Key Points
Naturally occurring CD7 negative T cells are functional effector T cells that can be used for chimeric antigen receptor therapy.
CD7 negative T cells expressing a CD7-CAR have robust antitumor activity and bypass fratricide.
Abstract
Chimeric antigen receptor (CAR) T-cell therapy targeting T-cell acute lymphoblastic leukemia (T-ALL) faces limitations such as antigen selection and limited T-cell persistence. CD7 is an attractive antigen for targeting T-ALL, but overlapping expression on healthy T cells leads to fratricide of CD7-CAR T cells, requiring additional genetic modification. We took advantage of naturally occurring CD7− T cells to generate CD7-CAR (CD7-CARCD7−) T cells. CD7-CARCD7− T cells exhibited a predominantly CD4+ memory phenotype and had significant antitumor activity upon chronic antigen exposure in vitro and in xenograft mouse models. Based on these encouraging results, we next explored the utility of CD7− T cells for the immunotherapy of CD19+ hematological malignancies. Direct comparison of nonselected (bulk) CD19-CAR and CD19-CARCD7− T cells revealed that CD19-CARCD7− T cells had enhanced antitumor activity compared with their bulk counterparts in vitro and in vivo. Lastly, to gain insight into the behavior of CD19-CAR T cells with low levels of CD7 gene expression (CD7lo) in humans, we mined single-cell gene and T-cell receptor (TCR) expression data sets from our institutional CD19-CAR T-cell clinical study. CD19-CARCD7lo T cells were present in the initial CD19-CAR T-cell product and could be detected postinfusion. Intriguingly, the only functional CD4+ CD19-CAR T-cell cluster observed postinfusion exhibited CD7lo expression. Additionally, samples from patients responsive to therapy had a higher proportion of CD7lo T cells than nonresponders (NCT03573700). Thus, CARCD7− T cells have favorable biological characteristics and may present a promising T-cell subset for adoptive cell therapy of T-ALL and other hematological malignancies.
Introduction
Adoptive transfer of chimeric antigen receptor (CAR) T cells has been shown to be effective for the treatment of B-cell malignancies, leading to its Food and Drug Administration approval.1-3 However, expanding the therapeutic application of CAR T cells for other hematological malignancies has been challenging. This is largely due to the lack of an ideal antigen, especially in the context of T-cell malignancies where there is a widespread overlap in antigen expression on T-cell acute lymphoblastic leukemia (T-ALL) blasts and healthy T cells,4 resulting in T-cell fratricide that compromises product effectiveness.
CD7 is a transmembrane protein highly expressed on T-cell malignancies, including most T-ALL blasts,5,6 T-cell lymphomas, and a subset of peripheral T-cell lymphomas.7 Canonically, CD7 plays a role in T-cell activation and is found on thymocytes, mature T cells, natural killer cells, and monocytes.8,9 Because of it ubiquitous expression, it is a promising target for T-ALL. However, to develop an effective CD7-specific CAR T-cell strategy, it is essential to bypass fratricide. Investigators have used CD7 disruption techniques, including gene and base editing using Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9),10-12 and have also sequestered CD7 internally using a protein expression blocker to create fratricide-resistant CD7-CAR T cells.13 Some of these approaches are being tested in early-phase clinical trials, with encouraging results (#NCT04004637, #ChiCTR2000034762, #NCT03690011; clinicaltrials.gov).14,15
Rather than genetically engineering T cells to render them CD7−, naturally occurring CD7− T cells16 might be a promising cell source for generating CD7-CAR T cells. These CD7− T cells have a predominantly CD4+ memory phenotype and typically display a Th0/Th2 phenotype in post-transplant or other immunodeficiency settings.16,17 Here, we evaluate if expressing CD7-CAR on the CD7− T-cell subset could bypass fratricide without the need for further genetic engineering while maintaining robust antitumor activity. In addition, we determine the antitumor activity of CD7− T cells expressing CD19-CARs in a B cell acute lymphoblastic leukemia (B-ALL) xenograft model and investigate the fate of T cells with low CD7 gene expression (CD7lo) in pre- and postinfusion samples of our institutional CD19-CAR T-cell therapy clinical study (#NCT03573700; clinicaltrials.gov). Our results demonstrate that CD7− T cells have favorable biological characteristics, warranting further active exploration of this T-cell subset for adoptive cell therapies.
Methods
Full methods can be found in supplemental Materials (available on the Blood Web site).
Cytotoxicity, repeat stimulation, and cytokine multiplex assays
Cytotoxicity, repeat stimulation, and cytokine multiplex assays are described in detail in supplemental Methods.
Single-cell gene expression analysis
We mined previously analyzed single-cell gene expression data of CD19-CAR T cells from good manufacturing practice–laboratory CAR T-cell products, and post-infusion samples were obtained from a St. Jude investigator-initiated clinical study evaluating the safety and efficacy of autologous CD19-CAR T cells in pediatric and young adult patients with relapsed/refractory CD19+ B-ALL (#NCT03573700; clinicaltrials.gov).18,19 These data were analyzed within the Seurat framework20 using the published cluster annotations. The VlnPlot and DotPlot functions were used to generate violin plots and dot plots, respectively.
Bulk TCR library preparation and sequencing
T-cell receptor (TCR) library preparation and sequencing methods are described in detail in supplemental Methods.
Processing bulk TCR sequencing
Demultiplexing and contig assembly of paired-end FASTQ reads was accomplished using migec (v1.2.9).21 VDJ junction mapping and clonotype assembly and annotation using the assembled contigs was done with mixcr (v3.0.13).22 The vdjtools v1.2.123 FilterNonFunctional, Correct, and Decontaminate functions were used on the filtered clonotype table outputs from mixcr for additional quality control to remove erroneous clonotypes due to PCR errors and crosscontaminating sequences between samples. The Immunarch (v0.6.6) package (ImmunoMind Team 2019)24 for R was used to measure sample diversity and clonality. The Top10 index, calculated as the cumulative relative abundance of the 10 largest clones in the bulk repertoire sample, was used to indicate clonality in Figure 4C. The donor 4, CD19-CARBulk, time point 3 repertoire sample is an outlier; it contained only 20 sequences, resulting in an inflated value.
Xenograft model
In vivo experiments were performed under a protocol approved by the St. Jude Institutional Animal Care and Use Committee. See supplemental Methods for full description.
Statistics
Data were summarized using descriptive statistics. Comparisons of continuous variables among ≥3 groups were made by one-way analysis of variance (ANOVA), 2-way ANOVA, and repeated measures, whereas comparisons between 2 groups were made by t-test or Wilcoxon rank-sum test as appropriate. We adjusted for multiple comparisons using Holm-Šídák's method. Survival times from tumor cell injection in the mouse experiments were analyzed by the Kaplan-Meier curves with Mantel-Cox log-rank test method. GraphPad Prism 8 software (GraphPad software) and R 3.6.0 (Lucent Technologies, New Providence, NJ) were used for statistical analysis. P values <.05 were considered statistically significant.
Results
Generation of fratricide-resistant CD7− CAR T cells using selection technology
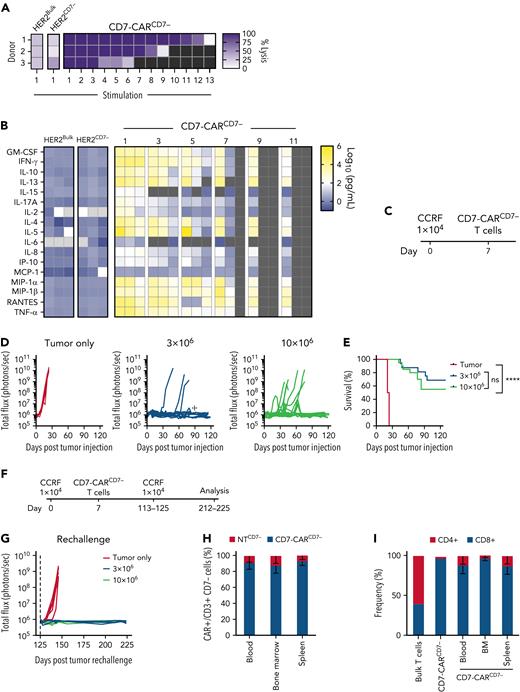
To assess the feasibility of generating fratricide-resistant CD7-CAR T cells using naturally occurring CD7− T cells, we first determined the frequency of CD7− T cells in the peripheral blood of healthy donor and patients with B-ALL or T-ALL at diagnosis and after at least 2 cycles of chemotherapy (Figure 1A-B). CD7− T-cell populations were present in all samples, ranging from 0.72% to 19.5% in healthy donors and 3% to 12.5% in patients.
CD7− T cells are predominantly CD4+ memory cells and are resistant to fratricide when expressing CD7-CAR. (A-B) Flow cytometric analysis of surface expression of CD3 and CD7 in (A) bulk peripheral blood mononuclear cells (PBMCs) from healthy donors: CD3+CD7+ (mean, 94.37 ± 4.55%) vs CD3+CD7− (mean, 5.18 ± 4.51%) (N = 50, P < .0001, paired t test) and (B) B-ALL patients: CD3+ CD7+ (mean, 93.4 ± 3.8%) and CD3+ CD7− (mean, 6.5 ± 3.8%) (N = 5, P < .0001, paired t test) and T-ALL patients: CD3+ CD7+ (mean, 95.6 ± 3.6) and CD3+CD7− (mean, 4.4 ± 3.6%) (N = 10, P < .0001 paired t test). (C) Representative dot plots of PBMCs before and after CD7 depletion and CD3 enrichment (left) and efficiency of CD7 selection on day 10 post–activation of nontransduced (NT) T cells (CD7+: 8.27 ± 4.61% and CD7−: 91.36 ± 4.56%, range 78.2-96.4, N = 26, P < .0001, paired t test). (D) Schematic of CD7-CAR construct. (E) Expression of CD7-CAR by flow cytometry using F(ab’)2 on day 7 after transduction (mean transduction efficiency 65.9 ± 16.7%, N = 22, P < .0001, paired t test). Representative dot plot showing all remaining CD7+ T cells were eliminated post–CD7-CAR transduction. (F) Expansion kinetics (absolute cell count) and viability (trypan blue exclusion). CD7-CARCD7− T cells had similar expansion to NT T cells (N = 11, P = ns, 2-way ANOVA). Bulk CD7-CAR T cells did not expand (N = 11, P < .01, 2-way ANOVA) and had decreased viability compared with CD7-CARCD7− and NT T cells (N = 11, P < .01, 2-way ANOVA). (G) Immunophenotype of CD7-CARCD7− T cells on day 7 posttransduction (CD4 vs CD8, effector memory [EM]: CCR7−, CD45RA−; central memory [CM]: CCR7+, CD45RA−; naïve-like: CCR7+CD45RA+; EM T cells reexpress CD45RA, EMRA: CCR7−, CD45RA+) (N = 11, 2-way ANOVA, bulk T cells vs CD7− T cells: CD4 vs 8 P < .0001, EM P < .0001, Naïve P < .0001, EMRA P < .01). (H) Luciferase-based cytotoxicity assay of CD7-CARCD7− T cells against CCRF (CD7+) and control BV173 (CD7−) target cells at 7 different effector:target (E:T) ratios (N = 8, P < .001, 2-way ANOVA at all ratios). (I) CD7-CARCD7− T cells were cocultured with media, BV173 (CD7lo target), CCRF or MOLT3 (CD7+ targets) at a 2:1 E:T ratio. Supernatants were harvested after 24 hours and analyzed for IFN-γ or IL-2 by ELISA (IFN-γ: N = 7-9, IL-2 N = 7-9, 1-way ANOVA, ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001). IgG1 Fc, immunoglobulin 1 fragment crystallizable; scFv, single-chain variable fragment; ELISA, enzyme-linked immunosorbent assay; ns, not significant.
CD7− T cells are predominantly CD4+ memory cells and are resistant to fratricide when expressing CD7-CAR. (A-B) Flow cytometric analysis of surface expression of CD3 and CD7 in (A) bulk peripheral blood mononuclear cells (PBMCs) from healthy donors: CD3+CD7+ (mean, 94.37 ± 4.55%) vs CD3+CD7− (mean, 5.18 ± 4.51%) (N = 50, P < .0001, paired t test) and (B) B-ALL patients: CD3+ CD7+ (mean, 93.4 ± 3.8%) and CD3+ CD7− (mean, 6.5 ± 3.8%) (N = 5, P < .0001, paired t test) and T-ALL patients: CD3+ CD7+ (mean, 95.6 ± 3.6) and CD3+CD7− (mean, 4.4 ± 3.6%) (N = 10, P < .0001 paired t test). (C) Representative dot plots of PBMCs before and after CD7 depletion and CD3 enrichment (left) and efficiency of CD7 selection on day 10 post–activation of nontransduced (NT) T cells (CD7+: 8.27 ± 4.61% and CD7−: 91.36 ± 4.56%, range 78.2-96.4, N = 26, P < .0001, paired t test). (D) Schematic of CD7-CAR construct. (E) Expression of CD7-CAR by flow cytometry using F(ab’)2 on day 7 after transduction (mean transduction efficiency 65.9 ± 16.7%, N = 22, P < .0001, paired t test). Representative dot plot showing all remaining CD7+ T cells were eliminated post–CD7-CAR transduction. (F) Expansion kinetics (absolute cell count) and viability (trypan blue exclusion). CD7-CARCD7− T cells had similar expansion to NT T cells (N = 11, P = ns, 2-way ANOVA). Bulk CD7-CAR T cells did not expand (N = 11, P < .01, 2-way ANOVA) and had decreased viability compared with CD7-CARCD7− and NT T cells (N = 11, P < .01, 2-way ANOVA). (G) Immunophenotype of CD7-CARCD7− T cells on day 7 posttransduction (CD4 vs CD8, effector memory [EM]: CCR7−, CD45RA−; central memory [CM]: CCR7+, CD45RA−; naïve-like: CCR7+CD45RA+; EM T cells reexpress CD45RA, EMRA: CCR7−, CD45RA+) (N = 11, 2-way ANOVA, bulk T cells vs CD7− T cells: CD4 vs 8 P < .0001, EM P < .0001, Naïve P < .0001, EMRA P < .01). (H) Luciferase-based cytotoxicity assay of CD7-CARCD7− T cells against CCRF (CD7+) and control BV173 (CD7−) target cells at 7 different effector:target (E:T) ratios (N = 8, P < .001, 2-way ANOVA at all ratios). (I) CD7-CARCD7− T cells were cocultured with media, BV173 (CD7lo target), CCRF or MOLT3 (CD7+ targets) at a 2:1 E:T ratio. Supernatants were harvested after 24 hours and analyzed for IFN-γ or IL-2 by ELISA (IFN-γ: N = 7-9, IL-2 N = 7-9, 1-way ANOVA, ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001). IgG1 Fc, immunoglobulin 1 fragment crystallizable; scFv, single-chain variable fragment; ELISA, enzyme-linked immunosorbent assay; ns, not significant.
To select naturally occurring CD7− T cells, we used bulk healthy donor PBMCs and performed 2-step magnetic bead separation, starting with CD7 depletion and followed by CD3 enrichment to obtain CD3+ CD7− T cells (supplemental Figure 1A). After CD7 selection, we consistently recovered >78% CD7− T cells, indicating a highly efficient selection protocol (Figure 1C). We then transduced CD7− T cells to express a previously described10 second-generation CD7-CAR (CD7-CARCD7−) (Figure 1D) or a control CAR (HER2-CAR; supplemental Figure 1B). Resulting CD7-CAR expression ranged from 31.4% to 91.2%, and any remaining CD7+ T cells present after selection were eliminated via fratricide mediated by the CD7-CAR (Figure 1E). CD7-CARCD7− T cells exhibited similar expansion kinetics viability and expansion compared with NTCD7− and NTBulk T cells, whereas bulk T cells expressing CD7-CAR (CD7-CARBulk) failed to expand due to fratricide (Figure 1F; supplemental Figure 1C-E). Flow cytometry immunophenotyping of cells on day 7 showed that CD7− T cells (NTCD7−, HER2-CARCD7−, or CD7-CARCD7−) had a predominantly CD4+ memory phenotype in comparison with bulk NT T cells (Figure 1G; supplemental Figure 1F). There was no increase in the expression of PD1, TIM3, or LAG3 immune checkpoint receptors in CD7-CARCD7− T cells as assessed by flow cytometry (supplemental Figures 1G-J and 2).
CD7-CARCD7− recognize and lyse target cells in an antigen-dependent manner
To determine antigen specificity of CD7-CARCD7− T cells, we used CD7+ (CCRF, MOLT3) or CD7lo (BV173) tumor cell lines (supplemental Figure 3A). We examined the antitumor activity of CD7-CARCD7− T cells using a luciferase-based cytotoxicity assay against luciferase-expressing target cells at multiple E:T ratios. When cocultured with CCRF cells (CD7+), CD7-CARCD7− T cells were the only CAR T-cell population that showed robust antigen-dependent antitumor activity (Figure 1H). In contrast, only background killing of BV173 (CD7lo) targets was observed by CD7-CARCD7− T cells, which was not significantly different from control CAR T cells (Figure 1H; supplemental Figure 3B-C). Antigen specificity was confirmed by determining cytokine production (interferon γ [IFN-γ], interleukin 2 [IL-2]) after coculturing CD7-CARCD7− T cells with CD7+ targets (CCRF or MOLT3) or CD7lo target cells (BV173) at an E:T ratio of 2:1. Although CD7-CARCD7− T cells produced significant amounts of IFN-γ and IL-2, control HER2-CAR T cells did not (Figure 1I; supplemental Figure 3D-E).
CD7-CARCD7− T cells retain their cytolytic activity after repeat exposure to T-ALL cells
We next set out to determine the capability of CD7-CARCD7− T cells to repeatedly kill tumor cells in a sequential stimulation assay that mimics chronic antigen exposure. Antitumor activity in the CD7-CARCD7− T cell group was donor dependent and lasted for 6 to 12 stimulations in the presence of CD7+ CCRF (Figure 2A). In contrast, CD7-CARCD7− T cells did not persist beyond the first stimulation in the presence of CD7− BV173 (supplemental Figure 3F). Supernatant was collected 24 hours after odd-numbered stimulations, and cytokine concentration was determined by Multiplex assay. CD7-CARCD7− T cells produced increased Th1/Tc1 cytokines (GM-CSF, IFN-γ, TNF-α) that decreased after each stimulation (Figure 2B). CD7-CARCD7− T cells also produced Th2/Tc2 cytokines (IL-4, IL-5, IL-10, and/or IL-13), albeit in a donor-dependent fashion. Of note, CD7-CARCD7− T cells were also able to sustain antitumor activity for more stimulations when compared with CD7-CARCD7KO T cells. (supplemental Figures 5-7).
CD7-CARCD7− T cells have potent anti–T-ALL activity. (A-B) Serial stimulation assay with CD7-CARCD7−, HER2-CARBulk, or HER2-CARCD7− T cells and CCRF target cells. (A) CCRF cells at a 1:1 E:T ratio and fresh target cells were added every 72 hours if a luciferase-based cytotoxicity assay demonstrated >50% killing (N = 3, at stimulation 1, P < .001, 1-way ANOVA, CD7-CARCD7− between stimulation 1 and 5, ns, unpaired t test). Double-gradient heat map (purple, 100% target cell lysis; white, 0% target cell lysis). (B) Multiplex analysis of cytokine production by CD7-CARCD7− T cells or HER2-CARBulk and HER2-CARCD7− against CCRF cells at a 1:1 E:T ratio. At stimulation 1 by 2-way ANOVA: CD7-CARCD7− vs HER2-CARBulk, P < .05 for granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-13, IL-4, IL-5, IL-8, MCP-1, Regulated on Activation, Normal T cell Expressed and Secreted (RANTES), tumor necrosis factor alpha (TNF-α) ; P < .01 for IL-17A, IP-10; P < .0001 for IFN-γ. CD7-CARCD7− vs HER2-CARCD7−P < .05 for IFN-γ, IL-4, and tumor necrosis factor α (TNF-α) P < .05. Double-gradient heat map (blue, 100 pg/mL; white, 102 pg/mL; yellow, 106 pg/mL). (C-I) In vivo testing of CD7-CARCD7− T cells. (C) Schematic of xenograft experiments: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were injected IV via tail vein with 1 × 104 CCRF cells on day 0 and a single IV dose of 3 × 106 (N = 16) or 10 × 106 (N = 20) CD7-CARCD7− T cells on day 7; mice injected with tumor-only served as a control (N = 20). Mice were monitored via IVIS imaging and tracked for (D) bioluminescence (total flux photons per second) and (E) survival (P < .0001, Mantel-Cox log-rank, P = ns between dose levels). (F) Schematic of tumor rechallenge experiments. (G) Mice surviving and having no detectable tumor burden after 125 days of treatment with CD7-CARCD7− T cells, received a second dose of 1 × 104 CCRF cells (N = 5 for 3 × 106 group and N = 4 for 10 × 106 group); naïve age-matched mice served as control (N = 5) (day 146, P = .007, 1-way ANOVA). (H-I) Peripheral blood, spleen, and bone marrow (N = 9) was collected from mice on days 212 to 225 (∼100 days postrechallenge). All surviving animals had <0.1% residual disease in the marrow. Flow cytometry analysis of CD3, CD4, CD8, and F(ab’)2 fragments to determine the presence of CAR T cells and their immunophenotype. Bulk T cells and CD7-CARCD7− T cells served as an immunophenotype reference (CAR+ vs CAR− T cells, P < .0001, 2-Way ANOVA, CD4 vs CD8 P < .01, 2-way ANOVA). ☩, one mouse that died of ischemia-associated tubular necrosis and one that died due to unknown cause; ns, not significant.
CD7-CARCD7− T cells have potent anti–T-ALL activity. (A-B) Serial stimulation assay with CD7-CARCD7−, HER2-CARBulk, or HER2-CARCD7− T cells and CCRF target cells. (A) CCRF cells at a 1:1 E:T ratio and fresh target cells were added every 72 hours if a luciferase-based cytotoxicity assay demonstrated >50% killing (N = 3, at stimulation 1, P < .001, 1-way ANOVA, CD7-CARCD7− between stimulation 1 and 5, ns, unpaired t test). Double-gradient heat map (purple, 100% target cell lysis; white, 0% target cell lysis). (B) Multiplex analysis of cytokine production by CD7-CARCD7− T cells or HER2-CARBulk and HER2-CARCD7− against CCRF cells at a 1:1 E:T ratio. At stimulation 1 by 2-way ANOVA: CD7-CARCD7− vs HER2-CARBulk, P < .05 for granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-13, IL-4, IL-5, IL-8, MCP-1, Regulated on Activation, Normal T cell Expressed and Secreted (RANTES), tumor necrosis factor alpha (TNF-α) ; P < .01 for IL-17A, IP-10; P < .0001 for IFN-γ. CD7-CARCD7− vs HER2-CARCD7−P < .05 for IFN-γ, IL-4, and tumor necrosis factor α (TNF-α) P < .05. Double-gradient heat map (blue, 100 pg/mL; white, 102 pg/mL; yellow, 106 pg/mL). (C-I) In vivo testing of CD7-CARCD7− T cells. (C) Schematic of xenograft experiments: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were injected IV via tail vein with 1 × 104 CCRF cells on day 0 and a single IV dose of 3 × 106 (N = 16) or 10 × 106 (N = 20) CD7-CARCD7− T cells on day 7; mice injected with tumor-only served as a control (N = 20). Mice were monitored via IVIS imaging and tracked for (D) bioluminescence (total flux photons per second) and (E) survival (P < .0001, Mantel-Cox log-rank, P = ns between dose levels). (F) Schematic of tumor rechallenge experiments. (G) Mice surviving and having no detectable tumor burden after 125 days of treatment with CD7-CARCD7− T cells, received a second dose of 1 × 104 CCRF cells (N = 5 for 3 × 106 group and N = 4 for 10 × 106 group); naïve age-matched mice served as control (N = 5) (day 146, P = .007, 1-way ANOVA). (H-I) Peripheral blood, spleen, and bone marrow (N = 9) was collected from mice on days 212 to 225 (∼100 days postrechallenge). All surviving animals had <0.1% residual disease in the marrow. Flow cytometry analysis of CD3, CD4, CD8, and F(ab’)2 fragments to determine the presence of CAR T cells and their immunophenotype. Bulk T cells and CD7-CARCD7− T cells served as an immunophenotype reference (CAR+ vs CAR− T cells, P < .0001, 2-Way ANOVA, CD4 vs CD8 P < .01, 2-way ANOVA). ☩, one mouse that died of ischemia-associated tubular necrosis and one that died due to unknown cause; ns, not significant.
CD7-CARCD7− have antitumor activity in vivo
To evaluate the antitumor activity of CD7-CARCD7− T cells in vivo, we used a CCRF (CD7+) NSG T-ALL xenograft model. Mice were injected IV with 1 × 104 CCRF.GFP.firefly luciferase (CCRF.ffLuc) cells and on day 7 received a single IV dose of 3 × 106 or 10 × 106 CD7-CARCD7− T cells (Figure 2C). Mice receiving only tumor served as controls. Control mice displayed marked disease progression, whereas mice receiving CD7-CARCD7− T cells exhibited potent antitumor activity against CD7+ targets at both dose levels (Figure 2D; supplemental Figure 4A). This resulted in a substantial survival advantage for the mice treated with CD7-CARCD7− T cells when compared with controls (Figure 2E). However, there was not a statistically significant difference between CD7-CARCD7− T cell dose levels. Treated mice maintained stable weights throughout the experiment (supplemental Figure 4B).
CD7-CARCD7− T cells were still detected by flow cytometry in the peripheral blood of surviving mice on days 104 to 107 post–tumor injection (supplemental Figure 4C). To evaluate the functionality of persistent CAR T cells, mice were rechallenged with a second IV dose of 1 × 104 CCRF.GFP.ffluc cells on days 113 to 125 (Figure 2F). Age-matched controls were injected with tumor only. Mice that had previously received either 3 × 106 or 10 × 106 CD7-CARCD7− T cells were able to control disease, whereas treatment-naïve mice receiving leukemia cells only succumbed to disease (Figure 2G; supplemental Figure 4D). Between days 212 and 225, the surviving mice were killed, and peripheral blood, bone marrow, and spleen were analyzed by flow cytometry for the presence of CD7-CARCD7− T cells (Figure 2H-I; supplemental Figure 4E-G). Less than 1.5% tumor cells were detected in the blood, bone marrow, or spleen, whereas CD7-CAR T cells were detected in each compartment and were mainly CD4+, illustrating that the CD7-CARCD7− T cells persisted past leukemia rechallenge. Further, there were no significant differences in complete blood counts between mice treated with CD7-CARCD7− T cells and age-matched, healthy NSG mice (data not shown).
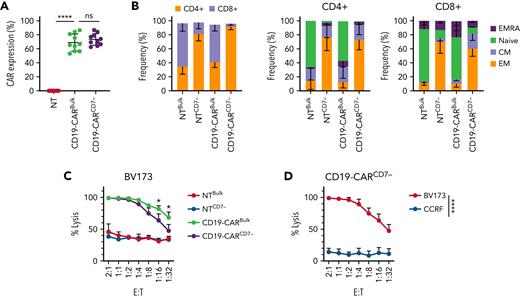
CD19-CARCD7− T cells have robust antitumor activity in vitro and maintain antitumor activity after repeated antigen stimulation
Having demonstrated that CD7-CARCD7− T cells have potent antitumor activity and persist in mice, we next explored whether CARCD7− T cells targeting another antigen have favorable biological characteristics. We generated CD19-CARCD7− using the same CD7− T-cell selection process and bulk CD19-CAR (CD19-CARBulk) T cells. Although there was a transient, minor decrease in the viability of CD19-CARCD7− T cells after transduction, there was no significant difference in expansion between bulk and CD7− CAR T-cell populations (supplemental Figure 8A-C). Both CD19-CARCD7− and CD19-CARBulk T cells exhibited transduction efficiencies > 50% (Figure 3A). Similar to the CD7-CARCD7−, the CD4+ subset was significantly enriched among the CD19-CARCD7− T cells (Figure 3B). Extended phenotyping revealed no significant differences in populations expressing 2 checkpoint receptors simultaneously (eg, PD-1+/LAG3+, PD1+/TIM3+, LAG3+/TIM3+) at 7 and 15 days posttransduction in either the CD4+ or CD8+ subset (supplemental Figure 9). In coculture assays, CD19-CARCD7− T cells demonstrated significant antitumor activity at multiple E:T ratios against BV173 (CD19+; supplemental Figure 10A) target cells with no significant differences to their CD19-CARBulk T-cell counterparts at E:T ratios above 1:8 (Figure 3C). Notably, CD19-CARCD7− T cells showed lower antitumor activity than CD19-CARBulk T-cells at rations below 1:16. In contrast, no statistical difference was observed between either CAR T-cell population when targeting CCRF (CD19−) cells (Figure 3D; supplemental Figure 10B). Specificity was confirmed by antigen-specific IFN-γ and IL-2 production, with no differences observed between CAR T-cell populations (supplemental Figure 10C).
CD19-CARCD7− T cells maintain a predominantly CD4+ phenotype and have comparable antitumor activity to CD19-CARBulk T cells. (A) CD19-CAR expression on bulk T cells and CD7− T cells (N = 10, 68.97 ± 12.20% vs 73.75 ± 8.27%, P < .01 compared with NT). (B) Immunophenotype of CD19-CAR–transduced T cells on day 7 posttransduction as determined by flow cytometry (CD4 vs CD8, effector memory (EM): CCR7−, CD45RA−; central memory (CM): CCR7+, CD45RA−; naïve-like: CCR7+CD45RA+; EM T cells reexpress CD45RA, EMRA: CCR7−, CD45RA+) (N = 3-6, 2-way ANOVA, values between bulk T cells and CD7− T cells were significant, P < .01, CD4+ and CD8+ EM bulk vs CD7− T cells P < .05, CD8+ naïve-like NTBulk vs NTCD7−P < .01, NTBulk vs CD19CD7−P < .05, NTCD7− vs CD19Bulk−P < .05). (C-D) Luciferase-based cytotoxicity assay to determine antitumor activity. (C) CD19-CAR T cells against BV173 (CD19+) for 24 hours, T-cell controls: NTBulk and NTCD7− T cells (N = 4, ns between CD19-CARBulk or CD7−, P < .05 at all ratios compared with NT T cells, 2-way ANOVA). (D) CD19-CARCD7− T cells were cocultured with BV173 (CD19+) or CCRF cells (CD19−) (N = 4, P < .05 at all ratios, 2-way ANOVA). ns, not significant.
CD19-CARCD7− T cells maintain a predominantly CD4+ phenotype and have comparable antitumor activity to CD19-CARBulk T cells. (A) CD19-CAR expression on bulk T cells and CD7− T cells (N = 10, 68.97 ± 12.20% vs 73.75 ± 8.27%, P < .01 compared with NT). (B) Immunophenotype of CD19-CAR–transduced T cells on day 7 posttransduction as determined by flow cytometry (CD4 vs CD8, effector memory (EM): CCR7−, CD45RA−; central memory (CM): CCR7+, CD45RA−; naïve-like: CCR7+CD45RA+; EM T cells reexpress CD45RA, EMRA: CCR7−, CD45RA+) (N = 3-6, 2-way ANOVA, values between bulk T cells and CD7− T cells were significant, P < .01, CD4+ and CD8+ EM bulk vs CD7− T cells P < .05, CD8+ naïve-like NTBulk vs NTCD7−P < .01, NTBulk vs CD19CD7−P < .05, NTCD7− vs CD19Bulk−P < .05). (C-D) Luciferase-based cytotoxicity assay to determine antitumor activity. (C) CD19-CAR T cells against BV173 (CD19+) for 24 hours, T-cell controls: NTBulk and NTCD7− T cells (N = 4, ns between CD19-CARBulk or CD7−, P < .05 at all ratios compared with NT T cells, 2-way ANOVA). (D) CD19-CARCD7− T cells were cocultured with BV173 (CD19+) or CCRF cells (CD19−) (N = 4, P < .05 at all ratios, 2-way ANOVA). ns, not significant.
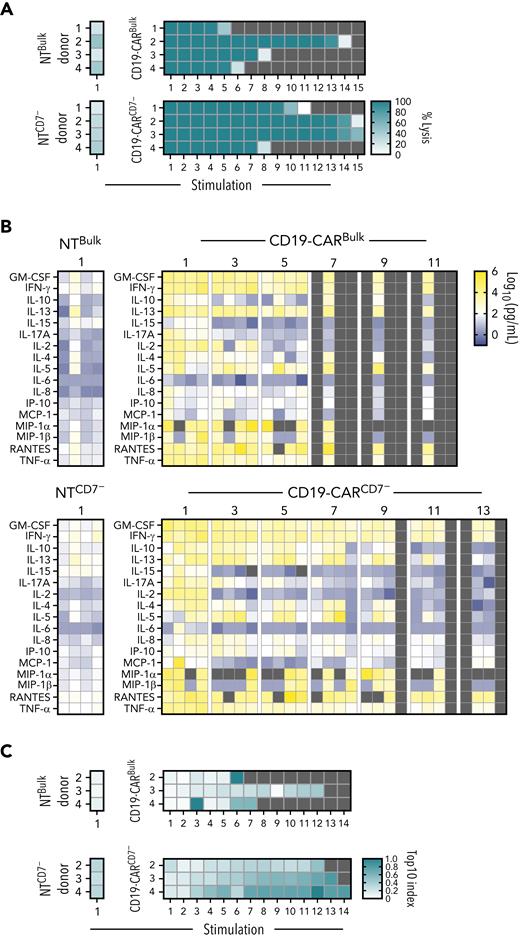
CD19-CARCD7− T cells maintain antitumor activity and secrete proinflammatory cytokines after repeated antigen exposure. (A-B) Serial stimulation analysis of effector cells in the presence of BV173 at a 1:1 E:T ratio. (A) Tumor cell lysis was evaluated every 72 hours by a luciferase-based assay, and in cases where lysis was >50%, fresh leukemia cells were added to the coculture (N = 4; stimulation 1, P < .001 between NT and CD19-CAR groups, ns between Bulk and CD7 at stimulation 1 and stimulation 5; 1-way ANOVA and nonpaired t test). Double-gradient heat map (teal, 100% target cell lysis; white, 0% target cell lysis). (B) Supernatant from odd-numbered stimulations starting at stimulation 1 was collected 24 hours after the addition of leukemia cells. Multiplex analysis of cytokine production of CAR T cells after 24 hours of restimulation (N = 4). Double-gradient heat map (blue, 10° pg/mL; white, 102 pg/mL; yellow, 106 pg/mL). CD19-CARBulk vs CD19-CARCD7− at stimulation 1 and 5, data ns by 2-way ANOVA. (C) Clonality as measured by next-generation sequencing of NTBulk, NTCD7−, CD19-CARBulk, and CD19-CARCD7− TCR repertoires throughout repeated exposure to antigen; samples were collected every 72 hours. Double-gradient heat map showing the Top10 index values (see “Methods”). Values closer to 1.0 (teal) indicate higher clonality, whereas values closer to 0.0 indicate a more even repertoire. ns, not significant.
CD19-CARCD7− T cells maintain antitumor activity and secrete proinflammatory cytokines after repeated antigen exposure. (A-B) Serial stimulation analysis of effector cells in the presence of BV173 at a 1:1 E:T ratio. (A) Tumor cell lysis was evaluated every 72 hours by a luciferase-based assay, and in cases where lysis was >50%, fresh leukemia cells were added to the coculture (N = 4; stimulation 1, P < .001 between NT and CD19-CAR groups, ns between Bulk and CD7 at stimulation 1 and stimulation 5; 1-way ANOVA and nonpaired t test). Double-gradient heat map (teal, 100% target cell lysis; white, 0% target cell lysis). (B) Supernatant from odd-numbered stimulations starting at stimulation 1 was collected 24 hours after the addition of leukemia cells. Multiplex analysis of cytokine production of CAR T cells after 24 hours of restimulation (N = 4). Double-gradient heat map (blue, 10° pg/mL; white, 102 pg/mL; yellow, 106 pg/mL). CD19-CARBulk vs CD19-CARCD7− at stimulation 1 and 5, data ns by 2-way ANOVA. (C) Clonality as measured by next-generation sequencing of NTBulk, NTCD7−, CD19-CARBulk, and CD19-CARCD7− TCR repertoires throughout repeated exposure to antigen; samples were collected every 72 hours. Double-gradient heat map showing the Top10 index values (see “Methods”). Values closer to 1.0 (teal) indicate higher clonality, whereas values closer to 0.0 indicate a more even repertoire. ns, not significant.
To further compare the effector function of CD19-CARCD7− and CD19-CARBulk T cells in a chronic antigen exposure setting, we set up a serial stimulation assay. CD19-CARBulk T cells had antitumor activity for 4 to 13 stimulations, whereas CD19-CARCD7− T cells were able to kill target cells for 7 to 14 stimulations depending on donor (Figure 4A). None of the controls (NT T cells [Bulk and CD7−] or either of the CD19-CAR T-cell products cultured with CD19− CCRF cells) showed antitumor activity after the first stimulation (Figure 4A; supplemental Figure 11). In addition, 24 hours after the addition of fresh target cells, we collected culture supernatant to measure cytokine secretion during alternating stimulations (ie, stimulation 1, 3, 5, etc). Between the first and fifth stimulations, CD19-CARBulk and CD19-CARCD7− T cells exhibited no difference in Th1/Tc1 (IFN-γ, TNF-α, GM-CSF, IL-2) or Th2/Tc2 (IL-4, IL-5, IL-6, IL-10, IL-13) cytokine secretion profiles (Figure 4B). CD19-CARCD7− T cells maintained high levels of secretion (>1000 pg/mL) of GM-CSF to stimulation 9 and IFN-γ through the 11th stimulation, whereas concentrations began to decrease in bulk populations at stimulation 7 or 5 for GM-CSF and IFN-γ, respectively.
To differentiate whether the antitumor activity was due to the predominantly CD4+ immunophenotype of CD19 CARCD7− T cells or due to the presence of CD7− T cells, we compared CD19Bulk and CD19CD7− with CD19CD4+ and CD19CD4+CD7− CAR T cells. CD19CD7− and CD19CD4+CD7− CAR T cells were able to demonstrate antitumor activity until stimulations 8 to 10, whereas CD19CD4+ CAR T cells only lasted until stimulation 6 to 7 (supplemental Figures 12-14).
Given that CD7− T cells have been implicated in certain lymphoproliferative diseases,25,26 we wanted to evaluate whether there was clonal focusing in the CD19-CARCD7− T cells. We performed endogenous TCR repertoire sequencing of CD19-CARCD7− and CD19-CARBulk after repeated stimulations. T cells were collected after each stimulation of BV173 cells, and TCRs were analyzed by next-generation sequencing. Despite starting as slightly more diverse clonal populations and growing less diverse across successive stimulations, CD19-CARCD7− T cells exhibited comparable clonality to CD19-CARBulk cells over time (Figure 4C).
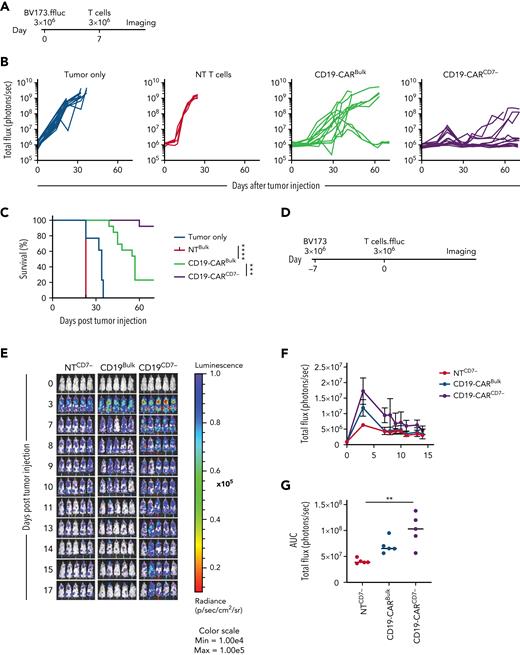
CD19-CARCD7− T cells outperform CD19-CARBulk T cells in vivo
To compare the antitumor activity of CD19-CARCD7− and CD19-CARBulk in vivo, we used our established BV173.GFP.ffluc xenograft model.27 Mice were injected with 3 × 106 BV173.GFP.ffluc cells IV and on day 7 received a single IV dose of 3 × 106 CD19-CARCD7− or CD19-CARBulk T cells (Figure 5A). CD19-CARBulk and CD19-CARCD7− T cells significantly reduced tumor burden compared with tumor-only and NT T-cell controls. However, CD19-CARCD7− T cells had significantly greater antitumor activity compared with CD19-CARBulk T cells as judged by bioluminescence imaging and survival of treated mice (Figure 5B-C; supplemental Figure 15). This improved antitumor activity correlated with increased expansion of CD19-CARCD7− T cells compared with CD19-CARBulk T cells in the same animal model using BV173 cells and GFP.ffluc-expressing CAR T cells to track their in vivo fate (Figure 5D-G).
CD19-CARCD7− T cells outperform CD19-CARBulk in vivo. (A) Schematic of BV173 xenograft model: NSG mice were injected IV via tail vein with 3 × 103 BV173.ffluc cells on day 0 and a single dose of 3 × 106 T cells on day 7. (B) Bioluminescence data (total flux = photons per second) tumor only (N = 15), NT T cells (N = 5), CD19-CARBulk (N = 13), and CD19-CARCD7− (N = 13). (C) Survival curve (∗∗∗P < .001; ∗∗∗∗P < .0001, Mantel-Cox log-rank test). (D) Schematic of in vivo persistence experiment: NSG mice were injected with 3 × 103 BV173 cells 7 days prior to a single dose of 3 × 106 CAR.ffluc T cells. (E) Corresponding bioluminescence imaging. (F) Quantitative bioluminescence data. (G) Area under the curve (AUC) of total flux in photons per second of bioluminescence (N = 5 per group; P < .01 between NTCD7− and CD19-CARCD7−; all other comparisons, ns; 1-way ANOVA). ns, not significant.
CD19-CARCD7− T cells outperform CD19-CARBulk in vivo. (A) Schematic of BV173 xenograft model: NSG mice were injected IV via tail vein with 3 × 103 BV173.ffluc cells on day 0 and a single dose of 3 × 106 T cells on day 7. (B) Bioluminescence data (total flux = photons per second) tumor only (N = 15), NT T cells (N = 5), CD19-CARBulk (N = 13), and CD19-CARCD7− (N = 13). (C) Survival curve (∗∗∗P < .001; ∗∗∗∗P < .0001, Mantel-Cox log-rank test). (D) Schematic of in vivo persistence experiment: NSG mice were injected with 3 × 103 BV173 cells 7 days prior to a single dose of 3 × 106 CAR.ffluc T cells. (E) Corresponding bioluminescence imaging. (F) Quantitative bioluminescence data. (G) Area under the curve (AUC) of total flux in photons per second of bioluminescence (N = 5 per group; P < .01 between NTCD7− and CD19-CARCD7−; all other comparisons, ns; 1-way ANOVA). ns, not significant.
CD19-CAR T cells expressing CD7 at low levels mark functional CD4+ effector CAR T cells in patients with refractory/relapsed B-ALL
Having established that CARCD7− T cells have potent antitumor activity in preclinical models, we wanted to gain insight into the behavior of CARCD7− T cells in patients. To accomplish this, we took advantage of published single-cell expression data from clinical-grade CD19-CAR T-cell products and postinfusion samples from our investigator-initiated clinical study for patients with refractory/relapsed B-ALL.19 Unsupervised clustering of 184 791 CD19-CAR+ T cells led to the identification of 21 transcriptional clusters that were functionally annotated based on key expression markers (supplemental Table 1, adapted from Wilson et al18). Upon assessing CD7 expression across all pre- and postinfusion samples, we noted clusters 4, 13, 14, and 20 as exhibiting the overall lowest relative CD7 expression and the fewest cells with any expression of CD7 (Figure 6A-B). Cluster 14 constituted the only functional CD4+ effector population within the dataset. Low CD7 expression also marked uncharacterizable mixed phenotypes (cluster 4) and dysfunctional (cluster 13) or dying (cluster 20) CD8+ subsets, respectively. In addition, we identified distinctly enriched CD7lo populations within CD8+ functional effector clusters 3 and 8, particularly after infusion; interestingly, CD7 expression often decreased over time within these functional T-cell populations (Figure 6C). Corresponding flow cytometric analysis at the above time points supported an enrichment of CD7− T-cell subsets in pre- and postinfusion samples (supplemental Figure 16). When evaluating relative expression of CD7 across the previously established functional annotations, we determined that the populations with the lowest CD7 expression were either CD4+ effector or dysfunctional T cells (Figure 6D).
CD19-CAR T cells expressing low levels of CD7 mark functional CD4+ effector CAR T cells in patients with refractory/relapsed B-ALL. (A) Violin plot of single-cell CD7 expression across 21 transcriptional clusters of clinical-grade CD19-CAR T-cell products and postinfusion samples from 16 patients with refractory/relapsed B-ALL. Data represent 184 791 transcriptionally identified CD19-CAR T cells. Transcriptional clusters were annotated using several key markers outlined in supplemental Table 1. (B) Dot plot of relative CD7 expression across transcriptional clusters from panel A. Dot size corresponds to the percentage of cells expressing any amount of CD7 as indicated. (C) Violin plot of single-cell CD7 expression from select clusters from panel A across sampling timepoints and clinical-grade CD19-CAR T-cell products as indicated by color. (D) Dot plot of relative expression of CD7 across functional annotations group as shown in (A). Dot size corresponds to the percentage of cells expressing any amount of CD7 as indicated. (E) Violin plot comparing expression of CD7 within the CAR+ CD4+ cytotoxic effector cells (cluster 14) between patients with complete response to therapy at 4 weeks after infusion (Responders) and patients without complete response to therapy (Nonresponders). Wilcoxon rank-sum with Bonferroni correction, adjusted P = 2.309236e-106. (F) Boxplot representing the proportion of CAR+ CD4+ cytotoxic effector cells (cluster 14) with no detected CD7 transcripts across patients with and without response to therapy. Boxplots indicate the median (middle bar), the 25th, and the 75th percentiles (lower and upper hinges, respectively) and 1.5 times the interquartile range from each hinge (whiskers). Wilcoxon rank-sum exact test, P = 0.03.
CD19-CAR T cells expressing low levels of CD7 mark functional CD4+ effector CAR T cells in patients with refractory/relapsed B-ALL. (A) Violin plot of single-cell CD7 expression across 21 transcriptional clusters of clinical-grade CD19-CAR T-cell products and postinfusion samples from 16 patients with refractory/relapsed B-ALL. Data represent 184 791 transcriptionally identified CD19-CAR T cells. Transcriptional clusters were annotated using several key markers outlined in supplemental Table 1. (B) Dot plot of relative CD7 expression across transcriptional clusters from panel A. Dot size corresponds to the percentage of cells expressing any amount of CD7 as indicated. (C) Violin plot of single-cell CD7 expression from select clusters from panel A across sampling timepoints and clinical-grade CD19-CAR T-cell products as indicated by color. (D) Dot plot of relative expression of CD7 across functional annotations group as shown in (A). Dot size corresponds to the percentage of cells expressing any amount of CD7 as indicated. (E) Violin plot comparing expression of CD7 within the CAR+ CD4+ cytotoxic effector cells (cluster 14) between patients with complete response to therapy at 4 weeks after infusion (Responders) and patients without complete response to therapy (Nonresponders). Wilcoxon rank-sum with Bonferroni correction, adjusted P = 2.309236e-106. (F) Boxplot representing the proportion of CAR+ CD4+ cytotoxic effector cells (cluster 14) with no detected CD7 transcripts across patients with and without response to therapy. Boxplots indicate the median (middle bar), the 25th, and the 75th percentiles (lower and upper hinges, respectively) and 1.5 times the interquartile range from each hinge (whiskers). Wilcoxon rank-sum exact test, P = 0.03.
To further explore the potential implications for CD7 expression in the context of patient response to CAR T-cell therapy, we investigated the variation in gene expression among the cytotoxic CD4+ effector population (cluster 14). Of the 16 patients profiled in this cohort, 15 were infused, with 12 achieving complete response by week 4 postinfusion.18,19 Compared with cluster 14 cells from nonresponders, the CD4+ effector cells from patients responding to therapy exhibited significantly less CD7 expression (Figure 6E). Concordantly, we detected CD7 expression in 76% of nonresponders’ CD4+ effector cells but only detected CD7 expression in 50.1% of responders’ CD4+ effector cells. Because our dataset included substantially more cytotoxic CD4+ T cells for responders than nonresponders, we also compared the proportion of CD7− to CD7+ cells within the CD4+ effectors across these patient subsets. As predicted, the proportion of CD7− cells among CD4+ effectors was significantly higher for patients who responded to treatment than for nonresponders (Figure 6F).
CD19-CAR CD4+CD7− T cells upregulate T-cell development, proliferation, and activation pathways
To further establish differences between CD4+ CD7−and CD7+ CD19-CAR T cells, we performed bulk RNA sequencing analysis. Differential gene expression analysis revealed 392 genes significantly upregulated in the CD7+ population and 493 genes significantly upregulated in the CD7− population (supplemental Table 2). Gene set enrichment analysis with hallmark gene sets showed upregulation in IL-2 STAT5 signaling, MTORC1 signaling, MYC targets, phosphatidylinositol 3-kinase AKT MTOR signaling, and glycolysis pathways in the CD7− population relative to the CD7+ population (supplemental Table 3). In contrast, the CD7+ population upregulated pathways involved in TNF-α signaling via NFkB, IFN-α response, and IFN-γ response. Notably, IFN-γ expression exhibited an opposite effect, with a significant increase observed in the CD7− population compared with the CD7+ population (log2 fold change = −1.5, adjusted P < .05; supplemental Table 2).
Discussion
Here, we successfully isolated naturally occurring CD7− T cells using a 2-step magnetic bead separation technique. We show that we can genetically modify this T-cell subset to express CD7-CARs that have robust antitumor activity against T-ALL cell lines. We then compare CD7− T cells expressing CD19-CARs to bulk T cells expressing CD19 CARs and determine that they have improved antitumor activity. To evaluate the presence of CD7 low cells by gene expression analysis (CD7lo), we investigated pre- and postinfusion samples from a cohort of patients with relapsed/refractory pre–B ALL that received CD19-CAR T cells and identified enriched CD7lo populations within the CD8+ and CD4+ compartments.
As gene editing plus CAR transduction can complicate the manufacturing process and increase the resources needed, our approach focused on harnessing naturally occurring T cells that lack surface expression of CD7. This subpopulation of CD7− T cells has previously been identified in biopsies of patients with diseases such as mycosis fungoides and peripheral T-cell lymphoma.25,26 CD7− T cells have also been noted to contribute to a defective immune response in patients with rheumatoid arthritis or patients that have undergone hematopoietic stem cell transplant.28-30 In addition, an increase in CD4+/CD7− T-cell count has been identified as a predictor of HIV infection progressing to AIDS.31 In contrast, reactive CD8+/CD7− T-cell populations have been detected in patients as a self-limited proliferative response to Epstein-Barr virus and cytomegalovirus infections.32 In addition, increased CD4+CD7− T cells have been correlated with improved prognosis in ovarian carcinoma.33 Here, we show that a subset of CD7− T cells can also have potent antitumor activity when expressing CARs redirected to hematological malignancies.
Selection techniques are widely used in hematopoietic stem cell transplant to enrich cellular products for CD34+ cells and deplete T cells.23,24 Additionally, selection protocols, such as CD4+/CD8+ selection, have been incorporated into CAR T-cell manufacturing, resulting in a change in the immunophenotypic and effector function of the T-cell product.3,25-27 Here, we used an efficient 2-step bead separation process to obtain a highly pure CD7− T-cell population with <10% CD7+ T cells. CD7 is transcriptionally regulated and lacks CD7 messenger RNA, allowing for a stable population of enriched CD7− T cells.25 We successfully generated CD7-CARCD7− T cells and observed that their immunophenotype was predominantly CD4+ effector memory and CD4+ terminally differentiated effector memory cells with expansion and viability on par with NT T cells, confirming that we had bypassed fratricide. Bulk T cells transduced with the CD7-CAR resulted in fratricide; as a result, there was decreased viability and cell number. Therefore, we were, unfortunately, unable to directly compare bulk T cells to CD7− selected T cells.
We show that CD7− T cells expressing CD7-CARs have robust and sustained antitumor activity against CD7+ leukemia in vitro and in vivo. Of note, although not evidenced in our experiments, CD7 downregulation after treatment with CD7-CAR T cells has been described,10,14 and needs to be considered as immunotherapeutic approaches are developed. In addition, our findings are in contrast to one other study that reported that naturally occurring CD7− T cells expressing CD7-CARs have limited effector function.34 These discrepant results are most likely explained by differences in the manufacturing process of CD7-CARCD7− T cells, including not only selection, activation, and transduction but also culture conditions. Due to the low frequency of CD7− T cells in the peripheral blood and the impact that T-cell selection, expansion, and overall manufacturing strategy can have on the viability of the approach, several feasibility studies involving apheresis products would be needed prior to implementing a clinical trial. In addition, it is likely that a donor-derived allogeneic arm in the posttransplant setting would need to be included. This donor-derived approach has been previously reported to have low incidence of graft-versus-host disease, comparable to that of donor-lymphocyte infusion, which is widely used in the clinic.35-37
To investigate if CD7− T cells targeting other antigens also have potent effector function, we generated CD19-CARCD7− T cells. This also provided an opportunity to directly compare the effector function of CD19-CARCD7− to bulk CD19-CAR T cells. Intriguingly, CD19-CARCD7− T cells exhibited enhanced and sustained antitumor activity both in vitro and in vivo when compared with bulk CD19-CAR T cells. Of note, the use of xenograft models precluded the evaluation for potential risks for cytokine release syndrome (CRS) and neurotoxicity. The used CD7− and CD19-CARs not only targeted different antigens but also had different costimulatory domains (CD7-CAR: CD28; CD19-CAR: 41BB), suggesting that both costimulatory domains are suitable for the generation of CARCD7− T cells with potent antitumor activity. Future studies should focus on delineating the contribution of CD28 and 4-1BB costimulation to CARCD7− T-cell function, similarly to studies performed with bulk CAR T cells.38-40 These findings corroborate our preclinical studies and provide strong impetus to further study CD7− T-cell biology in the context of adoptive cell therapies. What defines the subset of CD7− T cells that have high effector function has not been clearly delineated. Although we found differences in pathway activation, further exploration is needed to fully outline the differences between CD7− and CD7+ T cells. In addition, we hypothesize that cells that lack CD7 on their cell surface have different activation dynamics than CD7+ T cells, and this may have an impact on CAR T-cell function.
In conclusion, we demonstrate here that CARCD7− T cells derived from naturally occurring CD7− T cells have promising effector function as judged by their ability to expand, persist, and elicit potent antitumor activity against CD7+ T-ALL and CD19+ B-ALL. Thus, naturally occurring CD7− T cells present a promising T-cell subset not only for CAR-redirected CD7+ T-cell therapy but also other hematological malignancies.
Acknowledgments
The authors thank Hiro Watanabe for his help providing guide messenger RNA (mRNA) and reagents. The visual abstract and supplemental Figure 1A were generated using Biorender.com.
Surgeries and preclinical imaging were performed by the Center for In Vivo Imaging and Therapeutics, which is supported in part by National Institutes of Health (NIH) grants P01CA096832 and R50CA211481. This work was supported by NIH/NCI grants CA021765 and CA197695, the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities (ALSAC).
Authorship
Contribution: A.F., J.T.Z., S.G., and M.P.V. contributed to conceptualization; A.F., J.T.Z., J.C.C., A.V., S.A.S., S.G., and M.P.V. contributed to data analysis; A.F., J.T.Z., J.C.C., S.A.S., A.V., S.L.P., and D.M.L. contributed to investigation; J.C.C., J.A.M., M.K., H.I., J.M.K., C.G.M., G.K., P.J.C., S.N., M.M., E.A.O., and P.G.T. contributed to resources; A.F., J.T.Z., J.C.C., J.A.M., S.G., P.G.T., and M.P.V. contributed to formal analysis; A.F., J.T.Z., S.G., and M.P.V. contributed to writing the original draft; A.F., J.T.Z., J.C.C., A.V., J.A.M., S.A.S., S.L.P., M.K., H.I., J.M.K., C.G.M., G.K., P.J.C., S.N., D.M.L., M.M., E.A.O., S.G., P.G.T., and M.P.V. contributed to writing, reviewing, and editing; M.P.V. contributed to funding acquisition; and M.P.V. contributed to supervision.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Conflict-of-interest disclosure: A.F., J.T.Z., J.C.C., A.V., P.C., G.K., D.L., M.M., P.G.T., S.G., and M.P.V. have patent applications in the field of immunotherapy. S.G. has a research collaboration with TESSA Therapeutics, is a DSMB member of Immatics, and is on the scientific advisory board of Tidal. P.T. is on the scientific advisory boards of Immunoscape, Cytoagents, and Mirror Biologics, has a collaborative research agreement with Elevate Bio in the area of immunotherapy, and serves as a consultant to Johnson and Johnson. C.G.M. receives fees from Amgen and Illumina, consulting fees from Beam therapeutics, and research funding from AbbVie and Pfizer. J.C.C. reports other support from 10X Genomics and Illumina outside the submitted work. The remaining authors declare no competing financials interests.
Correspondence: M. Paulina Velasquez, Department of Bone Marrow Transplant and Cellular Therapy, St. Jude Children’s Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105; e-mail: paulina.velasquez@stjude.org.
References
Author notes
Send data sharing requests via e-mail to the corresponding author M. Paulina Velasquez (paulina.velasquez@stjude.org).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
A.F. and J.T.Z. contributed equally to this study.


![CD7− T cells are predominantly CD4+ memory cells and are resistant to fratricide when expressing CD7-CAR. (A-B) Flow cytometric analysis of surface expression of CD3 and CD7 in (A) bulk peripheral blood mononuclear cells (PBMCs) from healthy donors: CD3+CD7+ (mean, 94.37 ± 4.55%) vs CD3+CD7− (mean, 5.18 ± 4.51%) (N = 50, P < .0001, paired t test) and (B) B-ALL patients: CD3+ CD7+ (mean, 93.4 ± 3.8%) and CD3+ CD7− (mean, 6.5 ± 3.8%) (N = 5, P < .0001, paired t test) and T-ALL patients: CD3+ CD7+ (mean, 95.6 ± 3.6) and CD3+CD7− (mean, 4.4 ± 3.6%) (N = 10, P < .0001 paired t test). (C) Representative dot plots of PBMCs before and after CD7 depletion and CD3 enrichment (left) and efficiency of CD7 selection on day 10 post–activation of nontransduced (NT) T cells (CD7+: 8.27 ± 4.61% and CD7−: 91.36 ± 4.56%, range 78.2-96.4, N = 26, P < .0001, paired t test). (D) Schematic of CD7-CAR construct. (E) Expression of CD7-CAR by flow cytometry using F(ab’)2 on day 7 after transduction (mean transduction efficiency 65.9 ± 16.7%, N = 22, P < .0001, paired t test). Representative dot plot showing all remaining CD7+ T cells were eliminated post–CD7-CAR transduction. (F) Expansion kinetics (absolute cell count) and viability (trypan blue exclusion). CD7-CARCD7− T cells had similar expansion to NT T cells (N = 11, P = ns, 2-way ANOVA). Bulk CD7-CAR T cells did not expand (N = 11, P < .01, 2-way ANOVA) and had decreased viability compared with CD7-CARCD7− and NT T cells (N = 11, P < .01, 2-way ANOVA). (G) Immunophenotype of CD7-CARCD7− T cells on day 7 posttransduction (CD4 vs CD8, effector memory [EM]: CCR7−, CD45RA−; central memory [CM]: CCR7+, CD45RA−; naïve-like: CCR7+CD45RA+; EM T cells reexpress CD45RA, EMRA: CCR7−, CD45RA+) (N = 11, 2-way ANOVA, bulk T cells vs CD7− T cells: CD4 vs 8 P < .0001, EM P < .0001, Naïve P < .0001, EMRA P < .01). (H) Luciferase-based cytotoxicity assay of CD7-CARCD7− T cells against CCRF (CD7+) and control BV173 (CD7−) target cells at 7 different effector:target (E:T) ratios (N = 8, P < .001, 2-way ANOVA at all ratios). (I) CD7-CARCD7− T cells were cocultured with media, BV173 (CD7lo target), CCRF or MOLT3 (CD7+ targets) at a 2:1 E:T ratio. Supernatants were harvested after 24 hours and analyzed for IFN-γ or IL-2 by ELISA (IFN-γ: N = 7-9, IL-2 N = 7-9, 1-way ANOVA, ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001). IgG1 Fc, immunoglobulin 1 fragment crystallizable; scFv, single-chain variable fragment; ELISA, enzyme-linked immunosorbent assay; ns, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/25/10.1182_blood.2021015020/3/m_blood_bld-2021-015020-gr1a.jpeg?Expires=1769092569&Signature=jv3wAhZ9EbuMjjNFjaKymPYbPth6GqkS83JRZLZ2cw9NILm~NvKz0ZwopJnE60u1jFVk6VAt9MLcJHTWED~Rjj4lHgErld1bkEhJqLpRvuh70Bwth56HX62FhLPrktJIb-8VmLBoRPeFN~aKwpJZlAnSayvSKhovQaw45R7bErbo3qVWuZ0hDSfEWgHAoAOyeaC9aaX40sqHAiLLQnEL6IQlwBOpBIxScmQbHvB35wl6Nu-5mRa-eEPxB7ML5EPe7me9Z3h9IMLSQJDs7vxe3DDwZMSuB5LOdVLxsZ39~Zbt5wj84y8Yhfypexr6RZ4YPsz2fDFs94lqDeHmtRi6Dw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![CD7− T cells are predominantly CD4+ memory cells and are resistant to fratricide when expressing CD7-CAR. (A-B) Flow cytometric analysis of surface expression of CD3 and CD7 in (A) bulk peripheral blood mononuclear cells (PBMCs) from healthy donors: CD3+CD7+ (mean, 94.37 ± 4.55%) vs CD3+CD7− (mean, 5.18 ± 4.51%) (N = 50, P < .0001, paired t test) and (B) B-ALL patients: CD3+ CD7+ (mean, 93.4 ± 3.8%) and CD3+ CD7− (mean, 6.5 ± 3.8%) (N = 5, P < .0001, paired t test) and T-ALL patients: CD3+ CD7+ (mean, 95.6 ± 3.6) and CD3+CD7− (mean, 4.4 ± 3.6%) (N = 10, P < .0001 paired t test). (C) Representative dot plots of PBMCs before and after CD7 depletion and CD3 enrichment (left) and efficiency of CD7 selection on day 10 post–activation of nontransduced (NT) T cells (CD7+: 8.27 ± 4.61% and CD7−: 91.36 ± 4.56%, range 78.2-96.4, N = 26, P < .0001, paired t test). (D) Schematic of CD7-CAR construct. (E) Expression of CD7-CAR by flow cytometry using F(ab’)2 on day 7 after transduction (mean transduction efficiency 65.9 ± 16.7%, N = 22, P < .0001, paired t test). Representative dot plot showing all remaining CD7+ T cells were eliminated post–CD7-CAR transduction. (F) Expansion kinetics (absolute cell count) and viability (trypan blue exclusion). CD7-CARCD7− T cells had similar expansion to NT T cells (N = 11, P = ns, 2-way ANOVA). Bulk CD7-CAR T cells did not expand (N = 11, P < .01, 2-way ANOVA) and had decreased viability compared with CD7-CARCD7− and NT T cells (N = 11, P < .01, 2-way ANOVA). (G) Immunophenotype of CD7-CARCD7− T cells on day 7 posttransduction (CD4 vs CD8, effector memory [EM]: CCR7−, CD45RA−; central memory [CM]: CCR7+, CD45RA−; naïve-like: CCR7+CD45RA+; EM T cells reexpress CD45RA, EMRA: CCR7−, CD45RA+) (N = 11, 2-way ANOVA, bulk T cells vs CD7− T cells: CD4 vs 8 P < .0001, EM P < .0001, Naïve P < .0001, EMRA P < .01). (H) Luciferase-based cytotoxicity assay of CD7-CARCD7− T cells against CCRF (CD7+) and control BV173 (CD7−) target cells at 7 different effector:target (E:T) ratios (N = 8, P < .001, 2-way ANOVA at all ratios). (I) CD7-CARCD7− T cells were cocultured with media, BV173 (CD7lo target), CCRF or MOLT3 (CD7+ targets) at a 2:1 E:T ratio. Supernatants were harvested after 24 hours and analyzed for IFN-γ or IL-2 by ELISA (IFN-γ: N = 7-9, IL-2 N = 7-9, 1-way ANOVA, ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001). IgG1 Fc, immunoglobulin 1 fragment crystallizable; scFv, single-chain variable fragment; ELISA, enzyme-linked immunosorbent assay; ns, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/25/10.1182_blood.2021015020/3/m_blood_bld-2021-015020-gr1b.jpeg?Expires=1769092569&Signature=Za8YrchVYSZELzRCmW6TKsDiaNV-hfApbHfYYnGmpBWcP1igAij9aDPmBF9tU1FtMwWQL3aQBOmKAKDl6cijy9~NVN8nVJjkPlypDmhqVyuzqpEpsSnzynfWUEneIutXi8W2e75dmi-70heXI9EbQ-XgMHfhdMik0HjU4wjucr2Kc5wQCYBbJuw3OYXbJBuK~V5TlwzW72VQjyKQmoZY3BN~5wmFuIbrn3IIYZrZRr21d21yTfwKaaeNRBOyVBK~kt38dPOIxRqwc9RyTZJvgUtkuE2H~hekQgb1Wmi~Uc41JFpfkXK6j0ioDzqqnsg5~KhqrMvGypoWwWzFYC7OUQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal