC frequency in CD117+CD82+ cells is 1 in 25 978 (95% confidence interval [CI], 1 in 7616-88 604) and 1 in 87 326 (95% CI, 1 in 27 516-277 145), in CD117+CD82− cells is 1 in 221 246 (95% CI, 1 in 55 495-882 056) and 1 in 498 332 (95% CI, 1 in 70 566-3 519 195) for CSS15501 and CSS13693 samples, respectively, and in NOT CD117+CD82+ and CD117−CD82− cells is >1 in 146 876 (95% CI, 1 in 66 888-3 277 606) for both leukemias by extreme limiting dilution analysis. Taken together, these data support the conclusion that CD117 and CD82 may identify LIC-enriched cell subsets, which are considerably heightened in patients with T-ALL with MRD.
Coexpression of β-catenin, FOXO3, and CDK4 identify LIC-enriched CD117+CD82+cell subsets in MRD of human T-ALLs. (A) T-distributed stochastic neighbor embedding (tSNE) plots based on the scRNA-seq and AbSeq data from 3 primary T-ALL samples; CSS13693, CSS15501, and CSS20705, at the time of diagnosis (day 0) and 30 days after the start of therapy (MRD day 30) without passaging into immunocompromised mice. In the map, leukemia cells at days 0 and 30 are colored in grey and red, respectively. (B) tSNE plot as in panel A, but with each Phenograph cluster depicted in a different color. There was a total of 32 different phenotypic clusters identified by the Phenograph algorithm. The clusters with the highest expression of FOXO3, CTNNB1, and CDK4 transcripts are indicated in red. Dimensional reduction tSNE plots of scRNA-seq data with cells colored according to their assigned sample (C) or Phenograph cluster (D). (E) tSNE plot as in panel C, but with cells colored by their assigned meta-cluster (MC). The MC with the highest expression of FOXO3, CTNNB1, and CDK4 transcripts is indicated in red. (F) Expression heat map and hierarchical clustering of selected surface markers (GO:0016021) together with FOXO3, CTNNB1, and CDK4 genes, which resulted heterogeneously transcriptionally expressed in each of the 32 Phenograph clusters and hierarchically clustered into 15 MC groups. (G) Flow cytometry plots of CSS13693, CSS15501, and CSS20705 samples at the time of diagnosis (day 0) and MRD (day 30), showing the protein expression of CD117 and CD82 cell markers. Tumor cells were gated based on the expression of CD3, CD99, and CD7 surface markers and identified as CD3−CD99+CD7+ cells. (H) Survival of recipient NSG mice after transplantation with FACS-sorted CD117+CD82+, CD117+CD82−, CD117−CD82− or double negative plus single positive (NOT CD117+CD82+) subsets from CSS13693 and CSS15501 primary samples of MRD at day 30. The cell doses for each recipient animal are indicated in brackets. The calculated LIC frequency in CD117+CD82+ cells is 1 in 25 978 (95% CI, 1 in 7616-88 604) and 1 in 87 326 (95% CI, 1 in 27 516-277 145), in CD117+CD82− cells is 1 in 221 246 (95% CI, 1 in 55 495-882 056) and 1 in 498 332 (95% CI, 1 in 70 566-3 519 195) for CSS15501 and CSS13693 samples, respectively, and in NOT CD117+CD82+ and CD117−CD82− cells is >1 in 146 876 (95% CI, 1 in 66 888-3 277 606) for both leukemias.
Coexpression of β-catenin, FOXO3, and CDK4 identify LIC-enriched CD117+CD82+cell subsets in MRD of human T-ALLs. (A) T-distributed stochastic neighbor embedding (tSNE) plots based on the scRNA-seq and AbSeq data from 3 primary T-ALL samples; CSS13693, CSS15501, and CSS20705, at the time of diagnosis (day 0) and 30 days after the start of therapy (MRD day 30) without passaging into immunocompromised mice. In the map, leukemia cells at days 0 and 30 are colored in grey and red, respectively. (B) tSNE plot as in panel A, but with each Phenograph cluster depicted in a different color. There was a total of 32 different phenotypic clusters identified by the Phenograph algorithm. The clusters with the highest expression of FOXO3, CTNNB1, and CDK4 transcripts are indicated in red. Dimensional reduction tSNE plots of scRNA-seq data with cells colored according to their assigned sample (C) or Phenograph cluster (D). (E) tSNE plot as in panel C, but with cells colored by their assigned meta-cluster (MC). The MC with the highest expression of FOXO3, CTNNB1, and CDK4 transcripts is indicated in red. (F) Expression heat map and hierarchical clustering of selected surface markers (GO:0016021) together with FOXO3, CTNNB1, and CDK4 genes, which resulted heterogeneously transcriptionally expressed in each of the 32 Phenograph clusters and hierarchically clustered into 15 MC groups. (G) Flow cytometry plots of CSS13693, CSS15501, and CSS20705 samples at the time of diagnosis (day 0) and MRD (day 30), showing the protein expression of CD117 and CD82 cell markers. Tumor cells were gated based on the expression of CD3, CD99, and CD7 surface markers and identified as CD3−CD99+CD7+ cells. (H) Survival of recipient NSG mice after transplantation with FACS-sorted CD117+CD82+, CD117+CD82−, CD117−CD82− or double negative plus single positive (NOT CD117+CD82+) subsets from CSS13693 and CSS15501 primary samples of MRD at day 30. The cell doses for each recipient animal are indicated in brackets. The calculated LIC frequency in CD117+CD82+ cells is 1 in 25 978 (95% CI, 1 in 7616-88 604) and 1 in 87 326 (95% CI, 1 in 27 516-277 145), in CD117+CD82− cells is 1 in 221 246 (95% CI, 1 in 55 495-882 056) and 1 in 498 332 (95% CI, 1 in 70 566-3 519 195) for CSS15501 and CSS13693 samples, respectively, and in NOT CD117+CD82+ and CD117−CD82− cells is >1 in 146 876 (95% CI, 1 in 66 888-3 277 606) for both leukemias.
CD117 and CD82 markers identify a distinct T-ALL subgroup, enriched in MRD
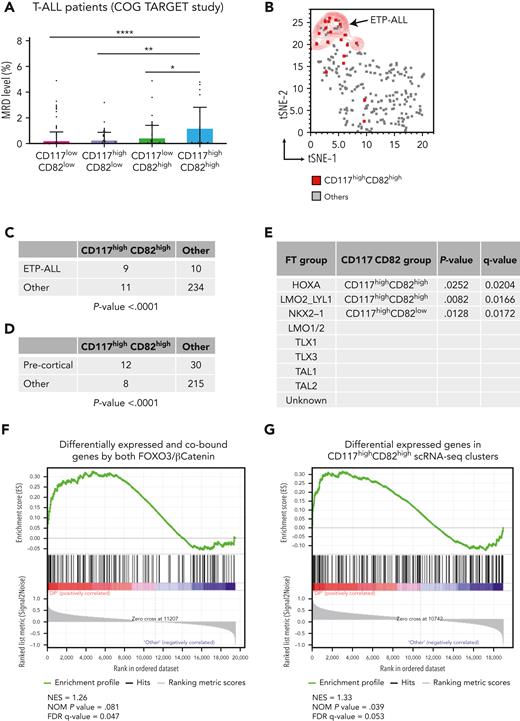
To assess the relevance of reported findings in human T-ALL, we evaluated the transcriptional level of both CD117 and CD82 cell markers in a cohort of 264 diagnostic T-ALL samples from the COG TARGET study.50 Strikingly, cases in the highest 25th percentile of transcriptional levels of CD117 and CD82 genes exhibited significantly greater values of MRD compared with other samples (Figure 5A), supporting the association between high mRNA expression of CD117 and CD82 and more aggressive clinical disease. Furthermore, we determined whether high CD117 and CD82 gene expression was enriched in patients with T-ALL originating from early T-cell precursors (ETPs), which present a poor prognosis after intensive chemotherapy.51 Interestingly, we found that patients with high CD117 and CD82 were also significantly associated with the group of ETP-ALL as well as leukemias at the precortical differentiation stage (Figure 5B-D; supplemental Table 11). We also assessed any other enrichment of high CD117 and CD82 cases within specific T-ALL subgroups.50 Interestingly, patients with high CD117 and CD82 expression were positively associated with the HOXA and LMO2/LYL1 subgroups, whereas cases with high CD117 and low CD82 mRNA expression were significantly associated with the NKX2-1 subgroup (Figure 5E). Finally, to further characterize the highlighted patients with T-ALL with high CD117 and CD82 mRNA expression at the transcriptional level, we performed the gene set enrichment analysis using 2 different gene signatures: the first was derived from the integrated analysis of RNA-seq and ChIP-seq data sets of FOXO3null CTNNB1null PF382 cells, modulated by the FOXO3 (TM)–active mutant in combination with β-catenin (ΔGSK) stable isoform (Figures 3B and 5F; supplemental Table 14); the second gene list resulted from the differential expression analysis of scRNA-seq data from 3 highlighted CD117highCD82high Phenograph clusters vs the others (Figure 5G; supplemental Figure 28). Strikingly, we found that patients with high levels of CD117 and CD82 showed a significant enrichment for the genes modulated by FOXO3 and β-catenin in PF382 cell line, as well as the differentially expressed genes in the LIC-enriched CD117+CD82+ cell subsets of primary T-ALLs. These data highlight patients with high CD117 and high CD82 mRNA expression as a distinct subgroup of ETP-ALL, bearing a gene signature dependent on the activity of both FOXO3 and β-catenin transcription factors.
High levels of CD117 and CD82 gene expression demarcates an early stage subgroup, enriched in MRD of human T-ALL. (A) Fraction of MRD 29 days after the start of therapy in 252 patients with T-ALL from the COG TARGET study.50 All patients have been subdivided in CD117lowCD82low (n = 146), CD117highCD82low (n = 42), CD117lowCD82high (n = 44), and CD117highCD82high (n = 20) groups based on the highest 25th percentile of transcriptional level of CD117 and CD82 genes on RNA-seq data of each patient with T-ALL. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 (2-way analysis of variance). (B) tSNE plot of RNA-seq data from 252 patients with T-ALL (COG TARGET study).50 Each sample derived from the CD117highCD82high subgroup is indicated by red dot. Early T-cell precursor ALL are also highlighted by the red contour plot. Correlation between CD117/CD82 subgroups and early T-cell precursor ALL (C), precortical stage (D), or transcription factor/genetic subgroups (E) among 252 patients with T-ALL. Statistical P values were calculated by Fisher exact test, corrected for multiple comparisons by FDR using a 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. (F-G) Gene set enrichment analysis. (F) The gene signature was derived by the differential expression analysis of integrated RNA-seq and ChIP-seq data sets from FOXO3null CTNNB1null PF382 cells cotransduced with FOXO3-TM and β-catenin (ΔGSK) vs other conditions (n = 140 genes). (G) The gene list derives from the differential expression analysis of scRNA-seq data from 3 highlighted CD117highCD82high Phenograph clusters vs the others (n = 185 genes). All genes were ranked for differential expression in CD117highCD82high vs the rest of the patients with T-ALL from the COG TARGET study.50
High levels of CD117 and CD82 gene expression demarcates an early stage subgroup, enriched in MRD of human T-ALL. (A) Fraction of MRD 29 days after the start of therapy in 252 patients with T-ALL from the COG TARGET study.50 All patients have been subdivided in CD117lowCD82low (n = 146), CD117highCD82low (n = 42), CD117lowCD82high (n = 44), and CD117highCD82high (n = 20) groups based on the highest 25th percentile of transcriptional level of CD117 and CD82 genes on RNA-seq data of each patient with T-ALL. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001 (2-way analysis of variance). (B) tSNE plot of RNA-seq data from 252 patients with T-ALL (COG TARGET study).50 Each sample derived from the CD117highCD82high subgroup is indicated by red dot. Early T-cell precursor ALL are also highlighted by the red contour plot. Correlation between CD117/CD82 subgroups and early T-cell precursor ALL (C), precortical stage (D), or transcription factor/genetic subgroups (E) among 252 patients with T-ALL. Statistical P values were calculated by Fisher exact test, corrected for multiple comparisons by FDR using a 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli. (F-G) Gene set enrichment analysis. (F) The gene signature was derived by the differential expression analysis of integrated RNA-seq and ChIP-seq data sets from FOXO3null CTNNB1null PF382 cells cotransduced with FOXO3-TM and β-catenin (ΔGSK) vs other conditions (n = 140 genes). (G) The gene list derives from the differential expression analysis of scRNA-seq data from 3 highlighted CD117highCD82high Phenograph clusters vs the others (n = 185 genes). All genes were ranked for differential expression in CD117highCD82high vs the rest of the patients with T-ALL from the COG TARGET study.50
Discussion
β-Catenin is a dual function protein that is entirely involved in the growth and development of normal and cancer stem cells.18,52,53 In chronic myeloid leukemia, the deletion of β-catenin by treatment with imatinib (a tyrosine kinase inhibitor) impairs the leukemia stem cell (LSC) activity and delays the recurrence of tumor.54 Moreover, in mouse models of acute myelogenous leukemia, the canonical Wnt/β-catenin pathway is involved in the self-renewal of LSCs, derived from either hematopoietic stem cells or more differentiated granulocyte-macrophage progenitors.55 In T-cell malignancies, gain-of-function mutations in the gene encoding β-catenin (CTNNB1) have been reported in lymphomas of T-lineage origin56 and subsets of human T-ALL.57 High expression levels of β-catenin have also been observed in LSCs of both human leukemias and mouse models of T-ALL.12,14 Moreover, an upregulated canonical β-catenin signaling was detected in 85% of childhood patients with T-ALL, compared with normal human thymocytes.57 In the PTEN-null mouse model of T-cell leukemia, the LSC-enriched c-Kitmid CD3+ Lin− cell subsets express high levels of unphosphorylated β-catenin protein, and their frequency was significantly reduced after the CRE-mediated conditional deletion of the CTNNB1 gene.12 In the same PTEN-null mouse model, by scRNA-seq profiling, a β-catenin-SPI1-HAVCR2 regulatory circuit was also reported to maintain LSC activity.58 All these findings support the idea that targeting β-catenin by drug treatment may represent a more effective therapy for the eradication of LSCs in T-cell malignancies.
To explore the regulatory mechanisms underlying the β-catenin activity in T-ALL, our investigations have identified relevant protein interactions of β-catenin and have revealed a noncanonical interaction of β-catenin with the FOXO3 transcription factor, which has been described to promote T-cell survival and cell cycle progression.35,36 Furthermore, we demonstrated that β-catenin and FOXO3 functionally cooperate to modulate LIC activity in T-ALL by mutually inducing genes, such as CDK4, involved in leukemia progression and growth.45 Through scRNA-seq profiling of tumor cells derived from patients with T-ALL at the time of diagnosis and MRD 30 days after the start of the therapy and without any expansion into immunocompromised mice, we also report that β-catenin and FOXO3 genes are highly expressed in LIC-enriched CD117+CD82+ cell subsets, mostly enriched in MRD. Taken together, these findings suggest a model in which proliferative leukemia cells might be selected after drug treatment upon in vivo activation by the protective local tumor microenvironment.59 It was already reported that hypoxic niches support Wnt signaling and increase an Hif1a-dependent transcription of β-catenin, promoting the self-renewal of LSCs in T-ALL.14,60 In agreement with these findings, our data also suggest that the tumor microenvironment might modulate the maintenance and proliferation of tumor cells through regulation of Wnt signaling. Finally, we report that CD117 and CD82 markers identify a distinct T-ALL subgroup, significantly associated with ETP-ALL, which is characterized by a poor clinical outcome after intensive chemotherapy.51,61,62
Although the relevance of canonical Wnt/β-catenin signaling has been explored in T-ALL maintenance and progression,12,14,15 the identified link between β-catenin and FOXO3 is likely to be the most relevant in the context of PI3K/AKT inhibition.63,64 Alternatively, cell signaling events that promote β-catenin and FOXO3 activity may also affect LIC activity and be effective in MRD as compensatory response.65,66 These findings also suggest that noncanonical and TCF-independent functions of β-catenin, involving FOXO3, might be mostly active in T-cell leukemias at the early stage of differentiation, such as in the precortical leukemias or the ETP-ALL subgroup under stress conditions. In fact, in acute myeloid leukemias with mixed lineage leukemia translocations, it has been reported that the expression of FOXO3 is protective against DNA damage and oxidative stress.67 Moreover, in acute myelogenous leukemia cell subsets, the deletion of FOXO1, FOXO3, and FOXO4 significantly reduces leukemic cell growth and LIC functions.68 A shift in the β-catenin activity between TCF- and FOXO-mediated signaling could indeed be related to the maintenance and progression of certain types of LICs, and thus, linked to the different clinical outcomes in patients with T-ALL. Nonetheless, further studies will be required to explore the intriguing possibility that therapies that inhibit β-catenin and FOXO3 expression and/or activity may antagonize LIC activity and thereby improve clinical outcomes in patients with T-ALL.
Acknowledgments
The authors thank Wolfgang Link (Spanish National Research Council and the Autonoma University of Madrid, Madrid, Spain) for providing FOXO reporter constructs. The authors are grateful to Rossella Di Paola and Tommaso Mazza for their help in the Illumina next-generation DNA sequencing and Giuseppe Merla for his support in the luciferase assays. The authors also thank Chiara Di Giorgio at the Fondazione IRCCS Casa Sollievo della Sofferenza in San Giovanni Rotondo (Italy) for proofreading the text.
This work was funded by grants from the Foundation with the South (Brain2South grant 2015-0251) (V.G.); the Italian Ministry of Health, “Ricerca Finalizzata” Young Researcher (GR-2016-02361287) (V.G.); the Italian Association for Cancer Research (AIRC), My First AIRC Grant (MFAG18487) (V.G.) and IG 2019 (project 23070) (V.G.); the Worldwide Cancer Research (20-0318) (V.G.); and the AIRC Fellowship for Italy (F.T.).
Authorship
Contribution: P.P., S.G., C.S., A.P.W., and V.G. conceptualized the study; G.R., A.M.C., C.S., A.P.W., and V.G. provided resources; P.P., M. Colucci, E.D.S., C.P., and V.G. curated data; P.P., M. Colucci, F.T., F.B., and V.G. performed formal analyses; P.P., M. Colucci, E.D.S., M.M., F.T., F.S., E.M., S.G., M. Ciavarella, E.A.C., and V.G. designed methodology; P.P., M. Colucci, F.T., F.S., S.G., and V.G. investigated; P.P., M. Colucci, F.T., and F.B. visualized the data; V.G. acquired funding; F.S. was the project administrator; F.B., G.R., A.M.C., C.S., A.P.W., and V.G. supervised the study; P.P., M.C., F.T., and V.G. wrote the original draft of the manuscript; and P.P., E.D.S., G.R., A.M.C., A.P.W., and V.G. reviewed and edited the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vincenzo Giambra, Hematopathology Laboratory, Institute for Stem Cell Biology, Regenerative Medicine and Innovative Therapies, Fondazione IRCCS Casa Sollievo della Sofferenza, viale Padre Pio 7, 71013 San Giovanni Rotondo, Italy; e-mail: v.giambra@operapadrepio.it.
References
Author notes
The normalized (rlog) data for messenger RNA expression reported in this study have been deposited in the National Center for Biotechnology Information, National Institutes of Health (accession number SRA PRJNA785416) and in supplemental Table 13. The single-cell RNA sequencing and chromatin immunoprecipitation data have been deposited in the National Center for Biotechnology Information, National Institutes of Health (accession numbers SRA PRJNA784728 and PRJNA785422, respectively). The mass spectrometry data can be found in supplemental Table 12.
Data are available on request from the corresponding author, Vincenzo Giambra (v.giambra@operapadrepio.it).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal