Key Points
NRX-0492 induced rapid and sustained degradation of wild-type and C481 mutant BTK at subnanomolar concentrations in primary CLL samples.
Orally administered NRX-0492 abrogated BTK-dependent signaling and demonstrated anti-CLL activity in patient-derived xenografts in vivo.
Abstract
Bruton tyrosine kinase (BTK) is essential for B-cell receptor (BCR) signaling, a driver of chronic lymphocytic leukemia (CLL). Covalent inhibitors bind C481 in the active site of BTK and have become a preferred CLL therapy. Disease progression on covalent BTK inhibitors is commonly associated with C481 mutations. Here, we investigated a targeted protein degrader, NRX-0492, that links a noncovalent BTK-binding domain to cereblon, an adaptor protein of the E3 ubiquitin ligase complex. NRX-0492 selectively catalyzes ubiquitylation and proteasomal degradation of BTK. In primary CLL cells, NRX-0492 induced rapid and sustained degradation of both wild-type and C481 mutant BTK at half maximal degradation concentration (DC50) of ≤0.2 nM and DC90 of ≤0.5 nM, respectively. Sustained degrader activity was maintained for at least 24 hours after washout and was equally observed in high-risk (deletion 17p) and standard-risk (deletion 13q only) CLL subtypes. In in vitro testing against treatment-naïve CLL samples, NRX-0492 was as effective as ibrutinib at inhibiting BCR-mediated signaling, transcriptional programs, and chemokine secretion. In patient-derived xenografts, orally administered NRX-0492 induced BTK degradation and inhibited activation and proliferation of CLL cells in blood and spleen and remained efficacious against primary C481S mutant CLL cells collected from a patient progressing on ibrutinib. Oral bioavailability, >90% degradation of BTK at subnanomolar concentrations, and sustained pharmacodynamic effects after drug clearance make this class of targeted protein degraders uniquely suitable for clinical translation, in particular as a strategy to overcome BTK inhibitor resistance. Clinical studies testing this approach have been initiated (NCT04830137, NCT05131022).
Introduction
B-cell receptor (BCR) signaling drives disease progression in chronic lymphocytic leukemia (CLL).1-4 Bruton tyrosine kinase (BTK) is essential for BCR signaling. BTK inhibitors (BTKis) have become a preferred treatment for patients with chronic lymphocytic leukemia (CLL) and other B-cell malignancies.1,3,5-10 Ibrutinib, the first in class covalent BTKi, binds to a cysteine residue (C481) in the active site of BTK, thereby achieving sustained inhibition of BCR signaling.11-13 The reversal of inhibition requires de novo synthesis of BTK.14 In CLL, ibrutinib induces high rates of response across genetic and clinical risk groups.15-20 Progression-free survival (PFS) with single agent ibrutinib in first-line was 70% at 5 years,21 even in patients with TP53 alterations.22
Patients with CLL with progressive disease on ibrutinib commonly harbor mutations that substitute a different amino for the C481 residue, thereby preventing covalent drug binding.23,24 Less common are mutations in BTK residues other than C481,25 and gain-of-function mutations in PLCG2.23,26,27 The emergence of BTK mutations in IBR-resistant CLL underscores the critical pathogenic role of BTK-dependent signals. Several noncovalent BTKis with activity against C481 mutant BTK have entered clinical studies.28-30 Among these, pirtobrutinib induced responses in 67% of patients with CLL with previous resistance to covalent BTKi.29 However, in patients progressing on pirtobrutinib, BTK mutations other than C481 were identified that restored BCR signaling despite continued therapy.31
Small molecule–induced BTK degradation is another promising approach to target mutant BTK.32,33 BTK degradation can be achieved by targeted protein degraders,32,33 and several BTK degraders have been developed. We have developed targeted protein degraders that link noncovalent BTK-binding “hooks” with “harnesses” that recruit cereblon, an adaptor protein of the E3 ubiquitin ligase complex.34-36 These molecules catalyze ubiquitylation and proteasomal degradation of BTK. NRX-0492 is a tool compound that represents the pharmacological mechanisms of NX-2127, a clinical compound that has been further optimized for improved drug-like properties. Here, we demonstrate the activity of NRX-0492 against primary CLL cells in vitro and in patient-derived xenografts.
Materials and methods
Reagents
NRX-0492, hook and harness were provided by Nurix.34 We purchased ibrutinib, lenalidomide, pomalidomide (Combi-blocks, Inc), MG132 (Enzo Life Sciences), and MLN4924 (Fisher Scientific).
BTK fluorescence resonance energy transfer (FRET) competition assay
Dose titrations of compound were incubated in a 20 mL reaction volume with 1 nM biontinylated BTK (full-length) wild-type (WT) or mutant proteins (Carna) preincubated with 3 nM terbium-coupled streptavidin (Cisbio) and EC50 of a Bodipy-FL labeled pan-kinase probe in a white 384-well microplate (Perkin Elmer Proxiplate). The final assay buffer was composed of 50 mM N-2-hydroxyethylpiperazine-N′-2-ethane sulfonic acid (HEPES), pH 7.5, 50 mM NaCl, 0.01% Triton X-100, 0.01% bovine serum albumin, and 1 mM dithiothreitol with 2% final dimethyl sulfoxide (DMSO). After 1-hour of incubation, plates were excited at 320 nm and time-resolved (TR) FRET signal read at 520 nm and 620 nm using an EnVision plate reader (Perkin Elmer). TR-FRET signal was calculated using a 520 by 620 nm ratio. Percentage of relative probe binding was calculated compared with DMSO control and fit in Prism (GraphPad) to calculate 50% inhibitory (or inhibition or infective) concentration (IC50).
Patient samples, cell lines, and in vitro studies
Peripheral blood mononuclear cells (PBMCs) were obtained from patients with CLL (supplemental Table 1; available on the Blood website) enrolled on observational or tissue collection studies (NCT00923507; NCT00071045). Patients treated with ibrutinib were enrolled on a phase 2 clinical trial (NCT01500733).15,23 Written informed consent was obtained in accordance with the Declaration of Helsinki under oversight by the National Institutes of Health (NIH) Institutional Review Board. Primary CLL cells were thawed and cultured in complete RPMI1640 (10% heat inactivated fetal bovine serum; 5% penicillin/ streptomycin) at 37°C for 24 hours before starting drug treatment. Healthy donor PBMCs were collected by the NIH blood bank. TMD8 cells were obtained from Tokyo Medical and Dental University.37 TMD8 cell lines expressing BTKC481S were generated by transfecting ribonucleoprotein [20 pmol Cas9 (Thermo Fisher, A36499) + 120 pmol single guide RNA (crispr RNA + transactivating crispr RNA annealed for 5 minutes at 95⁰C, IDT)] and 100 pmol single-stranded oligodeoxynucleotide into cells using a Lonza 4D Nucleofector with Solution SF and Program EO-100. Cells were recovered in full media + 0.25 mM NU-7441, and monoclonal cells were expanded for 2 weeks before DNA sequencing. Cells were treated with either ibrutinib or NRX-0492 at concentrations and times as indicated in the specific experiments, 10 μM NAEi, 10 μM harness, or DMSO. In drug washout experiments, cells were centrifuged at 1500 rpm for 5 minutes, washed twice with phosphate-buffered saline, and cultured in fresh medium.
Patient-derived xenograft (PDX) mouse model
Mouse experiments were approved by the Institutional Animal Care and Use Committee. NOD/SCID/IL2Rgnull (NSG) mice were purchased from Jackson Laboratory (stock #5557). Experiments in the PDX model were conducted as previously described,38 with minor modifications (supplemental Methods). Each mouse was injected with 20 × 106 cells intraperitoneally and 40 × 106 cells intravenously. Three days after cell inoculation, human cells in the peripheral blood were enumerated using flow cytometry. A single dose of 30 mg/kg NRX-0492 was administered by oral gavage on day 1, and for continuous dosing, added to drinking water with 5% hydroxypropyl beta cyclodextrin at 0.2 mg/mL. Mice were bled on day 8 and killed on day 22. The spleens were removed on day 22 and mashed through a cell strainer. Cells were suspended in ammonium chloride potassium buffer (Thermo Fisher) on ice for 10 minutes and analyzed by flow cytometry.
Cellular degradation and proteomic assays
Cells were treated with compounds and lysed in radioimmunoprecipitation assay buffer. Lysates were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (Thermo Fisher), transferred to polyvinylidene difluoride membranes, and incubated with primary antibodies (Cell Signaling Technology, #8547, anti-BTK, 1:1000; #5174, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 1:3000; #4695, Erk1/2, 1:1000; #4370, p-ErkThr202/Tyr204, 1:1000). Membranes were probed with horseradish peroxidase–conjugated secondary antibodies (Thermo Fisher). Images were acquired using an Amersham Imager 680 (Cytiva). Image J software was used for immunoblot quantification. In complementary experiments, cells were incubated with compounds for 4 hours and BTK levels were determined using total-BTK homologous time–resolved fluorescence (HTRF) kit according to manufacturer’s protocol (Cisbio). Half maximal degradation concentration (DC50) and DC90 were calculated using 4-parameter nonlinear regressions using GraphPad software. For proteomics, cells were treated with DMSO or degrader (50 nM) in triplicate. After 6 hours of treatment, cells were harvested, washed twice with phosphate-buffered saline, and stored as frozen cell pellets. Proteomic analysis was performed using tandem mass tagging by mass spectrometry Bioworks.
Flow cytometry
As indicated, cells were stained with a Fixable Violet Dead Cell Stain Kit (Thermo Fisher), or annexin V apoptosis detection kit (Invitrogen), CD45-fluorescein isothiocyanate, CD5-APCCy7, CD19-PECy7, CD69-phycoerythrin, CD3-PECy7, CD19-PECy5 (BD Biosciences), and CD5-AF700 (BioLegend), fixed and permeabilized with eBioscience Foxp3/transcription factor staining buffer set (Invitrogen, #00-5523-00). BTK-AF647 staining was done with BTK primary antibody (Cell Signaling, #8574S, 1:200) at 4°C for 40 minutes, and AF647 goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch, #111-605-144). For Ki67 and p-ERK1/2 staining cells were fixed, permeabilized, and stained with phycoerythrin-pERK1/2(Thr202/Tyr204) (BioLegend, catalog # 369506). CLL cells were gated on CD45+/CD19+/CD5+ using an LSRFortessa cell analyzer and FlowJo software (BD Biosciences).
In vitro stimulation, RNA sequencing, and chemokine measurements
PBMCs were depleted of CD3+ T cells using MACS cell separation columns (Miltenyi Biotec), treated with DMSO, 1 μM IBR, 2 nM hook, or 2 nM NRX-0492 for 18 hours followed by 20 μg/mL F(ab’) 2 antihuman immunoglobulin M (IgM; Jackson ImmunoResearch Laboratories) stimulation at 37°C. Total RNA was extracted from CLL cells using an RNeasy Kit (Qiagen). Sequencing libraries were constructed from 100 to 500 ng of RNA using Illumina’s TruSeq stranded total RNA kit with Ribo-Zero (catalog # 20040529). The fragment size of RNAseq libraries was verified using an Agilent 2100 Bioanalyzer (Agilent) and concentrations were determined using a Qubit instrument (LifeTech). The libraries were loaded onto an Illumina Novaseq 6000 for 2 × 50 bp paired end read sequencing.
To measure chemokine ligand 3 (CCL3) and chemokine ligand 4 (CCL4) secretion, CLL cells were seeded in 24-well plates at 1×106 cells per mL and treated as described for RNA sequencing. Supernatant was collected at 48 hours after IgM activation and centrifuged at 2000 rpm for 10 minutes. CCL3 and CCL4 concentrations were measured by enzyme-linked immunosorbent assay (Invitrogen, catalog #, 88-7035 and 88-7034, respectively).
RNAseq data analysis
Statistical analysis of gene expression was done in the R environment (v.3.5.2). Ensembl Gene IDs were mapped to gene symbols using biomaRt (v2.46.3). For a full description, please see supplemental Data.
Statistical analysis
To compare measurements between different treatment in the same patients, the paired t test was used, otherwise the unpaired t test was used (GraphPad Prism 8.0.1).
Results
Identification of NRX-0492 and biochemical characterization
To identify an efficient BTK degrader, we conjugated a pyrazine-carboxamide BTK-binding hook, with a harness based upon the cereblon (CRBN) binder thalidomide, bridged by a methylpyrrolidine linker (Figure 1A, supplemental Scheme 1).34,36 To characterize the interaction of NRX-0492 with target and ligase proteins, we examined the binding of NRX-0492 to BTK and CRBN. Using a FRET competition assay, the IC50 with BTK WT protein was determined to be 1.2 nM (Figure 1B). The IC50 with CRBN was 9 nM (supplemental Figure 1), similar to the IC50 of 53 nM observed with pomalidomide. Binding affinities of NRX-0492 to BTK proteins harboring 2 acquired inhibitor-resistance mutations (C481S or T474I) were also determined using the FRET competition assay. The IC50 values were 2.7 nM and 1.2 nM, respectively (Figure 1B). A ligand bound crystal structure of the BTK-binding hook of NRX-0492 with kinase domain of the WT BTK was solved at 1.6 Å (Figure 1C). A classical donor-acceptor-donor motif engages in 3 hydrogen bonds with main chain atoms of hinge residues, E475 and M477. An additional hydrogen bond is formed with backbone amine of C481. Common mutations at this hotspot are not expected to affect this hydrogen bond. No significant interactions are observed between the ligand and T474. The structural analysis of the binding provided the molecular basis for why NRX-0492 retains high affinity with mutations known to confer resistance to clinical-grade BTK inhibitors. Together, binding and structural analysis showed that NRX-0492 binds tightly to both BTK and CRBN and shows minimal loss of affinity to inhibitor-resistant forms of BTK.
NRX-0492 selectively degrades BTK via a cereblon (CRBN)- and proteasome-dependent mechanism. (A) Chemical structure of NRX-0492. (B) Binding of NRX-0492 to BTK WT, C481S, and T474I mutant BTK measured in a FRET competition assay. Data represent average values and standard deviations from 3 experiments. (C) Crystal structure of kinase domain of WT BTK in gray cartoon bound to the “hook” of NRX-0492 in yellow sticks. Functional kinase motives are colored for orientation: c-Helix in pink, P-loop in orange, activation loop in green, and gate/hinge in cyan. Mainchain atoms that engage in hydrogen bonding interaction are shown in sticks and the corresponding hinge region residues are labeled in black. The hydrogen bonding interactions are indicated with dashed black lines. Sidechains of mutational hotspots T474 and C481 are shown in sticks and labeled in red. (D) TMD8 cells were pretreated with MLN4924, MG132, harness, or hook at the indicated concentrations for 1 hour. Cells were then treated with NRX-0492 for an additional 4 hours, and BTK levels were quantified by total-BTK homologous time–resolved fluorescence. Each experiment was performed in triplicate. Graphs represent mean ± SD. (E) TMD8 cells were treated for 6 hours with 50 nM NRX-0492 or DMSO in triplicate. Samples were analyzed with tandem mass tag mass spectrometry. Results are graphed in a volcano plot with a statistical significance threshold of P < .01 indicated by the horizontal dotted line.
NRX-0492 selectively degrades BTK via a cereblon (CRBN)- and proteasome-dependent mechanism. (A) Chemical structure of NRX-0492. (B) Binding of NRX-0492 to BTK WT, C481S, and T474I mutant BTK measured in a FRET competition assay. Data represent average values and standard deviations from 3 experiments. (C) Crystal structure of kinase domain of WT BTK in gray cartoon bound to the “hook” of NRX-0492 in yellow sticks. Functional kinase motives are colored for orientation: c-Helix in pink, P-loop in orange, activation loop in green, and gate/hinge in cyan. Mainchain atoms that engage in hydrogen bonding interaction are shown in sticks and the corresponding hinge region residues are labeled in black. The hydrogen bonding interactions are indicated with dashed black lines. Sidechains of mutational hotspots T474 and C481 are shown in sticks and labeled in red. (D) TMD8 cells were pretreated with MLN4924, MG132, harness, or hook at the indicated concentrations for 1 hour. Cells were then treated with NRX-0492 for an additional 4 hours, and BTK levels were quantified by total-BTK homologous time–resolved fluorescence. Each experiment was performed in triplicate. Graphs represent mean ± SD. (E) TMD8 cells were treated for 6 hours with 50 nM NRX-0492 or DMSO in triplicate. Samples were analyzed with tandem mass tag mass spectrometry. Results are graphed in a volcano plot with a statistical significance threshold of P < .01 indicated by the horizontal dotted line.
The mechanism of action of NRX-0492–mediated BTK degradation was assessed in TMD8 cells (Figure 1D). To confirm BTK degradation with NRX-0492 was dependent on CRBN complexed Cullin Ring E3 ubiquitin ligase 4 (CRL4) and proteasome activity, TMD8 cells were pretreated for 1 hour with the proteasome inhibitor MG132 or the Neddylation activating enzyme inhibitor, MLN4924. Cells were then treated with NRX-0492 for 4 hours, and BTK levels were assessed by total-BTK homologous time–resolved fluorescence. Inhibition of CRL4CRBN or the proteasome precluded NRX-0492–mediated BTK degradation. In competition experiments, excess harness (CRBN ligand) or hook (BTK ligand) prevented BTK degradation (Figure 1D). These results confirm that NRX-0492 activity is proteasome-dependent and requires direct engagement with CRBN and BTK.
To determine the ability of NRX-0492 to direct degradation of CRBN neosubstrates, we quantified IKZF1 (Ikaros) and IKZF3 (Aiolos) levels in TMD8 and MOLT4 cells treated with NRX-0492.39-43 24 hours of NRX-0492 treatment resulted in dose-dependent decreases in IKZF1 and IKZF3 levels in both cell lines (supplemental Figure 2).
NRX-0492–mediated degradation of BTK is highly selective
The selectivity of NRX-0492–mediated protein degradation was assessed by tandem mass tag (TMT) proteomics in TMD8 cells treated with 50 nM NRX-0492 or DMSO for 6 hours. Levels of BTK protein were significantly and robustly decreased in NRX-0492–treated samples compared with DMSO control samples (Figure 1E, P < .0001). Levels of other proteins were minimally affected. We also found that NRX-0492 promotes minimal interleukin-2-inducible T-cell kinase degradation at concentrations at which it promotes complete BTK degradation (supplemental Figure 2B). Taken together, NRX-0492–mediated degradation is highly selective for BTK.
NRX-0492 induces rapid and complete degradation of both WT and C481S mutant BTK
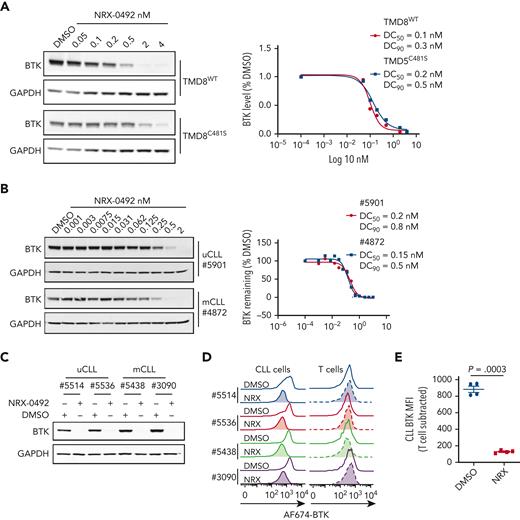
To examine dose-response and kinetics of BTK degradation, we treated TMD8 cells expressing WT or C481S mutant BTK with increasing concentrations of NRX-0492 for 4 hours and quantified BTK levels using Western blot (Figure 2A). Degradation of both WT and C481S mutant BTK was achieved at DC50 of 0.1 nM and 0.2 nM, and DC90 of 0.3 nM and 0.5 nM, respectively. BTK degradation in primary CLL cells was also achieved at subnanomolar concentrations (Figure 2B). In time course experiments with 0.2 nM NRX-0492, ≥50% reduction in BTK was achieved at 4 hours, and BTK was no longer detectable at 24 hours (supplemental Figure 3).
NRX-0492 induces degradation of WT and C481S mutant BTK. Degradation of BTK by NRX-0492 was assessed by Western blot in TMD8 (A) and primary CLL cells (B) and DC50 and DC90 were calculated based on immunoblot quantification. Comparative analysis of BTK degradation in PBMCs from 4 patients exposed to 2nM NRX-0492 for 4 hours using (C) immunoblotting and (D) flow cytometry. NRX-0492 reduced BTK staining intensity in CLL cells but not T cells. (E) Mean fluorescent intensity of anti-BTK AF647 in T cells was used as background control; comparison by paired t test. MFI, mean fluorescent intensity.
NRX-0492 induces degradation of WT and C481S mutant BTK. Degradation of BTK by NRX-0492 was assessed by Western blot in TMD8 (A) and primary CLL cells (B) and DC50 and DC90 were calculated based on immunoblot quantification. Comparative analysis of BTK degradation in PBMCs from 4 patients exposed to 2nM NRX-0492 for 4 hours using (C) immunoblotting and (D) flow cytometry. NRX-0492 reduced BTK staining intensity in CLL cells but not T cells. (E) Mean fluorescent intensity of anti-BTK AF647 in T cells was used as background control; comparison by paired t test. MFI, mean fluorescent intensity.
Next, we developed a flow cytometry assay to quantify BTK expression (supplemental Figure 4A). In CLL PBMCs treated with NRX-0492, flow cytometry and Western blotting equally detected BTK degradation (Figure 2C-D) and provided identical DC50 and DC90 concentration estimates (supplemental Figure 4B-C). Staining in T cells, which do not express BTK,44 was not affected by NRX-0492 and was used as background control (Figure 2E, supplemental Figure 4A).
NRX-0492 induces sustained E3 ligase-dependent BTK degradation in primary CLL cells
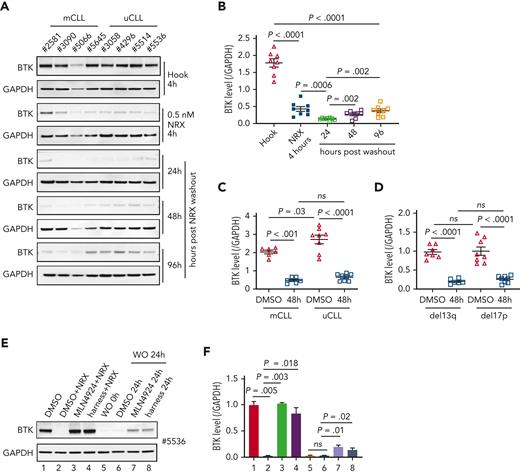
To test NRX-0492 across genetic risk groups in CLL and assess the kinetics of BTK recovery after drug exposure, we performed pulse-chase experiments. First, we treated PBMCs from 8 patients with CLL, 4 with mutated CLL (mCLL) and 4 with unmutated CLL (uCLL) with 0.5 nM NRX-0492 for 4 hours. After drug washout, BTK levels continued to decline reaching their lowest level 24 hours after washout, with only minimal recovery by 96 hours (Figure 3A-B). Pretreatment median BTK levels were ∼60% higher in uCLL than in mCLL, but BTK degradation was equally achieved in both subsets and in samples with deletion 13q or 17p (Figures 3C-D).
BTK degradation by NRX-0492 in CLL cells is sustained and equally achieved in high and standard-risk disease. (A) Immunoblots of CLL PBMCs that were treated with 0.5 nM hook or 0.5 nM NRX-0492 for 4 hours and then cultured for the indicated times after drug washout, with (B) quantification of BTK relative to loading control (GAPDH) over time. (C, D) Mean (± SEM) BTK levels in CLL PBMCs 48 hours after drug washout of vehicle (DMSO) or NRX-0492 with samples divided by (C) IGHV status into mutated CLL and unmutated CLL or (D) cytogenetic risk group, del13q (low risk), del17p (high risk). Comparisons over time by paired t test, comparisons between treatment subgroups by unpaired t test; ns, not significant. (E) Representative immunoblot showing BTK in CLL PBMCs treated as indicated: lane 1 DMSO only; lanes 2 to 4, cells were treated with DMSO, MLN4924, or harness for 1 hour followed by 2 nM NRX-0492 treatment for 4 hours. Lanes 5 to 8, cells were treated with NRX-0492 for 4 hours followed by drug washout (WO). 24 hours post washout, DMSO, MLN4924, or harness were added for an additional 24 hours. (F) Summary of experiments using cells from 3 different patients, statistics by paired t test.
BTK degradation by NRX-0492 in CLL cells is sustained and equally achieved in high and standard-risk disease. (A) Immunoblots of CLL PBMCs that were treated with 0.5 nM hook or 0.5 nM NRX-0492 for 4 hours and then cultured for the indicated times after drug washout, with (B) quantification of BTK relative to loading control (GAPDH) over time. (C, D) Mean (± SEM) BTK levels in CLL PBMCs 48 hours after drug washout of vehicle (DMSO) or NRX-0492 with samples divided by (C) IGHV status into mutated CLL and unmutated CLL or (D) cytogenetic risk group, del13q (low risk), del17p (high risk). Comparisons over time by paired t test, comparisons between treatment subgroups by unpaired t test; ns, not significant. (E) Representative immunoblot showing BTK in CLL PBMCs treated as indicated: lane 1 DMSO only; lanes 2 to 4, cells were treated with DMSO, MLN4924, or harness for 1 hour followed by 2 nM NRX-0492 treatment for 4 hours. Lanes 5 to 8, cells were treated with NRX-0492 for 4 hours followed by drug washout (WO). 24 hours post washout, DMSO, MLN4924, or harness were added for an additional 24 hours. (F) Summary of experiments using cells from 3 different patients, statistics by paired t test.
We previously found that ∼15% of BTK is newly synthesized in 24 hours.14 Therefore, we were surprised by the slow recovery of BTK levels after transient exposure to NRX-0492. To investigate whether there is a sustained drug effect after washout, we used inhibitors of E3 ligase–mediated protein degradation (Figure 3E-F). As expected, the Nedd8 E1 inhibitor MLN4924 or the E3 harness prevented NRX-0492–induced BTK degradation (Figure 3E-F; lanes 1-4). In PBMCs treated with NRX-0492 for 4 hours, followed by drug washout, BTK levels were ∼2% of the pretreatment levels at 24 hours. With the addition of MLN4924 or the E3 harness, BTK levels increased to 18% and 11% of pretreatment levels, respectively (Figure 3E-F; lanes 5-8), suggesting that intracellular NRX-0492 was still present and active.
In summary, NRX0492 was equally potent in uCLL and mCLL and in samples with high-risk cytogenetics. NRX0492-mediated BTK degradation was sustained after drug washout for at least 24 hours.
NRX-0492 has minimal direct cytotoxic effects
At concentrations achieved in vivo, in vitro treatment of CLL cells with IBR only modestly decreases cell viability.45,46 Here we compared CLL cell viability in PBMCs exposed to IBR or NRX-0492 in vitro over 3 days. Only at 72 hours, did we observe a statistically significant decrease in cell viability over controls (P = .02; supplemental Figure 5A). By Annexin V or propidium iodide staining, a slight increase in the percent of apoptotic CLL cells exposed to ibrutinib or NRX-0492 for 48 hours was not statistically significant (supplemental Figure 5B-D).
NRX-0492 inhibits BCR and NF-κB signaling
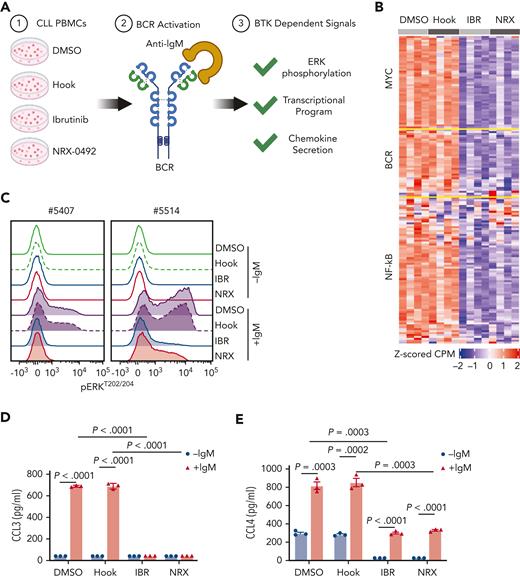
We previously characterized transcriptional changes in CLL cells from patients treated with BTKi demonstrating inhibition of BCR, NF-κB, and MYC signaling as pharmacodynamic measures of ontarget effects.12-14 To compare the effects of NRX-0492 and ibrutinib on the tumor transcriptome, we performed RNA sequencing of CLL cells from 4 patients treated for 18 hours in vitro with either 1 μM ibrutinib or 2 nM NRX-0492 or the controls, DMSO or hook, respectively, followed by anti-IgM stimulation for 6 hours (Figure 4A-B). Flow cytometry for CD69 and Western blot for BTK were used to confirm IgM-mediated activation of CLL cells and the expected drug effects (supplemental Figure 6A-B). In NRX and ibrutinib treated conditions, we observed strong downregulation of BCR, NF-κB, and MYC regulated genes consistent with effective inhibition of BTK-dependent signaling (Figure 4B; supplemental Table 2).
BTK inhibition and BTK degradation have comparable effects on CLL biology. (A) In vitro assays using CLL PBMCs to investigate the impact of different drugs on BTK-dependent signaling in response to BCR engagement. (B) CLL cell transcriptome assessed by RNA sequencing of CLL cells from 4 patients treated with DMSO, hook, 1 μM ibrutinib (IBR), or 2 nM NRX-0492, DMSO for 18 hours followed by 20 μg/mL anti-IgM stimulation for 6 hours. Median centered heatmap scaled as indicated depicts changes in defined gene signatures representing MYC, BCR, and NF-κB regulated genes. Each row represents a gene and each column represents a sample. supplemental Table 2 lists the genes in the same sequence as shown in the heat map together with the expression value across the different conditions. (C) Histogram depicting flow cytometry staining of p-ERK1/2Thr202/Tyr204 in CLL cells from 2 representative patients, drug treatment as in B, anti-IgM stimulation for 15 minutes. (D) CCL3 and (E) CCL4 concentrations by enzyme-linked immunosorbent assay in supernatant of CLL cells treated as in B, cell culture supernatants collected 48 hours after stimulation. ERK, extracellular signal-regulated kinase.
BTK inhibition and BTK degradation have comparable effects on CLL biology. (A) In vitro assays using CLL PBMCs to investigate the impact of different drugs on BTK-dependent signaling in response to BCR engagement. (B) CLL cell transcriptome assessed by RNA sequencing of CLL cells from 4 patients treated with DMSO, hook, 1 μM ibrutinib (IBR), or 2 nM NRX-0492, DMSO for 18 hours followed by 20 μg/mL anti-IgM stimulation for 6 hours. Median centered heatmap scaled as indicated depicts changes in defined gene signatures representing MYC, BCR, and NF-κB regulated genes. Each row represents a gene and each column represents a sample. supplemental Table 2 lists the genes in the same sequence as shown in the heat map together with the expression value across the different conditions. (C) Histogram depicting flow cytometry staining of p-ERK1/2Thr202/Tyr204 in CLL cells from 2 representative patients, drug treatment as in B, anti-IgM stimulation for 15 minutes. (D) CCL3 and (E) CCL4 concentrations by enzyme-linked immunosorbent assay in supernatant of CLL cells treated as in B, cell culture supernatants collected 48 hours after stimulation. ERK, extracellular signal-regulated kinase.
Next, we assessed ERK phosphorylation by flow cytometry and Western blotting in CLL cells pretreated with ibrutinib or NRX-0492 for 24 hours followed by anti-IgM stimulation (supplemental Figure 6C-D). Anti-IgM–induced phosphorylation of ERK was strongly inhibited by both ibrutinib and NRX-0492 (Figure 4C). Finally, we measured CCL3 and CCL4 secretion in the supernatant of CLL cells cultured in vitro (Figure 4D-E).47 As expected, BCR engagement stimulated CCL3 and CCL4 secretion in control treated cells (P < .001) which was greatly reduced by both ibrutinib and NRX-0492 (P < .001). Notably, both drugs also abolished baseline secretion of CCL4. Taken together, NRX-0492 is as effective as ibrutinib in downregulating BTK-dependent signaling, transcriptional programs, and chemokine secretion.
NRX-0492 inhibits treatment-naïve CLL cell proliferation and activation in vivo
Next, we tested NRX-0492 in a well-established PDX model,38,48-50 that we previously used to investigate covalent BTK inhibitors.38,51 For each experiment, 10 mice were injected with PBMCs from 1 patient and then equally divided to treatment with NRX-0492 or vehicle for 21 days (Figure 5A). A representative experiment is shown in detail in Figure 5. On day 8, BTK expression in peripheral blood cells was reduced in all mice treated with NRX-0492 compared with vehicle controls (Figure 5B-C; supplemental Figure 7A). On day 22, mice were euthanized. BTK levels were significantly lower in CLL cells from mice treated with NRX-0492 in both peripheral blood (P < .0001; Figure 5D) and spleen (P = .004; Figure 5E).
BTK degradation and anti-CLL activity of NRX-0492 in PDX model. In vivo activity was tested in the PDX model of CLL. Ten NSG mice were inoculated with PBMCs from patient #5539 with CLL. (A) Schematic display of the experimental schedule. (B) Representative histogram of AF647-BTK staining in CLL cells from peripheral blood (pb) on day1 before drug treatment (left) and after 7 days of treatment with NRX-0492 (day 8, right). (C-E) Mean (± SEM) CLL AF647-BTK MFI in peripheral blood on day 8 (C), day 22 (D), and spleen on day 22 (E). Each symbol represents 1 mouse. (F, G) Mean (± SEM) percentage of Ki67 expression in CLL cells in peripheral blood on day 8 (F), day 22 (G), and spleen on day 22 (H). (I, J) The percentage of CD69 positive CLL cells in peripheral blood (I) and spleen (J) on day 22. (K) Mean total cell counts (± SEM) of human cells (CD45+), T cells, and CLL cells in spleen on day 22. Comparisons are by unpaired student t test; ns, not significant. Veh, vehicle; NRX, NRX-0492; pb, peripheral blood; sp, spleen.
BTK degradation and anti-CLL activity of NRX-0492 in PDX model. In vivo activity was tested in the PDX model of CLL. Ten NSG mice were inoculated with PBMCs from patient #5539 with CLL. (A) Schematic display of the experimental schedule. (B) Representative histogram of AF647-BTK staining in CLL cells from peripheral blood (pb) on day1 before drug treatment (left) and after 7 days of treatment with NRX-0492 (day 8, right). (C-E) Mean (± SEM) CLL AF647-BTK MFI in peripheral blood on day 8 (C), day 22 (D), and spleen on day 22 (E). Each symbol represents 1 mouse. (F, G) Mean (± SEM) percentage of Ki67 expression in CLL cells in peripheral blood on day 8 (F), day 22 (G), and spleen on day 22 (H). (I, J) The percentage of CD69 positive CLL cells in peripheral blood (I) and spleen (J) on day 22. (K) Mean total cell counts (± SEM) of human cells (CD45+), T cells, and CLL cells in spleen on day 22. Comparisons are by unpaired student t test; ns, not significant. Veh, vehicle; NRX, NRX-0492; pb, peripheral blood; sp, spleen.
BCR signaling promotes proliferation and activation of CLL cells.3,4 Consistent with our previous study,38 the fraction of proliferating (Ki67+) and activated (CD69+) CLL cells was higher in the spleen than in the peripheral blood (supplemental Figure 7B-C). NRX-0492 greatly reduced the fraction of Ki67+ CLL cells in peripheral blood on day 8 (P = .002) and 22 (P = .003) and in the spleens on day 22 (P = .02; Figure 5F-H). There was no effect of NRX-0492 on T-cell proliferation (supplemental Figure 7D-F). NRX-0492 also significantly reduced the fraction of CD69+ CLL cells in the peripheral blood (P = .0003; Figure 5I) and spleen (P = .01; Figure 5J). Finally, CLL tumor burden in the spleens was reduced in NRX-0492–treated mice compared with controls (P = .001; Figure 5K).
The results from PDX experiments using PBMCs from 4 additional patients are summarized in Figure 6. Compared to vehicle treated mice, NRX-0492 consistently promoted BTK degradation in CLL cells in vivo in both blood and spleen (P < .0001; Figure 6A-B) and significantly reduced the fraction of Ki67+ CLL cells in both compartments (P < .0001; Figure 6C). In the PDX models of all 4 patients, total CLL tumor burden in the spleens was reduced by NRX-0492 treatment compared with vehicle controls (Figure 6D). In summary, NRX-0492 effectively degrades BTK in vivo resulting in inhibition of tumor proliferation and a decrease in tumor burden in the CLL PDX model of treatment- naïve CLL.
Summary of NRX-0492 testing in CLL PDX model. Experimental design as in Figure 5. PBMCs from 4 different patients (represented by unique symbols) were inoculated into NSG mice that were divided to vehicle control (Veh) and NRX-0492 (NRX) treatment groups. Each symbol represents 1 mouse. (A) Mean (± SEM) MFI of AF647-BTK in CLL cells in peripheral blood of mice after 7 days of NRX-0492 treatment. (B) Mean (± SEM) MFI of AF647-BTK in CLL cells from spleens on day 22. (C) Mean (± SEM) percentage of CLL cells expressing Ki67 in peripheral blood before treatment start (Day 1) and after 1 week of treatment and in cells from spleens on day 22. (D) Mean (± SEM) percentage of live CLL cells among CD45+ cells in the spleens on day 22. Statistical analysis by unpaired student t test; ns, not significant.
Summary of NRX-0492 testing in CLL PDX model. Experimental design as in Figure 5. PBMCs from 4 different patients (represented by unique symbols) were inoculated into NSG mice that were divided to vehicle control (Veh) and NRX-0492 (NRX) treatment groups. Each symbol represents 1 mouse. (A) Mean (± SEM) MFI of AF647-BTK in CLL cells in peripheral blood of mice after 7 days of NRX-0492 treatment. (B) Mean (± SEM) MFI of AF647-BTK in CLL cells from spleens on day 22. (C) Mean (± SEM) percentage of CLL cells expressing Ki67 in peripheral blood before treatment start (Day 1) and after 1 week of treatment and in cells from spleens on day 22. (D) Mean (± SEM) percentage of live CLL cells among CD45+ cells in the spleens on day 22. Statistical analysis by unpaired student t test; ns, not significant.
NRX-0492 degrades C481S mutant BTK in CLL cells from patients progressing on IBR
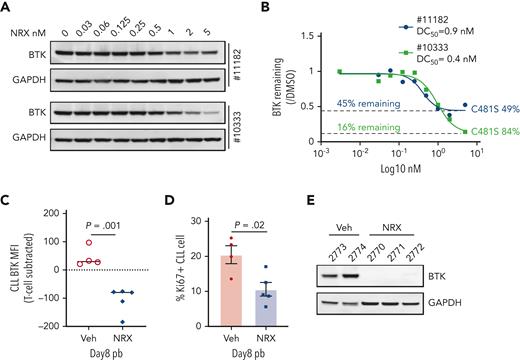
Patients with CLL who progress on IBR commonly harbor BTK C481 mutations that prevent covalent drug binding.23,24 We tested NRX-0492 in vitro using PBMCs from 2 patients with progressive disease and C481S mutant BTK with cancer cell fractions of 49% and 84%, respectively (Figure 7A-B). BTK degradation was achieved with DC50 <1nM in both patients but remained incomplete (Figure 7A). As both patients were still on active therapy with IBR at the time of sample collection, we reasoned that the fraction of WT BTK was covalently bound by IBR and thus protected from being engaged by NRX-0492. Indeed, the amount of BTK remaining was the inverse of the C481S cancer cell fractions (Figure 7B). Thus, NRX-0492 effectively degraded C481S mutant BTK in primary CLL cells, whereas WT BTK complexed with ibrutinib was protected from degradation.
Activity of NRX-0492 against IBR-resistant CLL with BTK C481S mutation. (A) PBMCs from 2 patients with CLL with clinical progression on IBR and BTK C481S mutations were treated with NRX-0492 for 24 hours at the indicated concentrations and analyzed for BTK expression by Western blotting. (B) Estimates of DC50 and DC90 concentrations based on data shown in A. The fraction of residual BTK and the cancer cell fractions of C481S mutations for the samples tested is indicated (C-F) PBMCs from patient with CLL #12803 who progressed on IBR with BTK C481S were used to establish the PDX model. (C) BTK and (D) Ki67 expression by flow cytometry in peripheral blood CLL cells of vehicle (Veh) and NRX-0492 treated mice on day 8; (E) Western blotting for BTK in CLL cells, selected by CD3 negative selection, from spleens on day 20.
Activity of NRX-0492 against IBR-resistant CLL with BTK C481S mutation. (A) PBMCs from 2 patients with CLL with clinical progression on IBR and BTK C481S mutations were treated with NRX-0492 for 24 hours at the indicated concentrations and analyzed for BTK expression by Western blotting. (B) Estimates of DC50 and DC90 concentrations based on data shown in A. The fraction of residual BTK and the cancer cell fractions of C481S mutations for the samples tested is indicated (C-F) PBMCs from patient with CLL #12803 who progressed on IBR with BTK C481S were used to establish the PDX model. (C) BTK and (D) Ki67 expression by flow cytometry in peripheral blood CLL cells of vehicle (Veh) and NRX-0492 treated mice on day 8; (E) Western blotting for BTK in CLL cells, selected by CD3 negative selection, from spleens on day 20.
Next, we sought to test the activity of NRX-0492 against ibrutinib resistant CLL in the PDX model. PBMCs from a patient who had been on ibrutinib for over 7 years before progressing with CLL harboring C481S at 88%, cancer cell fractions were injected into 10 NSG mice that were equally divided to treatment with vehicle or NRX-0492. Drug-induced BTK degradation and inhibition of proliferation was demonstrated in peripheral blood CLL cells after 1 week of treatment with NRX-0492 using flow cytometry (Figure 7C-D). After 3 weeks of continuous therapy, spleen infiltrating CLL cells were purified from 5 mice and analyzed by Western blotting (Figure 7E). In drug treated mice, BTK was undetectable, consistent with complete degradation of both WT BTK, newly synthesized during the course of the experiment,14 and C481 mutant BTK by NRX-0492. Limited sample availability and poor engraftment of CLL PBMCs from patients after long-term therapy with ibrutinib precluded further testing in the PDX model.
Discussion
Degradation of mutant BTK represents a strategy to overcome resistance to BTKis. A key difference in the mechanism of action of NRX-0492 and BTKis is the required ontarget residence time. BTKis are most effective when virtually all BTK molecules are constantly occupied by drug and even small reductions in target occupancy can translate into more BTK-dependent signaling.14 Recently, short BTK residence time emerged as a likely explanation for the disappointing clinical activity of the noncovalent BTKi vecabrutinib.28 In contrast, NRX-0492 recruits the enzymatic activity of the cereblon E3 ligase complex leading to ubiquitination and proteasomal degradation of BTK. Thus, even with transient drug binding, BTK is irreversibly targeted for destruction. NRX-0492, therefore does not require sustained binding and can degrade both WT and C481 mutant BTK in vitro and in vivo. A ligand bound crystal structure of the hook of NRX-0492 with kinase domain of the WT BTK explains how NRX-0492 retains high affinity with mutations known to confer resistance to clinical-grade BTKis. Using FRET assays we confirmed no change in affinity with the gatekeeper mutation T474I or the C481S mutation compared with WT. In contrast, a substantial loss of binding of noncovalent BTKis to T474I has been reported.31
In a broad survey of kinases, BTK was identified as a target that can be readily degraded when brought into proximity of the cereblon E3 ligase complex by a small molecule,52 and several BTK degraders have been described.52,53 NRX-0492 belongs to a new class of targeted BTK degraders we developed combining a noncovalent BTK-binding hook with a cereblon recruiting harness.34-36 Depending on the structure of the chemical linker and the cereblon binding harness, these targeted protein degraders may also degrade cereblon neosubstrates such as IKZF1 and IKZF3. NRX-0492, NX-2127, and NX-5948 all degrade BTK at low or subnanomolar concentrations but differ in their activity against cereblon neosubstrates. NRX-0492 and NX-2127 catalyze the degradation of IKZF1 and IKZF3, whereas NX-5948 does not.35,36 Standout features of these molecules include >90% degradation of BTK at sub to low nanomolar concentrations, their oral bioavailability, and sustained pharmacodynamic effects after drug washout. Based on these favorable characteristics, these BTK targeted protein degraders are among the first in this new class of drugs to have entered clinical testing.54
Estimation of on-target activity of covalent BTKis has been facilitated by assays quantifying target occupancy. In patients with CLL treated with BTKis, steady state occupancy of BTK typically exceeds 90%.14 Commonly reported additional pharmacodynamic readouts are downregulation of BCR and NF-ĸB target genes and secretion of CCL3 and CCL4 chemokines.12-14,55,56 To assess on-target effects of NRX-0492, we tracked BTK protein expression and downstream signaling. In vitro, we observed rapid and sustained degradation of BTK at low nanomolar concentrations with NRX-0492, achieving > 90% reduction in BTK levels. In treatment-naïve CLL samples, IBR and NRX-0492 equally inhibited BCR-dependent transcriptional programs and chemokine secretion.
In PDX models, NRX-0492–induced BTK degradation in vivo and inhibited CLL cell activation, proliferation, and expansion. In vivo activity of NRX-0492 was comparable to our prior experience with IBR and acalabrutinib in the same model.38,51 We also tested NRX-0492 in the PDX model using PBMCs from a patient with CLL progressing on IBR with the classic C481S mutation. NRX-0492 effectively degraded mutant BTK and inhibited CLL cell activation and proliferation. Extension of these experiments to more patients was hampered by limited sample availability and poor engraftment of cells from these patients. Nevertheless, in aggregate our data show activity of NRX-0492 against WT and C481 mutant CLL in vitro and in vivo.
Although BTK degraders have only just entered clinical studies, the emerging experience with covalent BTKi provides valuable insights. First, CLL progressing on covalent BTKi remains dependent on BTK and retargeting C481 mutant CLL is a viable clinical strategy.29,30 Second, BTK mutations other than C481 emerge in patients progressing on noncovalent BTKi.31 Alternative strategies for BTK targeting will therefore gain in importance. Targeted protein degraders hold promise as a novel therapeutic strategy to overcome BTKi resistance, and clinical studies in B-cell malignancies are ongoing (NCT04830137, NCT05131022).
Acknowledgments
The authors thank the patients for donating samples to make this research possible. They acknowledge the NHLBI genomics core for RNA sequencing.
This work was funded by the Intramural Research Program of the NHLBI, NIH and Nurix Therapeutics.
Authorship
Contribution: S.E.M.H., D.W.R., M.N., A.W. were responsible for conceptualization; D.Z., H.M.H., J.C., J.J., G.J., A.K., J.M., A.T.-M., E.A., Y.S.T, S.G., M.C.C., S.Y. performed investigation; D.Z., H.M.H., J.C., J.J., M.P., S.S.S., E.G., M.M., C.U., S.G., M.C.C., D.W.R., M.N. were responsible for analysis; D.Z., J.C., J.J, A.W, M.N, H.L. performed visualization; E.G., M.M., C.U., I.E.A., C.S., J.M. provided reagents and patient samples; D.Z., A.W., M.K., H.L. wrote the original draft; A.W. acquired funding; and all the authors were involved with writing, reviewing and editing of the manuscript.
Conflict-of-interest disclosure: A.K., J.M., A.T.-M., E.A., Y.S.T., H.L., S.G., M.C.C., S.Y., D.W.R., and M.N. were or currently are full-time employees and shareholders of Nurix Therapeutics. A.W. received research funding from Pharmacyclics, Acerta Pharma, Merck, Verastem, Genmab, and Nurix Therapeutics. C.S. received research funding from Genmab. The remaining authors declare no competing financial interests.
Correspondence: Adrian Wiestner, Building 10, CRC 3-5140, 10 Center Drive, Bethesda, MD 20892-1202; e-mail: wiestnera@mail.nih.gov.
References
Author notes
This study was presented, in part, in abstract form at the 63rd annual meeting of the American Society of Hematology, Orlando, FL, 7 to 9 December 2019.
The RNAseq data are available in GEO with accession number GSE198992.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal