Key Points
Human Richter syndrome and murine CLL cells receive growth-promoting signals from B-cell receptor but not from Toll-like receptors in vivo.
Depletion of macrophages results in reduced growth of murine CLL and human Richter syndrome cells in vivo.
Abstract
A large amount of circumstantial evidence has accumulated suggesting that Toll-like receptor (TLR) signals are involved in driving chronic lymphocytic leukemia (CLL) cell proliferation, but direct in vivo evidence for this is still lacking. We have now further addressed this possibility by pharmacologically inhibiting or genetically inactivating the TLR pathway in murine CLL and human Richter syndrome (RS) patient-derived xenograft (PDX) cells. Surprisingly, we show that pharmacologic inhibition of TLR signaling by treatment with an IRAK1/4 inhibitor delays the growth of the transplanted malignant cells in recipient mice, but genetic inactivation of the same pathway by CRISPR/Cas9-mediated disruption of IRAK4 or its proximal adaptor MyD88 has no effect. We further show that treatment with the IRAK1/4 inhibitor results in depletion of macrophages and demonstrate that these cells can support the survival and enhance the proliferation of both murine Eμ-TCL1 leukemia and human RS cells. We also show that genetic disruption of the B-cell receptor (BCR) by CRISPR/Cas9 editing of the immunoglobulin M constant region gene inhibits the growth of human RS-PDX cells in vivo, consistent with our previous finding with murine Eμ-TCL1 leukemia cells. Finally, we show that genetic disruption of IRAK4 does not result in negative selection of human CLL cell lines xenografted in immunodeficient mice. The obtained data suggest that TLR signals are unlikely to represent a major driver of CLL/RS cell proliferation and provide further evidence that signals from macrophages and the BCR promote the growth and survival of CLL and RS cells in vivo.
Introduction
B-cell receptor (BCR) signals generated by binding to external autoantigens or cell-autonomous BCR-BCR interactions play a major role in the pathogenesis of both human chronic lymphocytic leukemia (CLL) and murine models of the disease.1-4 Such signals have been shown to provide a competitive advantage to the malignant B cells by increasing their apoptosis resistance and by promoting their homing to the lymph nodes, where they receive additional nourishing signals from the microenvironment.5,6 However, BCR signals are unable to induce the proliferation of CLL cells on their own and require either costimulatory signals from the microenvironment or certain genetic lesions that result in loss of cell cycle control, such as inactivating genetic lesions in cell cycle inhibitors that frequently occur during Richter transformation.7-9
The microenvironmental signals that drive CLL cell proliferation have still not been fully characterized. In vitro, CLL cell proliferation can be induced efficiently by certain T cell– or Toll-like receptor (TLR)–derived signals, such as the combination of CD40L/IL-4/IL-21 or the TLR9 ligand CpG-DNA, respectively.10-16 However, the extent to which such signals contribute to CLL cell proliferation in vivo is still unknown. Currently, there is substantial evidence that T cells play an important role, as CLL cells in lymph node proliferation centers are frequently seen in direct contact with CD4 T cells and display increased expression of genes that can be induced by T cell–derived stimuli.12,17-19 Moreover, experiments in immunodeficient NOD/SCID/Il2Rγnull (NSG) mice have shown that activated autologous CD4 T cells are required for the engraftment and expansion of xenografted human CLL cells, further suggesting that T cells provide proliferative signals in vivo.20,21
Evidence for a role of TLR signals in driving CLL cell proliferation is more scarce and mainly includes data from gene expression profiling studies showing enrichment of TLR9-target genes in the proliferating lymph node compartment.22,23 Such enrichment is more prominent in CLL cells belonging to the IGHV-unmutated CLL (U-CLL) subset, which typically show a greater proliferative response upon stimulation with CpG-unmethylated DNA in vitro.23-25 This CLL subset is also characterized by frequent reactivity of the leukemic BCRs with DNA or DNA-associated autoantigens, which could allow CLL cells to internalize DNA-containing complexes and activate the endosomally located TLR9.26-28 Additional support for a possible role for TLR9 signaling in CLL pathogenesis is provided by recent studies showing that apoptotic cells and CpG-unmethylated cell–free DNA are commonly present in lymphoid tissues and plasma of patients with CLL and that cell-free plasma DNA levels positively correlate with a more aggressive clinical course.29,30
In addition to signals from the microenvironment, the TLR pathway can be activated in a small proportion of CLL cases (2%-4%) by an L265P mutation in the adaptor protein MyD88. This adaptor protein interacts with activated TLRs to form a signaling complex that recruits the kinases IRAK4 and IRAK1, which then activate several downstream effector pathways.28 The MyD88 L265P mutation, which is also found in 95% of Waldenström macroglobulinemia (WM) and 30% of activated B-cell diffuse large B-cell lymphoma (ABC DLBCL) cases, increases the oligomerization propensity of the MyD88 TIR domain, resulting in TLR ligand–independent recruitment and activation of IRAK4.31 The pathogenic role of this mutation in WM and ABC DLBCL has been further supported by studies showing that IRAK1/4 inhibitors suppress the growth of WM and ABC DLBCL cell lines that carry the mutation but have no effect on the growth of cells with wild-type MyD88.32,33
Two recent studies showed that IRAK4 inhibitors can also reduce the viability and proliferation of CpG-DNA–stimulated human CLL cells in vitro and delay the growth of murine Eμ-TCL1 CLL cells in vivo, further suggesting that the TLR pathway is involved in the pathogenesis of CLL and could represent a potential therapeutic target in this disease.34,35 In addition, the BTK inhibitor ibrutinib has been shown to inhibit CpG-DNA–induced proliferation of CLL cells in vitro, indicating that part of the clinical activity of this drug could result from inhibition of TLR signaling.23,36,37
To further explore the relevance of the TLR pathway as a potential therapeutic target, we compared the effects of pharmacologic inhibition and genetic disruption of TLR signaling on the growth of murine Eμ-TCL1 CLL and patient-derived Richter syndrome (RS) cells transplanted in syngeneic or immunodeficient mice, respectively. Genetic disruption of the BCR was performed in parallel experiments in the human RS cells to compare the relevance of the 2 pathways in the same in vivo setting. We show that in contrast to genetic disruption of BCR signaling, disruption of TLR signaling does not negatively affect the expansion of the malignant cells. In addition, we show that IRAK4 inhibitor treatment in these in vivo models results in macrophage depletion with consequent deprivation of the malignant cells from macrophage-derived growth and survival signals, thus providing a potential explanation for the therapeutic activity of IRAK4 inhibitors in human and murine models of CLL and RS.
Materials and methods
CRISPR/Cas9 editing of murine CLL and human RS cells
Approval for the use of samples from CLL patients was obtained from the Institutional Review Board committee at the Catholic University Hospital (14563/15), whereas approval for the animal studies was obtained from the Italian Ministry of Health (no. 347/2017-PR).
CRISPR/Cas9-editing of murine Eμ-TCL1 leukemia cells was done as recently described.9 Briefly, cells were thawed and cultured in RPMI-1640 supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin, 0.1 mg/mL streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate (Invitrogen), and 1 μM CpG-1668 (InvivoGen) at a concentration of 5 × 106 cells per mL for 20 hours prior to CRISPR/Cas9 editing with the Alt-R system (Integrated DNA Technologies). To generate the Cas9 ribonucleoprotein (RNP) complex that targets the MyD88 gene, a predesigned MyD88 cr-RNAs (1.5 μM) was combined with 1.5 μM ATTO 550-labeled tracr-RNA, 0.75 μM recombinant Cas9 protein, and 1.5 μM Alt-R Cas9 electroporation enhancer in 5 μL nuclease-free duplex buffer (all from Integrated DNA Technologies). The Cas9 RNP complex was then electroporated using the Amaxa Nucleofector II device and the Z-001 program into 6 × 106 leukemic cells resuspended in 100 μL mouse B cell Nucleofector solution (Lonza).
For CRISPR/Cas9 editing of the human RS–patient-derived xenograft (PDX) tumors, cells were thawed, cultured in the same medium as was used for the murine cells at a concentration of 5 × 106 cells per mL, and stimulated with 1 μM CpG-2006 and 25 ng/mL human IL-15 for 20 hours prior to transfection of the Cas9 RNPs using the NEPA21 Super Electroporator (Nepagene, Chiba, Japan) and the parameters from supplemental Table 1, available on the Blood website. Control cells were nucleofected with Cas9 RNPs without cr-RNA. Following transfection, cells were cultured for 3 days with 3T3-msCD40L fibroblasts and 25 ng/mL human IL-4 prior to injection in NSG mice. Editing efficiency was evaluated by amplicon capillary electrophoresis on a 3500 Genetic Analyzer (Applied Biosystems) of polymerase chain reaction fragments spanning the region of genomic DNA around the targeted site, as described in more detail elsewhere.9 Cr-RNA and polymerase chain reaction primer sequences are provided in supplemental Tables 2 and 3.
In vivo experiments with murine models
All animal procedures were performed under a protocol approved by the Italian Ministry of Health (no. 347/2017-PR). CRISPR/Cas9-edited Eμ-TCL1 leukemia cells were transferred by intraperitoneal injection of 3 × 107 cells in 2- to 3-month-old NSG or C57BL/6 mice. The human RS-PDX cells were inoculated intraperitoneally (2 × 106 cells) and subcutaneously in the right flank of NSG mice (1.2 × 107 cells in 0.2 mL of a 50:50 mixture of cells and Matrigel Matrix). For the in vivo treatment experiments, C57BL/6 mice or NSG mice were fed with standard diet containing vehicle control or medicated chow containing 0.25 g/kg R221 (kindly provided by Rigel Pharmaceuticals, Inc). Additional information is provided in the supplemental Materials and Methods.
Results
Treatment with an IRAK1/4 inhibitor delays leukemia progression in Eμ-TCL1 adoptive transfer model
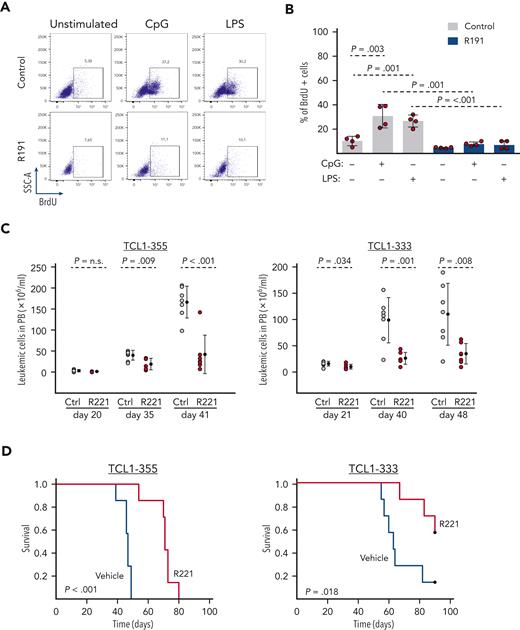
To further understand the role of TLR signaling in CLL cell growth and expansion, we first tested the capacity of R191, a dual inhibitor of the kinases IRAK4 and IRAK1, to inhibit TLR signaling in murine Eμ-TCL1–derived CLL cells and human RS-PDX cells. Experiments performed with primary leukemic cells isolated from 4 different Eμ-TCL1 mice showed that R191 completely inhibited proliferation induced by the TLR ligands CpG or LPS (Figure 1A-B). Treatment with R191 also inhibited the proliferation of human RS-PDX cells induced by stimulation with CpG while having only a modest effect on proliferation induced by CD40L/IL-4/IL-21 stimulation (supplemental Figure 1). R191 was also modestly cytotoxic against unstimulated TCL1 and RS-PDX cells, whereas it was not toxic for unstimulated human CLL cells. However, CpG-stimulated or CD40L/IL-4/IL-21–stimulated human CLL cells underwent apoptosis in the presence of R191 (supplemental Figure 2).
The IRAK1/4 inhibitorR191/R221 inhibits TLR signaling and delays the growth of murine Eμ-TCL1 CLL cells in vivo. (A-B) Effect of R191 on proliferation of TLR-stimulated Eμ-TCL1 CLL cells. The percentage of proliferating cells was determined by analysis of 5-bromo-2'-deoxyuridine (BrdU) incorporation in viable Eμ-TCL1 leukemia cells that were cultured for 28 hours in the presence or absence of CpG-1668 (1 μM), LPS (5 μg/mL), and R191 (1 μM) and then for additional 20 hours in the presence of BrdU (10 μM). One representative experiment with TCL1-333 cells is shown in panel A, and a summary of 4 experiments with CLL cells derived from 4 different Eμ-TCL1 transgenic mice is shown in panel B. Statistical analysis was done using 1-way repeated measures analysis of variance (ANOVA) with Tukey test for multiple comparisons. (C) Analysis of leukemia cell counts (CD5+/CD19+) in peripheral blood of mice inoculated with TCL1-355 or TCL1-333 leukemia cells and treated with R221 or vehicle control (n = 7 per group). Statistical analysis was done using the t test or Mann-Whitney test, as appropriate. (D) Survival analysis of mice treated with R221 or vehicle control. Treatment was started 3 days after tumor transfer. X-axis indicates days from tumor transfer. Survival curves were estimated using the Kaplan-Meier method, and curve differences were assessed using the log-rank test.
The IRAK1/4 inhibitorR191/R221 inhibits TLR signaling and delays the growth of murine Eμ-TCL1 CLL cells in vivo. (A-B) Effect of R191 on proliferation of TLR-stimulated Eμ-TCL1 CLL cells. The percentage of proliferating cells was determined by analysis of 5-bromo-2'-deoxyuridine (BrdU) incorporation in viable Eμ-TCL1 leukemia cells that were cultured for 28 hours in the presence or absence of CpG-1668 (1 μM), LPS (5 μg/mL), and R191 (1 μM) and then for additional 20 hours in the presence of BrdU (10 μM). One representative experiment with TCL1-333 cells is shown in panel A, and a summary of 4 experiments with CLL cells derived from 4 different Eμ-TCL1 transgenic mice is shown in panel B. Statistical analysis was done using 1-way repeated measures analysis of variance (ANOVA) with Tukey test for multiple comparisons. (C) Analysis of leukemia cell counts (CD5+/CD19+) in peripheral blood of mice inoculated with TCL1-355 or TCL1-333 leukemia cells and treated with R221 or vehicle control (n = 7 per group). Statistical analysis was done using the t test or Mann-Whitney test, as appropriate. (D) Survival analysis of mice treated with R221 or vehicle control. Treatment was started 3 days after tumor transfer. X-axis indicates days from tumor transfer. Survival curves were estimated using the Kaplan-Meier method, and curve differences were assessed using the log-rank test.
Next, to investigate the effects of inhibition of TLR signaling with this drug in vivo, we inoculated C57BL/6 mice with primary leukemic cells isolated from 2 different Eμ-TCL1 mice (TCL1-355 and TCL1-333) and treated them with the R191 prodrug R221 or vehicle control. Analysis of leukemia cell counts showed no differences until day 20 of treatment, but a significant reduction was observed in R221-treated mice after day 35 (Figure 1C). Consistent with the delayed leukemia progression, mice treated with R221 also exhibited a significantly longer survival compared with the control mice (Figure 1D).
Genetic disruption of MyD88 does not affect the growth of adoptively transferred Eμ-TCL1 leukemia cells
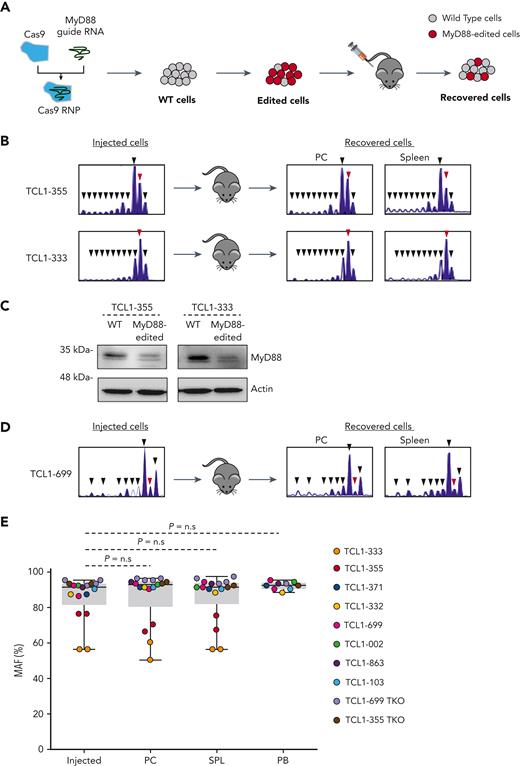
In addition to inhibiting its primary targets IRAK1 and IRAK4 (IC50 of 3 nmol/L), R191 has been reported to inhibit the kinase activity of 13 other kinases by >80% at concentrations ranging from 50 nmol/L to 250 nmol/L.38 Therefore, to determine whether the effect of R221 treatment was caused by inhibition of TLR signaling, we targeted the MyD88 gene in the Eμ-TCL1 leukemias TCL1-355 and TCL1-333 by CRISPR/Cas9 editing (Figure 2A). Targeting of the MyD88 gene was done by nucleofection-mediated delivery of a ribonucleoprotein complex containing recombinant Cas9 and a guide RNA against MyD88, resulting in an editing efficiency of 60% to >90% and a strong reduction of MyD88 protein expression (Figure 2B-C). Leukemic cells with >90% MyD88 knockout were resistant to TLR stimulation, as evidenced by the lack of an increase in BrdU incorporation or phosphorylation of the NF-kB subunit p65 following stimulation with CpG (supplemental Figure 3).
Murine CLL cells with disrupted MyD88 are not negatively selected in vivo. (A) Schematic representation of the procedure to investigate the impact of CRISPR/Cas9-mediated disruption of the MyD88 gene on the growth of adoptively transferred Eμ-TCL1 CLL cells. The scheme illustrates expected outcome in case TLR signals are required for leukemia growth in vivo. (B) Indel analysis by amplicon capillary electrophoresis of the targeted region of MyD88 in TCL1-355– and TCL1-333–injected and recovered leukemia cells isolated from peritoneal cavity (PC) and spleen. The wild type allele is indicated by a red arrow, and mutant alleles are indicated by black arrows. (C) Immunoblotting analysis of wild type (WT) and MyD88-edited TCL1-355 and TCL1-333 leukemia cells. (D) Amplicon capillary electrophoresis of the targeted region of MyD88 in TCL1-699–injected and recovered leukemia cells isolated from PC and spleen. Experiment was performed with the second MyD88 guide RNA. (E) MyD88 mutant allele frequency (MAF) in injected leukemia cells and cells isolated from the PC and spleen of mice 21 to 35 days after adoptive transfer. Seventeen independent experiments were performed with the 8 TCL1-derived CLL and the 2 TCL1-derived RS lines. MAF was calculated by dividing the area of the mutated alleles with the total area of all amplified alleles (mutant + WT) detected in the amplicon capillary electrophoresis. Statistical analysis was done using 1-way repeated measures ANOVA.
Murine CLL cells with disrupted MyD88 are not negatively selected in vivo. (A) Schematic representation of the procedure to investigate the impact of CRISPR/Cas9-mediated disruption of the MyD88 gene on the growth of adoptively transferred Eμ-TCL1 CLL cells. The scheme illustrates expected outcome in case TLR signals are required for leukemia growth in vivo. (B) Indel analysis by amplicon capillary electrophoresis of the targeted region of MyD88 in TCL1-355– and TCL1-333–injected and recovered leukemia cells isolated from peritoneal cavity (PC) and spleen. The wild type allele is indicated by a red arrow, and mutant alleles are indicated by black arrows. (C) Immunoblotting analysis of wild type (WT) and MyD88-edited TCL1-355 and TCL1-333 leukemia cells. (D) Amplicon capillary electrophoresis of the targeted region of MyD88 in TCL1-699–injected and recovered leukemia cells isolated from PC and spleen. Experiment was performed with the second MyD88 guide RNA. (E) MyD88 mutant allele frequency (MAF) in injected leukemia cells and cells isolated from the PC and spleen of mice 21 to 35 days after adoptive transfer. Seventeen independent experiments were performed with the 8 TCL1-derived CLL and the 2 TCL1-derived RS lines. MAF was calculated by dividing the area of the mutated alleles with the total area of all amplified alleles (mutant + WT) detected in the amplicon capillary electrophoresis. Statistical analysis was done using 1-way repeated measures ANOVA.
The edited cells were injected in the peritoneal cavity of recipient mice and recovered 5 weeks later for analysis of the proportion of mutant and wild type alleles. Surprisingly, no negative selection of MyD88-mutant alleles was observed, suggesting that cells with a disrupted TLR signaling pathway do not have a growth disadvantage in vivo (Figure 2B). To extend these findings and exclude possible off-target effects, a separate set of experiments using another MyD88 guide RNA and leukemia cells derived from 6 other Eμ-TCL1 transgenic mice was performed (Figure 2D). In addition, to investigate whether there is a difference in the behavior of cells from the CLL and RS phase of the disease, we included 2 recently established Eμ-TCL1–derived RS models (TCL1-355 TKO and TCL1-699 TKO).9 The results of 17 independent experiments with the 8 Eμ-TCL1–derived CLL and 2 Eμ-TCL1–derived RS lines are summarized in Figure 2E. Comparison of the mutant allele frequency of the injected and recovered cells again showed no negative selection of cells with MyD88-mutant alleles in any of the 3 investigated compartments (peritoneal cavity, spleen, and peripheral blood).
Different effects of pharmacologic inhibition and genetic disruption of IRAK4 on the growth of xenografted human RS cells
The results of the previous experiments confirmed reported findings that treatment with an IRAK1/4 inhibitor delays leukemia progression in the Eμ-TCL1 model35 but also suggested that this effect is not caused by disruption of TLR signaling in the malignant cells themselves. To validate these findings in a human setting, we repeated these experiments using 4 recently established RS-PDX models.39,40 These RS-PDX models cannot be propagated in vitro because of spontaneous apoptosis but grow efficiently in immunodeficient NSG mice, suggesting that they receive and are responsive to microenvironmental growth and/or survival signals in vivo.
To investigate whether TLR signals are required for the growth of these cells in vivo, we first tested the activity of R221 against the xenografted RS-PDX models RS9737 and RS1316. Because the malignant cells at different anatomic sites may receive different microenvironmental signals, these cells were transplanted both intraperitoneally and subcutaneously in each mouse. After 27 days of treatment, the total number of malignant B cells was evaluated in the subcutaneous tumor, peritoneal cavity, and spleen of vehicle and R221-treated mice. Interestingly, in both experiments a significant reduction in the number of malignant B cells was observed in the subcutaneous tumor and spleen but not in the peritoneal cavity of R221-treated mice, suggesting that IRAK1/4 inhibitor treatment differently affects the growth of the malignant cells in different anatomic compartments (Figure 3A).
R221 treatment and IRAK4 knockdown differently affect the growth of xenografted human RS cells. (A) Treatment with R221 results in reduced growth of RS1316 and RS9737 cells in subcutaneous tumor and spleen but not in PC of xenografted NSG mice. Graphs show absolute number of leukemic cells at each site. Statistical analysis was done using the t test or Mann-Whitney test, as appropriate. (B) Immunoblotting analysis of IRAK4 expression in WT and IRAK4-edited RS9737 and RS1316 cells. (C) Indel analysis by amplicon capillary electrophoresis of the targeted region of IRAK4 in injected and recovered leukemia cells isolated from subcutaneous tumor, PC, and spleen. The WT allele is indicated by a red arrow, and mutant alleles are indicated by black arrows. (D) IRAK4 MAF in injected leukemia cells and cells isolated from subcutaneous tumors, PCs, and spleens of NSG mice. The tumor cells were recovered between 27 and 34 days after transplantation. Eleven independent experiments were performed, 3 each with RS1316, RS9737, and IP867/17 and 2 with RS1050 cells. Statistical analysis was done using 1-way repeated measures ANOVA.
R221 treatment and IRAK4 knockdown differently affect the growth of xenografted human RS cells. (A) Treatment with R221 results in reduced growth of RS1316 and RS9737 cells in subcutaneous tumor and spleen but not in PC of xenografted NSG mice. Graphs show absolute number of leukemic cells at each site. Statistical analysis was done using the t test or Mann-Whitney test, as appropriate. (B) Immunoblotting analysis of IRAK4 expression in WT and IRAK4-edited RS9737 and RS1316 cells. (C) Indel analysis by amplicon capillary electrophoresis of the targeted region of IRAK4 in injected and recovered leukemia cells isolated from subcutaneous tumor, PC, and spleen. The WT allele is indicated by a red arrow, and mutant alleles are indicated by black arrows. (D) IRAK4 MAF in injected leukemia cells and cells isolated from subcutaneous tumors, PCs, and spleens of NSG mice. The tumor cells were recovered between 27 and 34 days after transplantation. Eleven independent experiments were performed, 3 each with RS1316, RS9737, and IP867/17 and 2 with RS1050 cells. Statistical analysis was done using 1-way repeated measures ANOVA.
To determine whether these effects are caused by disruption of IRAK1/4 signaling in the malignant B cells, we targeted exon 4 of the human IRAK4 gene by CRISPR/Cas9 and inoculated the malignant cells intraperitoneally and subcutaneously in each mouse (Figure 3B, and 3C top panels). Consistent with the previous experiments using murine Eμ-TCL1 leukemia cells, no negative selection of IRAK4-mutant alleles was observed in cells isolated from any of the 3 investigated anatomic compartments. A repeated experiment with 2 additional RS-PDX models (IP867/17 and RS1050) yielded identical results (Figure 3C bottom panels). Figure 3D summarizes the results of 11 independent experiments with the 4 RS-PDX models, showing no reduction in the MAF of injected cells versus those isolated from the PC, spleen, or subcutaneous tumor 4 to 5 weeks later.
To further validate our experimental approach, we targeted the human immunoglobulin M (IgM) constant region gene (IGHM) in RS9737, RS1316, IP867/17, and RS1050 cells and investigated changes in MAF following in vivo propagation. In contrast with the IRAK4-knockout experiments, we observed a significant reduction in the proportion of mutant IGHM alleles in all 3 investigated compartments, which was accompanied with selective loss of leukemic cells lacking surface IgM expression (Figure 4). A separate set of experiments using another IGHM guide RNA yielded identical results, providing further evidence that RS cells depend on BCR signals for their growth and/or survival (supplemental Figure 4).
RS-PDX cells with CRISPR/Cas9-disrupted IGHM gene are negatively selected in vivo. (A) Indel analysis of the targeted region of the IGHM gene in injected and recovered leukemia cells isolated from subcutaneous tumor, PC, and spleen of xenografted NSG mice. The wild type allele is indicated by a red arrow, and mutant alleles are indicated by black arrows. (B) Flow cytometry analysis of surface IgM expression in IGHM-edited tumor cells before injection and after recovery from subcutaneous tumor, PC, and spleen of xenografted NSG mice. Control refers to mock-transfected cells. (C) IGHM MAF in injected leukemia cells and cells isolated from subcutaneous tumors, PCs, and spleens of NSG mice. The tumor cells were recovered 30 to 31 days after transplantation. Twelve independent experiments were performed, 3 each with RS1316, RS9737, IP867/17, and RS1050 cells. Statistical analysis was done with 1-way repeated measures ANOVA with Tukey test for multiple comparisons.
RS-PDX cells with CRISPR/Cas9-disrupted IGHM gene are negatively selected in vivo. (A) Indel analysis of the targeted region of the IGHM gene in injected and recovered leukemia cells isolated from subcutaneous tumor, PC, and spleen of xenografted NSG mice. The wild type allele is indicated by a red arrow, and mutant alleles are indicated by black arrows. (B) Flow cytometry analysis of surface IgM expression in IGHM-edited tumor cells before injection and after recovery from subcutaneous tumor, PC, and spleen of xenografted NSG mice. Control refers to mock-transfected cells. (C) IGHM MAF in injected leukemia cells and cells isolated from subcutaneous tumors, PCs, and spleens of NSG mice. The tumor cells were recovered 30 to 31 days after transplantation. Twelve independent experiments were performed, 3 each with RS1316, RS9737, IP867/17, and RS1050 cells. Statistical analysis was done with 1-way repeated measures ANOVA with Tukey test for multiple comparisons.
To determine whether the findings from the human RS models are applicable to human CLL cells, we investigated the impact of IRAK4 and IGHM knockout in the human CLL cell lines MEC1, C1, and HG3 (Figure 5). The experiments with the CLL cell lines showed no negative selection of IRAK4-knockout cells following propagation in vivo but also did not show negative selection of IGHM-knockout cells, presumably because all CLL cell lines express the Epstein-Barr virus LMP2A protein, which is a BCR mimic that can substitute for the BCR.41
Analysis of changes in IGHM or IRAK4 MAF following in vivo propagation of human CLL cell lines MEC1, C1, and HG3. (A) Indel analysis of the targeted region of the IRAK4 gene or (B) IGHM gene in injected and recovered leukemia cells isolated from subcutaneous tumor, PC, and spleen of xenografted NSG mice. The wild type allele is indicated by a red arrow, and mutant alleles are indicated by black arrows. (C-D) IRAK4 and IGHM MAF in injected leukemia cells and cells isolated from subcutaneous tumors, PCs, and spleens of NSG mice. The tumor cells were recovered 24 to 37 days after transplantation. Each circle represents an independent experiment. Statistical analysis was done with 1-way repeated measures ANOVA with Tukey test for multiple comparisons.
Analysis of changes in IGHM or IRAK4 MAF following in vivo propagation of human CLL cell lines MEC1, C1, and HG3. (A) Indel analysis of the targeted region of the IRAK4 gene or (B) IGHM gene in injected and recovered leukemia cells isolated from subcutaneous tumor, PC, and spleen of xenografted NSG mice. The wild type allele is indicated by a red arrow, and mutant alleles are indicated by black arrows. (C-D) IRAK4 and IGHM MAF in injected leukemia cells and cells isolated from subcutaneous tumors, PCs, and spleens of NSG mice. The tumor cells were recovered 24 to 37 days after transplantation. Each circle represents an independent experiment. Statistical analysis was done with 1-way repeated measures ANOVA with Tukey test for multiple comparisons.
IRAK4 inhibitor treatment depletes macrophages in NSG and wild-type mice
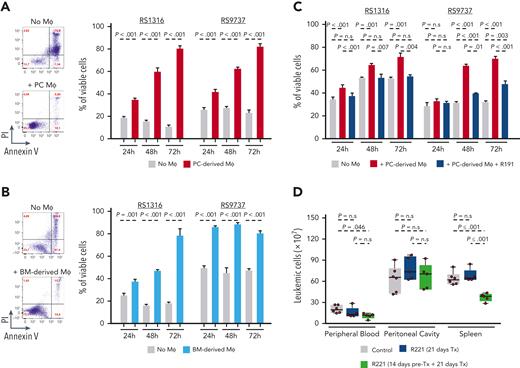
The previous experiments showed that treatment with R221 reduces the growth of the malignant B cells in vivo but also that this effect is not caused by disruption of TLR signaling in the malignant B cells themselves. Considering that the reduction in tumor growth occurred only after prolonged R221 treatment, we investigated whether it could be related to an effect on some other cellular subset present in the tumor microenvironment. We focused in particular on monocytes and macrophages, which rely for their activity on TLR-mediated signals and have been shown to support CLL cell survival in vitro and in vivo.42-46 A significant reduction in the absolute number of granulocytes (CD11b+F4/80−) and monocytes (CD11b+F4/80int) and almost complete disappearance of macrophages (CD11blowF4/80hi) was observed in the spleens of NSG mice xenografted with RS9737 cells that had been treated for 18 days with R221 (Figure 6A). A separate experiment with wild-type C57BL/6 mice that had not been inoculated with leukemia cells also showed depletion of macrophages and a significant reduction in the number of granulocytes in the spleen of R221-treated mice (Figure 6B and supplemental Figure 5). In contrast, treatment with R221 induced no change in the number or percentage of macrophages in the peritoneal cavity, where no effect on the growth of the malignant B cells had been observed (Figure 6C and supplemental Figure 6). In vitro experiments showed that both peritoneal cavity– and bone marrow–derived macrophages are susceptible to 1.0 μM R191, suggesting that the lack of an effect in vivo against peritoneal cavity macrophages is not because of different sensitivity to the drug (supplemental Figure 7).
Effects of R221 treatment on monocyte/macrophage populations in NSG and C57BL/6 mice. (A) Representative flow cytometry analysis showing the percentage of granulocytes (CD11b+F4/80−), monocytes (CD11b+F4/80int), and monocyte-derived macrophages (CD11blowF4/80hi) gated on viable CD45+ cells isolated from the spleen of R221- or vehicle control–treated NSG mice (n = 7 per group) xenografted with RS9737 cells. The box plot shows the absolute number of granulocytes (Gran), monocytes (Mo), and macrophages (Mφ) in the spleen of vehicle control– and R221-treated NSG mice. (B) Flow cytometry dot plots and box plot showing the percentage (left) and the absolute number (right) of Gran, Mo, and Mφ in the spleen of R221- or vehicle control–treated C57/BL6 mice. (C) Flow cytometry dot plots and box plot showing the percentage (left) and the absolute number (right) of monocyte-derived Mφ (CD11b+F4/80int) and tissue-resident Mφ (CD11b+F4/80hi) in the PC of R221 and vehicle control C57/BL6 mice. Cell populations were defined as described in Hanna et al44 Statistical analysis was done using the t test or Mann-Whitney test, as appropriate.
Effects of R221 treatment on monocyte/macrophage populations in NSG and C57BL/6 mice. (A) Representative flow cytometry analysis showing the percentage of granulocytes (CD11b+F4/80−), monocytes (CD11b+F4/80int), and monocyte-derived macrophages (CD11blowF4/80hi) gated on viable CD45+ cells isolated from the spleen of R221- or vehicle control–treated NSG mice (n = 7 per group) xenografted with RS9737 cells. The box plot shows the absolute number of granulocytes (Gran), monocytes (Mo), and macrophages (Mφ) in the spleen of vehicle control– and R221-treated NSG mice. (B) Flow cytometry dot plots and box plot showing the percentage (left) and the absolute number (right) of Gran, Mo, and Mφ in the spleen of R221- or vehicle control–treated C57/BL6 mice. (C) Flow cytometry dot plots and box plot showing the percentage (left) and the absolute number (right) of monocyte-derived Mφ (CD11b+F4/80int) and tissue-resident Mφ (CD11b+F4/80hi) in the PC of R221 and vehicle control C57/BL6 mice. Cell populations were defined as described in Hanna et al44 Statistical analysis was done using the t test or Mann-Whitney test, as appropriate.
Macrophages sustain the survival of murine TCL1 and human RS leukemia cells
To determine the capacity of murine macrophages to support the survival of human RS cells, we cocultured RS1316 and RS9737 cells with peritoneal cavity– or bone marrow–derived macrophages from NSG mice (Figure 7A-B). In both sets of experiments, coculture with macrophages significantly protected the malignant B cells from spontaneous apoptosis. Moreover, an independent experiment with the RS-PDX lines RS9737, RS1316, and IP867/17 and peritoneal cavity macrophages, which were used because of the ease of isolation, showed that the malignant B cells can be induced to proliferate and can be maintained in culture for extended periods (>10 days) if supplemented every 3 to 4 days with viable macrophages (supplemental Figure 8). The effects on survival were abrogated when the cells were cocultured with macrophages that had been pretreated with R191 (Figure 7C). Coculture experiments with peritoneal cavity macrophages from C57BL/6 mice and the murine leukemias TCL1-355 and TCL1-863 also showed a significant increase in the percentage of viable and proliferating CLL cells (supplemental Figure 9). The supportive effect of macrophages was primarily contact-dependent, as it was largely lost when RS-PDX cells were separated from the macrophages with well inserts to prevent them from directly interacting (supplemental Figure 10). However, a small increase in viability was observed when RS-PDX cells were cultured with recombinant BAFF and IL-15 but not with other soluble macrophage-derived factors that have been reported to support the survival of CLL cells in vitro, such as Wnt5a and CXCL12 (supplemental Figure 11).
Macrophages prolong the survival of PDX-RS cells in vitro, and their depletion delays the expansion of adoptively transferred TCL1 leukemia cells in vivo. (A) Representative annexin V/propidium iodide (PI) analysis of RS1316 cells cultured for 72 hours in the presence or absence of PC-isolated Mφ (left); summary of 3 independent experiments using RS1316 and RS9737 cells (right). (B) Representative annexin V/PI analysis of RS1316 cells cultured for 72 hours in the presence or absence of bone marrow (BM)-derived Mφ generated by differentiating BM mononuclear cells for 4 days in culture with M-CSF (left); summary of 3 independent experiments using RS1316 and RS9737 cells (right). (C) Percentage of viable RS1316 or RS9737 cells following coculture with peritoneal cavity–derived macrophages that had previously been exposed to 1μM R191 for 24 hours. Macrophages were washed to remove R191 prior to coculture with the RS-PDX cells. Viability was assessed by annexin V/PI staining. Graph represents summary of 3 independent experiments. (D) Analysis of leukemia cell counts (CD5+/CD19+) in peripheral blood, peritoneal cavity, and spleen of mice treated with vehicle control (n = 6), mice treated for 21 days with R221 (n = 5), or mice pretreated for 14 days with R221 prior to tumor transfer and then treated for additional 21 days (n = 5). Statistical analysis was done using 1-way ANOVA with Tukey test for multiple comparisons.
Macrophages prolong the survival of PDX-RS cells in vitro, and their depletion delays the expansion of adoptively transferred TCL1 leukemia cells in vivo. (A) Representative annexin V/propidium iodide (PI) analysis of RS1316 cells cultured for 72 hours in the presence or absence of PC-isolated Mφ (left); summary of 3 independent experiments using RS1316 and RS9737 cells (right). (B) Representative annexin V/PI analysis of RS1316 cells cultured for 72 hours in the presence or absence of bone marrow (BM)-derived Mφ generated by differentiating BM mononuclear cells for 4 days in culture with M-CSF (left); summary of 3 independent experiments using RS1316 and RS9737 cells (right). (C) Percentage of viable RS1316 or RS9737 cells following coculture with peritoneal cavity–derived macrophages that had previously been exposed to 1μM R191 for 24 hours. Macrophages were washed to remove R191 prior to coculture with the RS-PDX cells. Viability was assessed by annexin V/PI staining. Graph represents summary of 3 independent experiments. (D) Analysis of leukemia cell counts (CD5+/CD19+) in peripheral blood, peritoneal cavity, and spleen of mice treated with vehicle control (n = 6), mice treated for 21 days with R221 (n = 5), or mice pretreated for 14 days with R221 prior to tumor transfer and then treated for additional 21 days (n = 5). Statistical analysis was done using 1-way ANOVA with Tukey test for multiple comparisons.
Collectively, the previous data suggested that the antileukemic activity of R221 may be primarily caused by deprivation of the malignant B cells from macrophage-derived growth and survival signals rather than a direct effect on the malignant B cells. To further evaluate this possibility, we repeated the experiment described in Figure 1C, except that this time we included an additional group of mice in which macrophages were depleted by R221 pretreatment for 14 days prior to injection of TCL1-355 leukemia cells. Treatment was continued thereafter in both groups for 21 days, when mice were euthanized for analysis of tumor burden. In contrast to mice that had not received pretreatment, a significant reduction in the number of leukemic cells was observed in the peripheral blood and spleen of the R221-pretreated mice in comparison to the untreated mice (Figure 7D). Consistent with the lack of an effect on peritoneal cavity macrophages, no reduction in the number of leukemic cells at this site was observed.
Discussion
Over the past decades, numerous microenvironmental signals have been identified that can increase the survival or induce the proliferation of CLL cells in vitro, but the relevance of these signals in promoting the growth and survival of the leukemic cells in vivo has still not been fully established. In this article, we performed in vivo pharmacologic inhibition and CRISPR/Cas9 gene editing of murine and human PDX models to interrogate the role of the TLR pathway in the pathogenesis of CLL and RS. In contrast to our initial expectations, we find no evidence that TLR signals influence the growth of the malignant cells in any of the investigated in vivo models. Instead, our data provide additional genetic evidence for a major dependence of the malignant cells on BCR signals and further emphasize an important role for macrophages in supporting the growth and survival of CLL and RS cells.
The negative findings of our study cannot completely exclude the possibility for a less essential role of TLR signals in CLL pathogenesis or progression. In particular, considering that both the murine TCL1 and the human RS models are characterized by rapid tumor growth, a more subtle effect of TLR pathway disruption may have remained undetected. For example, a recent study by Kennedy et al suggested that mitochondrial DNA, which is present at higher levels in the plasma of CLL patients compared with healthy controls, increases by two-fold the propensity of the leukemic cells to migrate towards CXCL12.30 Such an effect is unlikely to have a major impact on the growth of rapidly proliferating tumors and would likely be missed in our experimental models. However, using the same experimental approach, we readily demonstrated strong negative selection of human IGHM-knockout RS-PDX cells, thus demonstrating that our experimental system is robust enough to identify signals that are essential for the growth of the malignant cells.
Another limitation of our study is that it cannot exclude the possibility that TLR9 signaling is involved in the pathogenesis of a subset of CLL cases that were not represented by our models. Such a possibility is supported by data from 2 recent studies that reported the presence of activated TLR9 complexes in lymph node CLL cells from only 2 of 14 biopsies investigated with the proximity ligation assay.23,47 In addition, it is worth noting that TLR9 activation depends on the availability of a BCR capable of recognizing and transporting DNA-containing complexes to the endosomal compartment where TLR9 is located, and such reactivity has been detected in only a fraction of cases.27 Future experiments with primary CLL xenografts, which were not possible in the current study because of insufficient editing efficiency, will be required to address this possibility.
Another unexpected finding of our study was the compartment-restricted effect of R191/R221 treatment on the growth of the transplanted malignant B cells. One possible explanation for this phenomenon is lower bioavailability of R191 in the peritoneal cavity and an off-target cytotoxic effect of the drug. R191 has been reported to downregulate MYC, CDK4, and CDK6 and induce cell cycle arrest and apoptosis in WM cell lines, suggesting that it could be selectively toxic for cycling cells.38 In line with this possibility, we observed that R191 reduces the viability of RS, TCL1, and CpG- or CD40L/IL-4/IL-21–stimulated human CLL samples, which all contain a substantial proportion of cycling cells, but has no impact on the viability on unstimulated, resting CLL cells. However, considering that the difference in tumor growth in vivo was seen only after prolonged R221 treatment and coincided with a reduction in the number of macrophages, another nonmutually exclusive possibility is that the effect of treatment was a consequence of reduced availability of macrophage-derived growth and survival signals. This possibility was additionally supported by our in vitro coculture experiments and by the study of Giménez et al, who, using another IRAK4 inhibitor, also observed a concomitant reduction in the number of monocytes, macrophages, and adoptively transferred murine TCL1 leukemia cells in the spleens of treated mice.35
The mechanism causing macrophage depletion in IRAK4 inhibitor–treated mice is currently unknown. In the study of Giménez et al,35 IRAK4 inhibitor treatment was associated with reduced monocyte expression of the chemokine receptor CCR2, which is essential for the CCL-2–mediated recruitment of monocytes by CLL cells.48 However, in our study the effect of IRAK4 inhibitor treatment was more direct, resulting in selective killing of monocytes and macrophages but not B and T cells present in the same compartment. Whether the cytotoxic effect of R191 against macrophages is a consequence of inhibition of TLR signals, which are required for the differentiation of blood monocytes into macrophages,49-51 or an unrelated pathway remains to be determined.
The capacity of macrophages to support CLL cell survival has been well established in numerous in vitro studies with primary CLL cells and has been further validated in in vivo studies with Eμ-TCL1–derived murine CLL cells or the human CLL cell line MEC1.43-46,52-55 The novelty of our findings in this respect is that the same effect was seen with nonimmortalized human RS-PDX cells, suggesting that therapeutic strategies based on interfering with macrophage-tumor cell interactions could provide a therapeutic benefit in this condition. In addition, coculture experiments showed enhanced ex vivo proliferation of the human RS-PDX cells in the presence of macrophages, whereas no proliferation was detected with nontransformed human CLL cells (supplemental Figure 8). This finding, together with previous observations that human RS cells, unlike human CLL cells, can be grown efficiently in the absence of T cells in immunodeficient mice,20,21,39,40 suggests that macrophage-derived signals, in combination with certain genetic lesions in cell cycle regulators,9 may substitute for T cell–derived signals in driving the proliferation of RS cells.
Acknowledgment
This research was supported by a grant from the Fondazione AIRC per la Ricerca sul Cancro (AIRC project IG 2020, ID no. 24566 [D.G.E.]).
Authorship
Contribution: C.M. and D.G.E. designed the study. C.M., S.C., M.V., and S.G. performed the experiments. T.V., S.D., L.L., and A.J.D. provided vital reagents and/or supervised certain experiments. C.M. and D.G.E. wrote the manuscript. All authors analyzed data and reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dimitar G. Efremov, Molecular Hematology Unit, ICGEB, Padriciano 99, 34149 Trieste, Italy; e-mail: efremov@icgeb.org.
References
Author notes
Data are available from the corresponding author, Dimitar G. Efremov (efremov@icgeb.org).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal