Key Points
ChAdOx1 nCoV-19 vaccine contains human TRex HEK293 cell-derived proteins and EDTA.
Vaccine components and PF4 form complexes on platelet surfaces to which VITT patient antibodies bind.
Abstract
SARS-CoV-2 vaccine ChAdOx1 nCoV-19 (AstraZeneca) causes a thromboembolic complication termed vaccine-induced immune thrombotic thrombocytopenia (VITT). Using biophysical techniques, mouse models, and analysis of VITT patient samples, we identified determinants of this vaccine-induced adverse reaction. Super-resolution microscopy visualized vaccine components forming antigenic complexes with platelet factor 4 (PF4) on platelet surfaces to which anti-PF4 antibodies obtained from VITT patients bound. PF4/vaccine complex formation was charge-driven and increased by addition of DNA. Proteomics identified substantial amounts of virus production-derived T-REx HEK293 proteins in the ethylenediaminetetraacetic acid (EDTA)-containing vaccine. Injected vaccine increased vascular leakage in mice, leading to systemic dissemination of vaccine components known to stimulate immune responses. Together, PF4/vaccine complex formation and the vaccine-stimulated proinflammatory milieu trigger a pronounced B-cell response that results in the formation of high-avidity anti-PF4 antibodies in VITT patients. The resulting high-titer anti-PF4 antibodies potently activated platelets in the presence of PF4 or DNA and polyphosphate polyanions. Anti-PF4 VITT patient antibodies also stimulated neutrophils to release neutrophil extracellular traps (NETs) in a platelet PF4-dependent manner. Biomarkers of procoagulant NETs were elevated in VITT patient serum, and NETs were visualized in abundance by immunohistochemistry in cerebral vein thrombi obtained from VITT patients. Together, vaccine-induced PF4/adenovirus aggregates and proinflammatory reactions stimulate pathologic anti-PF4 antibody production that drives thrombosis in VITT. The data support a 2-step mechanism underlying VITT that resembles the pathogenesis of (autoimmune) heparin-induced thrombocytopenia.
Introduction
Vaccination against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is an important countermeasure to fight the ongoing COVID-19 pandemic. The European Medicines Agency has approved 2 adenoviral (AV) vector-based vaccines: recombinant chimpanzee AV [ChAdOx1-S] vector encoding the spike glycoprotein of SARS-CoV-2, COVID-19 Vaccine AstraZeneca [ChAdOx1 nCoV-19; Vaxzevria]; and recombinant human AV type 26 vector encoding SARS-CoV-2 spike glycoprotein, COVID-19 Vaccine Janssen. ChAdOx1-S is propagated using T-REx HEK293 cells, a transformed human embryonic kidney cell line.1
Beginning in March 2021, cerebral venous sinus thrombosis (CVST), splanchnic vein thrombosis, and other unusual severe thrombotic events in combination with thrombocytopenia were reported in otherwise healthy individuals beginning 5 to 20 days following ChAdOx1 nCoV-19 vaccination. This novel disorder, “vaccine-induced immune thrombotic thrombocytopenia” (VITT; synonym, thrombosis with thrombocytopenia syndrome), has been associated with high-titer immunoglobulin G (IgG) class antibodies directed against the cationic platelet chemokine, platelet factor 4 (PF4).2 Anti-PF4 antibodies potently activate platelets with platelet activation greatly enhanced by PF4.2,3 Pathologic anti-PF4 antibodies were infrequently found in CVST patients prior to VITT, suggesting that the vaccine-induced antibodies drive these thrombotic complications.4
PF4 opsonizes negatively charged surfaces of microbial pathogens, facilitating binding of anti-PF4 antibodies.5 This is likely an evolutionary ancient immune defense mechanism.6-8 However, a misdirected strong anti-PF4 antibody response underlies the thromboembolic disorder immune heparin-induced thrombocytopenia (HIT; caused by anti-PF4/heparin antibodies) and its most severe presentation, autoimmune HIT.9-12 This latter subtype of HIT is characterized by the formation of high-avidity platelet-activating anti-PF4 antibodies that are reactive even in the absence of heparin.13
HIT proceeds by a 2-step mechanism: initially, PF4/heparin complexes expose a neoantigen on PF4 that stimulates B cells to produce high-affinity anti-PF4/heparin antibodies in the presence of proinflammatory costimuli. Five to 10 days following heparin exposure, sufficient quantities of these antibodies are present to activate cellular Fcγ receptors on platelets, monocytes, and granulocytes, culminating in life-threatening thrombosis. Major risk factors for forming anti-PF4 antibodies and for developing HIT are inflammation and tissue trauma. Both disease states provide immunologic “danger signals” that increase the likelihood and intensity of forming an anti-PF4 immune response.14,15 Marginal zone B cells mediate anti-PF4/heparin antibody production in HIT, and their activation depends on Notch-2 signaling.16-18 Although anti-PF4/heparin antibodies develop commonly after heparin exposure, only a small subset of heparin-sensitized patients develop thrombocytopenia and thrombosis. Despite decades of research, comprehensive insight into the factors that predispose to adverse heparin-induced immune thrombotic events has remained enigmatic.19
VITT closely mimics autoimmune HIT both clinically and serologically20; however, the nature of neoantigens that trigger pathologic anti-PF4 antibodies, the “danger signal(s)” that prime for adverse immune reactions and prothrombotic mechanisms, remains to be established in VITT. Here, we identify key components of VITT immunopathogenesis. The data suggest that VITT proceeds via a 2-step mechanism: (1) Vaccine components, including the AV hexon protein, form complexes with PF4, leading to neoantigen exposure on PF4. Vaccine components also have the capability to trigger proinflammatory responses that are “danger signals” known to amplify anti-PF4 antibody production in autoimmune HIT. (2) Between days 5-20 postvaccination, anti-PF4 antibodies from VITT patients activate platelets in a PF4- and polyanion-dependent manner. Additionally, anti-PF4 antibodies activate granulocytes in the presence of platelets to release procoagulant neutrophil extracellular traps (NETs) that are found in abundance in CVSTs of VITT patients. Together, the data highlight similarities in VITT and HIT pathogenesis and identify strategies to interfere with VITT-driven thrombotic events.
Methods
Detailed description of antibodies, reagents, and additional methods can be found in supplemental Methods (available on the Blood Web site).
3D super-resolution microscopy
For three-dimensional (3D) super-resolution microscopy imaging of platelets and immobilized ChAdOx1 nCoV-19 vaccine, coverslips were mounted inversely in Mowiol (Carl Roth, Karlsruhe, Germany) or Everspark buffer (Idylle Labs, Paris, France) for direct stochastic optical reconstruction microscopy (dSTORM) localization microscopy. 3D super-resolution microscopy imaging was either performed at NIPOKA GmbH, Greifswald, Germany on a Nikon N-SIM E system (Nikon Instruments, Tokyo, Japan) equipped with 488 and 640 nm laser lines and a 100x 1.35NA silicon immersion objective, or on a Zeiss Elyra PS.1 super-resolution system equipped with a 63x 1.4NA oil-immersion objective. Raw image stacks with 3 rotations and 5 shifts of the illumination grating were acquired and reconstructed as super-resolved 3D SIM datasets as described.21 Reconstructed data were saved as .nd or .czi files and imported to FIJI22 using the BioFormats importer. Chromatic aberration was corrected using analogous prepared reference slides coated with 100 nm Tetraspek beads diluted 1:2000 in ultrapure water. Data were analyzed using customized FIJI and NIS scripts. Full dSTORM super-resolution imaging methods are given in supplemental Methods.
Dynamic light scattering
All dynamic light scattering (DLS) measurements were performed in a fixed scattering angle Zetasizer Nano-S system (Malvern Instruments Ltd, Malvern, United Kingdom). The hydrodynamic diameter (nm) was measured at 25°C, and light scattering was detected at 173° using 3 repeating measurements consisting of 12 runs. Experimental data were collected from 4 independent experimental replicates. For all DLS measurements, ChAdOx1 nCoV-19 was diluted at a ratio of 1:10 in sterile-filtered 0.9% NaCl supplemented with 4 mg/mL D (+) saccharose (ribonuclease/DNase free; catalog no. 9097.1; Carl Roth GmbH, Germany). Assessment of changes in the hydrodynamic diameter of ChAdOx1 nCoV-19 vector in the presence of PF4 was performed by incubating 10 µg/mL of human PF4 (Chromatec, Greifswald, Germany) with ChAdOx1 nCoV-19 vector at room temperature (RT) for 2 minutes before DLS measurements. Anti-PF4 mouse monoclonal IgG (Clone RTO, catalog no. MA5-17639, Invitrogen) affinity-purified anti-PF4 IgG antibodies from VITT sera, and control human IgG from a healthy individual were purified by Protein G affinity purification and were used at 5 µg/mL final concentration. Double-stranded annealed DNA 25-mer was used at 0.5 µg/mL. Dissociation of complexes formed between ChAdOx1 nCoV-19 vector and added components was achieved by 100 IU/mL unfractionated heparin (UFH; Ratiopharm GmbH, Ulm, Germany).
Transmission electron microscopy
For transmission electron microscopy (TEM), the vaccine was incubated with biotinylated PF4 (10 ng/mL in phosphate-buffered saline [PBS]; PF4-biotin) for at least 1 hour at RT. The sample was transferred to formvar-coated TEM grids (400 mesh; Plano GmbH, Germany), washed with PBS, and blocked with bovine serum albumin (BSA) in PBS. Samples were labeled with an anti-AV mAb (B025/AD51, ab7428, 1:500; Abcam, Waltham, MA) for 1 hour at RT and detected with an anti-mouse gold conjugate secondary antibody (GMHL10, 10 nm, 1:50; BBI Solutions). Alternatively, for PF4-biotin staining, the same samples were labeled with a streptavidin-gold conjugate (10 nm, 1:10; Sigma) for 45 minutes at RT. All grids were stained with 1% phosphotungstic acid at pH 7.4 and analyzed with a Tecnai-Spirit (FEI, Eindhoven, The Netherlands) transmission electron microscope at an accelerating voltage of 80 kV. The same procedure was used for preparing control samples of vaccine alone and PF4-biotin. The vaccine was also incubated with heparin (100 IU/mL) for 30 minutes. After that, samples were transferred to formvar-coated TEM grids (400 mesh; Plano GmbH) and processed as above.
Composition of ChAdOx1 CoV-19 vaccine
ChAdOx1 CoV-19 vaccine was analyzed by mass spectrometry, 1H-nuclear magnetic resonance (NMR) spectroscopy, and 1-dimensional sodium dodecyl sulphate-polyacrylamide gel electrophoresis (1D SDS-PAGE) (detailed methods are given in supplemental Methods).
Platelet activation assay with washed platelets (PIPA test)
Platelet activation and aggregation by VITT patient serum were tested as previously described.2 In brief, washed platelets of at least 3 healthy donors were incubated in the absence or presence of either PF4 (10 mg/mL), DNA (ds 25-mer, 1 µg/mL), or EDTA (0.2 µM) and serum from VITT patients under stirring conditions. Therefore, 75 µL platelet suspension was mixed with 20 µL serum prior to stirring. The time to aggregation was measured up to 45 minutes. The test was determined to be positive if platelets aggregated within 30 minutes.
Immunofluorescence staining and confocal imaging of cerebral sinus vein thrombus
For visualization of NETs within cerebral sinus vein thrombi, 5 µm paraffin sections were deparaffinized and rehydrated prior to antigen retrieval using sodium citrate. Slides were blocked in 2% BSA and 0.1% Triton X-100 in PBS for 45 minutes at RT, followed by incubation with primary antibodies overnight at 4°C. After washing 3 times with PBS, sections were incubated with secondary antibodies for 60 minutes at RT before further washing and quenching autofluorescence with 0.1% Sudan Black B in 70% ethanol for 25 minutes. Sections were washed again and covered with Dako fluorescence mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI).
For visualization of NETs, 100 µm transverse vibratome sections of a cerebral sinus vein thrombus from a VITT patient and a nonvaccinated control patient were permeabilized using 0.5% Triton X-100 in PBS for 20 minutes at RT, followed by blocking with 3% BSA in PBSTx for 2 hours. Primary antibodies were incubated overnight at 4°C, washed 3 times with PBSTx, and subsequently incubated with secondary antibodies overnight at 4°C before further washing and mounting in Dako fluorescence mounting medium.
The following primary antibodies were used: antineutrophil elastase (NE; 1:100, ab68672; Abcam), antichromatin (antihistone H2A/H2B/DNA-complex, 5 µg/mL; Davids Biotechnologie GmbH), anti-VWF (1:100, A0082; Dako), and antimyeloperoxidase (MPO) (1:25, ab9535; Abcam). Secondary antibodies conjugated to AlexaFluor-488 and −594 were obtained from Jackson ImmunoResearch (all donkey, used in 1:200 dilution). Confocal tissue images represent maximum intensity projections of z stacks that were acquired using a Leica SP8 inverted confocal microscope with 10x HC PL APO CS and 63x HC PL APO Oil CS2 objectives and Leica LAS-X software.
Results
ChAdOx1 nCoV-19 vaccine constituents and PF4 form immunogenic complexes recognized by VITT patient anti-PF4 IgG
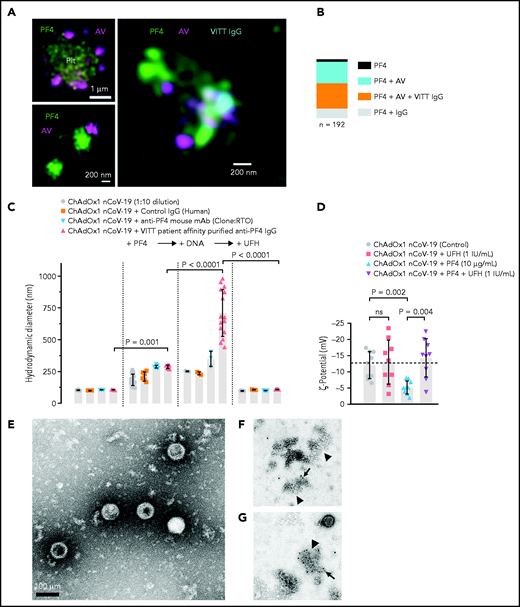
ChAdOx1 nCoV-19 vaccination is associated with delayed local and systemic reactions after the first administration indicating immunogenic reactions triggered by vaccine components.23 We analyzed interactions of vaccine constituents with blood by biophysical techniques. 3D super-resolution immunofluorescence microscopy visualized complexes formed between AV-derived hexon protein and platelets (Figure 1A, upper left panel), as well as between AV particles and PF4 (Figure 1A, lower left panel). These interactions led to complexes comprised of vaccine particles and PF4 to which VITT patient-derived anti-PF4 IgG bound on platelet surfaces (Figure 1A, right panel). Quantification of the components of the particles revealed that 45% contained ternary complexes of PF4, AV hexon proteins, and anti-PF4 antibodies, whereas ∼50% were positive for PF4/AV hexon protein or PF4/anti-PF4 antibody complexes only (Figure 1B). DLS (Figure 1C) confirmed formation of PF4/vaccine aggregates that were recognized by VITT patient antibodies: Addition of PF4 (diameter 5 nm24) to ChAdOx1 nCoV-19 vaccine increased the size of AV particles from 105.5 ± 2.8 nm to 185.2 ± 44.5 nm (mean diameter ± standard deviation; P < .001). Subsequently, addition of VITT patient-derived, affinity-purified anti-PF4 IgG or monoclonal anti-PF4 IgG (mAb clone RTO) further increased the diameter of PF4/ChAdOx1 nCoV-19 vaccine complexes to 288.4 ± 10.4 nm and 288.5 ± 13.9 nm, respectively. Adding DNA further increased the size of VITT patient antibody/PF4/ChAdOx1 nCoV-19 complexes to 710.4 ± 183 nm, whereas heparin (≥1 IU/mL) dissociated 98.5% of complexes. As an indication of charge-driven complex formation of vaccine components with PF4, surface ζ potential of the vaccine particles was negative (−12 ± 4.2 mV), and the addition of PF4, but not heparin, neutralized their negative charge (-5.2 ± 2.1 and −13 ± 6.8 mV, respectively; Figure 1D). Consistent with DLS, TEM visualized AV particles and small amorphous structures contained in the vaccine (Figure 1E). Larger aggregates formed upon incubation of the vaccine with PF4. The aggregates stained positive for AV hexon polypeptide and PF4 (Figure 1F-G; supplemental Figure 1). Both patient-purified and hybridoma-cell derived anti-PF4 antibodies specifically detected PF4 alone or PF4 in a mixture with the vaccine; however, there was no detectable signal with ChAdOx1 nCoV-19 vaccine alone. Similarly, anti-AV hexon protein antibodies stained the ChAdOx1 nCoV-19 vaccine alone or in mixture with PF4; however, they did not bind to PF4 alone (supplemental Figure 2). The data reveal charge-driven complex formation of PF4 and AV hexon proteins, to which VITT patient anti-PF4 IgG bound.
Complex formation of PF4, vaccine components, and anti-PF4 antibodies on platelet surfaces. (A) 3D super-resolution microscopy of PF4, AV and affinity-purified anti-PF4 antibodies (obtained from VITT patients) reveals complex formation. Upper left panel: PF4 (green) and AV hexon protein (purple) accumulation on platelet surfaces. Scale bar, 1 µm. Lower left panel: Localization microscopy (dSTORM) of PF4 (green) at ChAdOx1 vaccine aggregates (AV, purple). Scale bar, 200 nm. Right panel: colocalization of ChAdOx1 AV hexon protein (AV; purple), PF4 (green) and purified anti-PF4 IgG from VITT patient sera (blue). Scale bar, 200 nm. (B) Relative composition of 192 complexes analyzed. Approximately 44.5% of complexes investigated showed VITT anti-PF4 IgG bound to particles containing both PF4 and AV hexon proteins. (C) Analyses of ChAdOx1 nCoV-19 vaccine by DLS. Diameter of ChAdOx1 nCoV-19 vaccine aggregates before (left) and after addition of PF4 (+PF4, 10 µg/ml). Incubation of mouse anti-P F4 recombinant antibody (clone RTO) or purified anti-PF4 IgG from VITT patients increased the size of vaccine aggregates in the presence of PF4. Addition of DNA further increased the size of PF4/vaccine aggregates (+DNA). In contrast, addition of heparin (100 IU/mL) dissociated previously formed vaccine/PF4/anti-PF4 IgG complexes. Each data point represents 12 runs of n ≥3 individual measurements. Statistical assessment by ordinary 1-way analysis of variance (ANOVA) with Sidak's multiple comparisons test. (D) Negative charge indicated by surface ζ potential (ζ,13 mV) of ChAdOx1 nCoV-19 vaccine particles in the presence of buffer (control), UFH (1 IU/mL), PF4 (reduced the negative charge; −5 mV), and coapplication of PF4 and heparin. Statistical assessment by Brown-Forsythe and Welch ANOVA test followed by Dunnett's T3 multiple comparisons test. Transmission electron microscopy images of aggregates formed in the vaccine upon addition of PF4. (E) Vaccine without added PF4 shows the virion particles and multiple small amorphous structures. (F) Aggregate (arrowhead) formation in the vaccine following addition of PF4. Biotinylated PF4 (arrow) is labeled with 10 nm gold particles. (G) Aggregate (arrowhead) as in panel F. Here, AV capsid protein (arrow) is labeled with 10 nm gold particles. Scale bars, 100 nm.
Complex formation of PF4, vaccine components, and anti-PF4 antibodies on platelet surfaces. (A) 3D super-resolution microscopy of PF4, AV and affinity-purified anti-PF4 antibodies (obtained from VITT patients) reveals complex formation. Upper left panel: PF4 (green) and AV hexon protein (purple) accumulation on platelet surfaces. Scale bar, 1 µm. Lower left panel: Localization microscopy (dSTORM) of PF4 (green) at ChAdOx1 vaccine aggregates (AV, purple). Scale bar, 200 nm. Right panel: colocalization of ChAdOx1 AV hexon protein (AV; purple), PF4 (green) and purified anti-PF4 IgG from VITT patient sera (blue). Scale bar, 200 nm. (B) Relative composition of 192 complexes analyzed. Approximately 44.5% of complexes investigated showed VITT anti-PF4 IgG bound to particles containing both PF4 and AV hexon proteins. (C) Analyses of ChAdOx1 nCoV-19 vaccine by DLS. Diameter of ChAdOx1 nCoV-19 vaccine aggregates before (left) and after addition of PF4 (+PF4, 10 µg/ml). Incubation of mouse anti-P F4 recombinant antibody (clone RTO) or purified anti-PF4 IgG from VITT patients increased the size of vaccine aggregates in the presence of PF4. Addition of DNA further increased the size of PF4/vaccine aggregates (+DNA). In contrast, addition of heparin (100 IU/mL) dissociated previously formed vaccine/PF4/anti-PF4 IgG complexes. Each data point represents 12 runs of n ≥3 individual measurements. Statistical assessment by ordinary 1-way analysis of variance (ANOVA) with Sidak's multiple comparisons test. (D) Negative charge indicated by surface ζ potential (ζ,13 mV) of ChAdOx1 nCoV-19 vaccine particles in the presence of buffer (control), UFH (1 IU/mL), PF4 (reduced the negative charge; −5 mV), and coapplication of PF4 and heparin. Statistical assessment by Brown-Forsythe and Welch ANOVA test followed by Dunnett's T3 multiple comparisons test. Transmission electron microscopy images of aggregates formed in the vaccine upon addition of PF4. (E) Vaccine without added PF4 shows the virion particles and multiple small amorphous structures. (F) Aggregate (arrowhead) formation in the vaccine following addition of PF4. Biotinylated PF4 (arrow) is labeled with 10 nm gold particles. (G) Aggregate (arrowhead) as in panel F. Here, AV capsid protein (arrow) is labeled with 10 nm gold particles. Scale bars, 100 nm.
ChAdOx1 nCoV-19 vaccination-induced inflammatory reactions in mice and humans
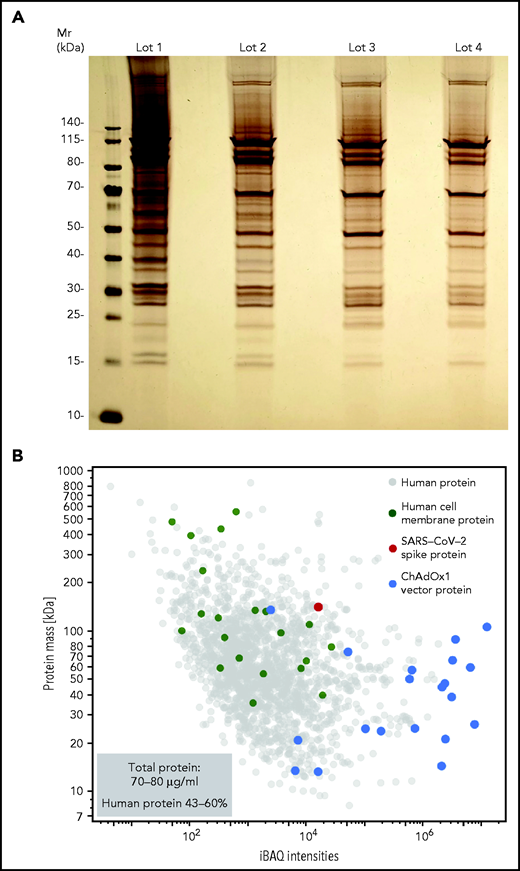
We hypothesized that ChAdOx1 nCoV-19 vaccine induces a proinflammatory “danger signal” that promotes pathologic anti-PF4 antibody production in autoimmune HIT and possibly in VITT.25,26 Accordingly, we sought to characterize the vaccine composition. Unexpectedly, we found 70-80 µg protein/mL in 4 independent ChAdOx1 nCoV-19 vaccine lots analyzed. Silver staining of ChAdOx1 nCoV-19 vaccine separated on SDS-PAGE showed numerous protein bands (Figure 2A). Proteomics identified AV vector proteins (blue dots), virus production-derived T-REx HEK293 (human) proteins (gray dots) (some constituting membrane proteins [green dots]), and the SARS-CoV-2 spike protein (red dot) in the ChAdOx1 nCoV-19 vaccine. Approximately 43% to 60% of the protein content of the vaccine (15-24 µg per dose) was assigned to T-REx HEK293 cell protein origin (Figure 2B; supplemental Table 1).
ChAdOx1 nCoV-19 vaccine contains multiple proteins originating from the AV production process. (A) Four distinct lots of ChAdOx1 nCoV-19 vaccine were separated by 1D SDS-PAGE and proteins were visualized by silver staining. (B) Proteomics of ChAdOx1 nCoV-19 vaccine: iBAQ protein intensities and theoretical molecular masses of identified proteins. Protein intensities of vaccine lot 3 were calculated using the iBAQ algorithm (≥3 unique peptides per protein) and plotted against theoretical molecular mass. Human proteins are indicated in gray and ChAdOx1 proteins in blue. Furthermore, green dots mark human membrane proteins (UniProt annotation) and the single red dot shows the SARS-CoV-2 spike protein. Total amount of protein determined in 5 different lots ranged between 70-80 µg/mL, with human proteins constituting ∼43% to 60% of total proteins.
ChAdOx1 nCoV-19 vaccine contains multiple proteins originating from the AV production process. (A) Four distinct lots of ChAdOx1 nCoV-19 vaccine were separated by 1D SDS-PAGE and proteins were visualized by silver staining. (B) Proteomics of ChAdOx1 nCoV-19 vaccine: iBAQ protein intensities and theoretical molecular masses of identified proteins. Protein intensities of vaccine lot 3 were calculated using the iBAQ algorithm (≥3 unique peptides per protein) and plotted against theoretical molecular mass. Human proteins are indicated in gray and ChAdOx1 proteins in blue. Furthermore, green dots mark human membrane proteins (UniProt annotation) and the single red dot shows the SARS-CoV-2 spike protein. Total amount of protein determined in 5 different lots ranged between 70-80 µg/mL, with human proteins constituting ∼43% to 60% of total proteins.
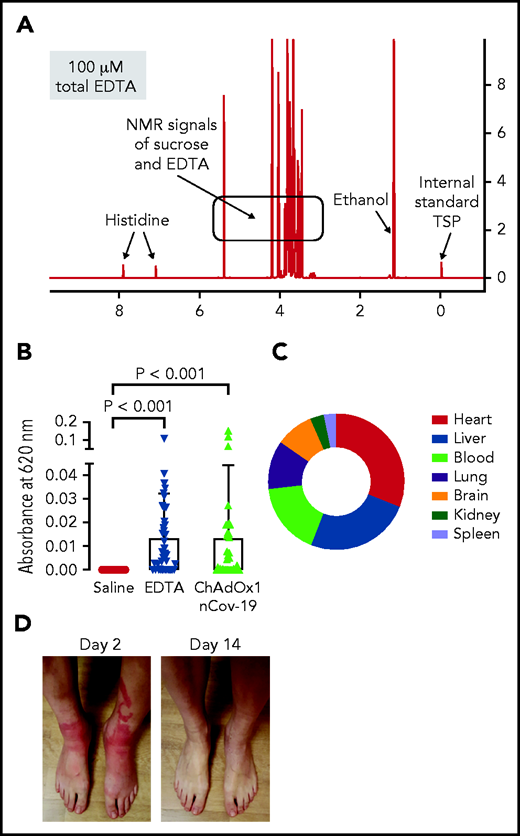
We further analyzed the ChAdOx1 nCoV-19 vaccine for small molecules by 1H-NMR spectrometry and identified EDTA (∼100 µM; Figure 3A). Leaky vessels are a hallmark of inflammation, and the Ca2+ chelator EDTA increases vascular leakage by vascular endothelial (VE)-cadherin endothelial junctional disassembly.27 We therefore analyzed ChAdOx1 nCoV-19 vaccine for inducing proinflammatory reactions in mice using the Miles edema model (Figure 3B). Intradermally injected vaccine triggered leakage in dermal vessels that was quantified by Evans Blue tracer extravasation. Increase of vascular permeability appeared mostly mediated by EDTA in the vaccine, as intradermal injection of EDTA alone (100 µM) stimulated vascular leakage to a similar extent as the vaccine. Moreover, reconstituting the ChAdOx1 nCoV-19 vaccine and EDTA with Ca2+ (100 µM) prior to intradermal injection prevented vaccine- and EDTA-triggered increases in permeability and vascular leakage (supplemental Figure 3). We assessed consequences of EDTA-induced local vascular leakage for dissemination of vaccine components in challenged mice. The vast majority of the intradermally-injected AV DNA remained at the injection site (>99.99% at 30 minutes postinjection, supplemental Figure 4). However, digital polymerase chain reaction (PCR) detected ChAdOx1 nCoV-19–specific sequences in multiple tissues including the brain, as well as the spleen, where B cells are enriched in the splenic marginal zones28,29 (Figure 3C).
ChAdOx1 nCoV-19 vaccine contains EDTA and induces inflammatory reactions in mouse edema models and vaccinated individuals. (A) 1H-NMR spectrum of ChAdOx1 nCoV-19 vaccine showed signals of sucrose, ethanol, histidine, and EDTA (∼100 µM). X-axis: NMR chemical shift signals in ppm relative to internal standard TSP; y-axis: relative signal intensity. (B) Skin edema in wild-type mice was induced by intradermal injection of 50 μL each saline, EDTA (100 μM), or ChAdOx1 nCoV-19. Evans blue dye was intravenously infused as a tracer, and after 30 minutes extravasated dye was quantified. Columns show mean ± SD, n = 41 per group. Paired 1-way ANOVA followed by Dunn’s multiple comparison test. (C) Digital PCR quantified AV DNA 30 minutes after intradermal injection of 50 μL ChAdOx1 nCoV-19 in wild-type mice. Segments give relative percentage in multiple organs of disseminated ChAdOx1 nCoV-19 AV copy numbers; n = 3 individual experiments. (D) Skin reaction 2 days after ChAdOx1 nCov-19 vaccination and following symptom resolution on day 14. D-Dimer was elevated at 4 and 6 days following ChAdOx1 nCoV-19 vaccination, with symptom resolution in following days.
ChAdOx1 nCoV-19 vaccine contains EDTA and induces inflammatory reactions in mouse edema models and vaccinated individuals. (A) 1H-NMR spectrum of ChAdOx1 nCoV-19 vaccine showed signals of sucrose, ethanol, histidine, and EDTA (∼100 µM). X-axis: NMR chemical shift signals in ppm relative to internal standard TSP; y-axis: relative signal intensity. (B) Skin edema in wild-type mice was induced by intradermal injection of 50 μL each saline, EDTA (100 μM), or ChAdOx1 nCoV-19. Evans blue dye was intravenously infused as a tracer, and after 30 minutes extravasated dye was quantified. Columns show mean ± SD, n = 41 per group. Paired 1-way ANOVA followed by Dunn’s multiple comparison test. (C) Digital PCR quantified AV DNA 30 minutes after intradermal injection of 50 μL ChAdOx1 nCoV-19 in wild-type mice. Segments give relative percentage in multiple organs of disseminated ChAdOx1 nCoV-19 AV copy numbers; n = 3 individual experiments. (D) Skin reaction 2 days after ChAdOx1 nCov-19 vaccination and following symptom resolution on day 14. D-Dimer was elevated at 4 and 6 days following ChAdOx1 nCoV-19 vaccination, with symptom resolution in following days.
Consistent with the dissemination of vaccine constituents in the mouse model, ChAdOx1 nCoV-19 vaccination triggered systemic inflammatory reactions in humans. Out of 22 healthy, vaccinated health care workers, 12 reported adverse effects including fever, chills, and joint pain starting 6-12 hours after vaccination, usually resulting in 1 to 2 days of sick leave. A representative example of skin inflammation associated with high levels of D-Dimer (1,115 ng/mL) indicating proinflammatory reactions at days 1-2 after vaccination is shown in Figure 3D. Taken together, the data are consistent with a model of neoantigen formation induced by vaccine constituents and PF4, which jointly stimulate pathologic anti-PF4 antibody formation facilitated by a vaccine-triggered inflammatory costimulus.
Pathologic anti-PF4 antibodies activate platelets and neutrophils, leading to thrombosis
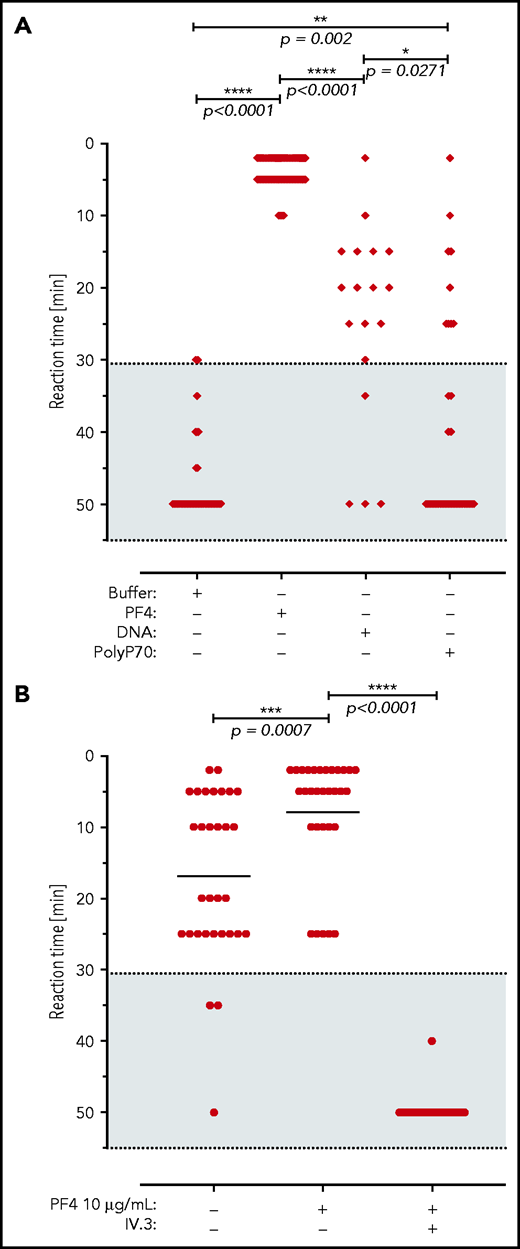
We next analyzed for thrombotic reactions triggered by VITT patient anti-PF4 antibodies. Consistent with our previously published data,2 all (14/14) sera of VITT patients analyzed, as well as their respective affinity-purified antibody fractions, showed strong reactivity toward immobilized PF4/heparin in an enzyme-linked immunosorbent assay (ELISA). In a washed platelet aggregation-based assay, the addition of PF4 amplified platelet activation triggered by VITT patient sera (up to a serum titer of 1:1000, Figure 4A). Similarly, addition of short-chain polyphosphate (a platelet-derived inorganic polymer,30 0.2 µg/mL) or synthetic DNA (1 µg/mL) increased VITT patient serum-initiated platelet activation, albeit to a lower extent compared with PF4. VITT patient serum/PF4-stimulated platelet aggregation was completely inhibited by FcγRIIA receptor blockade using IV.3 antibody (Figure 4B). Taken together, the data indicate a function of anti-PF4 IgG/PF4 complexes visualized in Figure 1 in mediating platelet activation.
VITT patient antibodies activate platelets in a PF4, FcγRIIA, and polyanion dependent manner. (A) Platelet activation by VITT sera in the presence of buffer (n = 60 independent experiments), PF4 (n = 78), DNA (n = 18), and short-chain polyphosphate (polyP70; n = 29). Numbers refer to total experiments with 14 VITT sera and healthy donor platelets. A shorter reaction time indicates stronger platelet activation. Patient sera and platelet donors were selected for these experiments by assessing those VITT sera that did not induce strong platelet activation in the presence of buffer to enable cross-reactivity testing. (B) Sera of VITT patients were incubated with washed platelets from healthy donors in the presence or absence of PF4 (10 µg/mL) and/or monoclonal antibody IV.3, which blocks the platelet FcγRIIA receptor (n = 31). Datasets were compared using Wilcoxon matched-pairs signed rank test.
VITT patient antibodies activate platelets in a PF4, FcγRIIA, and polyanion dependent manner. (A) Platelet activation by VITT sera in the presence of buffer (n = 60 independent experiments), PF4 (n = 78), DNA (n = 18), and short-chain polyphosphate (polyP70; n = 29). Numbers refer to total experiments with 14 VITT sera and healthy donor platelets. A shorter reaction time indicates stronger platelet activation. Patient sera and platelet donors were selected for these experiments by assessing those VITT sera that did not induce strong platelet activation in the presence of buffer to enable cross-reactivity testing. (B) Sera of VITT patients were incubated with washed platelets from healthy donors in the presence or absence of PF4 (10 µg/mL) and/or monoclonal antibody IV.3, which blocks the platelet FcγRIIA receptor (n = 31). Datasets were compared using Wilcoxon matched-pairs signed rank test.
Anti-PF4 antibodies of HIT patients activate neutrophils and are a major driver of thrombosis by the formation of procoagulant NETs (NETosis).31 We tested pathologic anti-PF4 antibodies of VITT patients for their potency to trigger neutrophil activation and procoagulant NET formation in the presence of PF4 or platelets. Incubation of isolated human neutrophils and platelets with VITT patient serum induced NET formation (Figure 5A) that was significantly increased by the addition of PF4 (Figure 5B,K). In contrast, healthy control serum did not trigger NET formation (Figure 5C,D,K). Similar to VITT patient serum, affinity-purified anti-PF4 antibodies from these respective samples triggered NETosis (Figure 5E,K). PF4 addition strongly enhanced VITT patient serum-stimulated NET formation, confirming that PF4 in anti-PF4 IgG VITT antibodies initiates procoagulant NET formation (Figure 5F,K). NETosis was virtually absent when neutrophils and platelets were coincubated in buffer, in the absence or presence of added PF4 (Figure 5G-H). In the absence of platelets, VITT patient serum failed to induce NETosis even in the presence of added PF4, suggesting that in VITT platelets play a key role in triggering NET formation (Figure 5I-J).
VITT patient anti-PF4 antibodies trigger NETosis. (A-B) Confocal laser scanning microscopy images of in vitro NETs generated by human neutrophils and platelets following incubation with VITT patient serum alone (A) or VITT patient serum in combination with PF4 (B). (C-D) NET formation induced in healthy control serum alone (C) or healthy serum in the presence of PF4 (D). (E-H) NET formation induced by affinity-purified (Affi.pur.) anti-PF4 IgG from VITT patients (E); combination of anti-PF4 IgG from VITT patients and PF4 (F); buffer; (G) or PF4 only (H). (I-J) Neutrophils (without [w/o] platelets) were incubated with VITT patient serum (I) or VITT patient serum and PF4 (J), and NET formation was measured. (K) Quantification of NETs (NETosis [%]) from confocal laser scanning microscopy images obtained from nuclear and extracellular DNA fluorescent channels was performed using at least 12 individual images for each condition, n = 3 experimental replicates. Statistical analysis was performed using the Welch and Brown-Forsythe ANOVA multiple comparisons test with post hoc 2-stage step-up method per Benjamini, Krieger, and Yekutieli, q = 0.05.
VITT patient anti-PF4 antibodies trigger NETosis. (A-B) Confocal laser scanning microscopy images of in vitro NETs generated by human neutrophils and platelets following incubation with VITT patient serum alone (A) or VITT patient serum in combination with PF4 (B). (C-D) NET formation induced in healthy control serum alone (C) or healthy serum in the presence of PF4 (D). (E-H) NET formation induced by affinity-purified (Affi.pur.) anti-PF4 IgG from VITT patients (E); combination of anti-PF4 IgG from VITT patients and PF4 (F); buffer; (G) or PF4 only (H). (I-J) Neutrophils (without [w/o] platelets) were incubated with VITT patient serum (I) or VITT patient serum and PF4 (J), and NET formation was measured. (K) Quantification of NETs (NETosis [%]) from confocal laser scanning microscopy images obtained from nuclear and extracellular DNA fluorescent channels was performed using at least 12 individual images for each condition, n = 3 experimental replicates. Statistical analysis was performed using the Welch and Brown-Forsythe ANOVA multiple comparisons test with post hoc 2-stage step-up method per Benjamini, Krieger, and Yekutieli, q = 0.05.
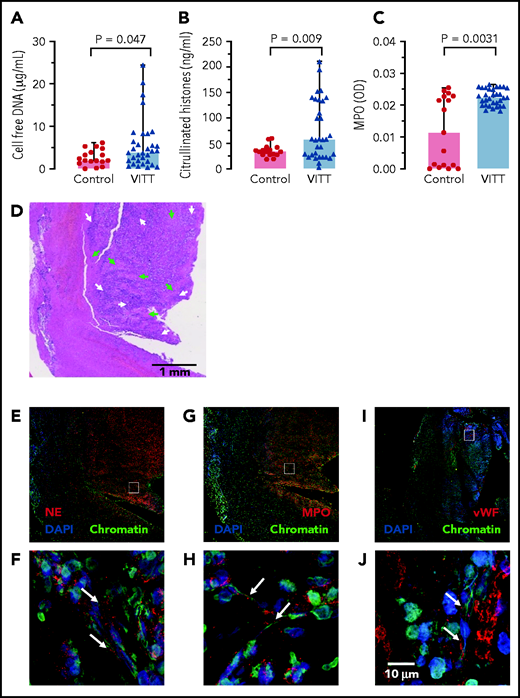
Supporting a role of NETs in VITT, 3 NET biomarkers: cell-free DNA, citrullinated histones, and MPO (marker for neutrophil activation), were elevated in VITT patient sera compared with healthy controls (Figure 6A-C). CVST is a hallmark complication reported in many VITT patients. We analyzed the composition of cerebral sinus vein thrombi obtained by thrombectomy from 1 VITT patient or by autopsy from a second VITT patient, respectively. Histology of the thrombectomy-derived tissue showed regions with amorphous material, likely representing fibrin deposition surrounded by nucleated cell-rich areas throughout the thrombi (Figure 6D). Immunohistochemistry revealed that these cell-rich areas contained abundant activated neutrophils and NETs. Antibodies against the NET biomarker NE (Figure 6E-F) and MPO (Figure 6G-H) visualized degranulated neutrophils releasing elongated DAPI- and chromatin-positive NETs into the platelet-rich (Figure 6I-J) thrombus. A highly similar NET staining pattern was observed in a cerebral sinus vein thrombus of a second VITT patient obtained at the time of autopsy, whereas cerebral sinus vein thrombus regions from a nonvaccinated control patient contained less neutrophils (supplemental Figure 5). Together, for the first time the data visualize NETs in VITT-induced cerebral sinus vein thrombosis, indicating a causal relationship of NET prothrombotic mechanisms and VITT.
NETs in VITT patient cerebral sinus vein thrombi. (A) Cell-free DNA serum levels of VITT patients and controls using fluorescent DNA-intercalating dye Sytox Green. Statistical comparison by unpaired Student t test; (B) Serum citrullinated histone H3 (CitH3) levels in VITT patients and healthy controls measured by ELISA. Statistical comparison by unpaired Student t test; (C) Serum MPO levels in VITT patients and healthy controls quantified using an ELISA. Mann-Whitney nonparametric test compared the 2 groups. (D) Hematoxylin and eosin (H&E) stained histologic section of a cerebral venous sinus thrombus of a VITT patient. Arrows indicate amorphous fibrin (green) and granulocyte-rich areas (white) in the thrombus core, respectively (E-J). Immunohistochemistry images of the VITT patient cerebral sinus vein thrombus shown in panel A. The section assessed in more detail in panels C, E, and G are given as small rectangles in panels B, D, and F. Markers for NETs and neutrophils include NE (red) in panels E and F, and MPO (red) in panels G and H. Von Willebrand factor (vWF, red) is shown in panels I and J for comparison. DAPI stains DNA in blue while chromatin (antihistone H2A/H2B/DNA-complexes) is shown in green.
NETs in VITT patient cerebral sinus vein thrombi. (A) Cell-free DNA serum levels of VITT patients and controls using fluorescent DNA-intercalating dye Sytox Green. Statistical comparison by unpaired Student t test; (B) Serum citrullinated histone H3 (CitH3) levels in VITT patients and healthy controls measured by ELISA. Statistical comparison by unpaired Student t test; (C) Serum MPO levels in VITT patients and healthy controls quantified using an ELISA. Mann-Whitney nonparametric test compared the 2 groups. (D) Hematoxylin and eosin (H&E) stained histologic section of a cerebral venous sinus thrombus of a VITT patient. Arrows indicate amorphous fibrin (green) and granulocyte-rich areas (white) in the thrombus core, respectively (E-J). Immunohistochemistry images of the VITT patient cerebral sinus vein thrombus shown in panel A. The section assessed in more detail in panels C, E, and G are given as small rectangles in panels B, D, and F. Markers for NETs and neutrophils include NE (red) in panels E and F, and MPO (red) in panels G and H. Von Willebrand factor (vWF, red) is shown in panels I and J for comparison. DAPI stains DNA in blue while chromatin (antihistone H2A/H2B/DNA-complexes) is shown in green.
Discussion
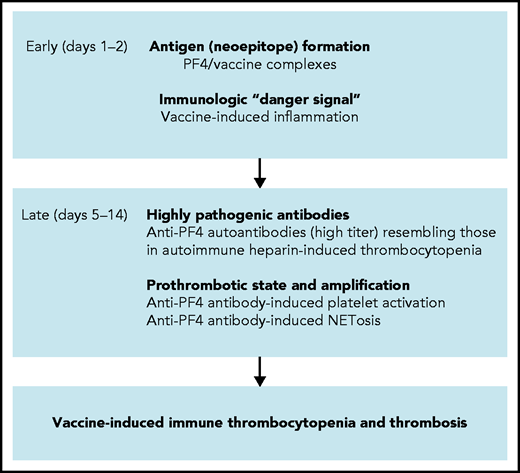
Based on a combination of biophysical imaging, mouse models, and analysis of VITT patient material, our study suggests a 2-step mechanism underlying VITT-driven thrombosis that is schematically shown in Figure 7. VITT pathogenesis is reminiscent of autoimmune HIT pathology. Initially, neoantigens are generated by interaction of PF4 with vaccine components. As visualized by TEM, 3D super-resolution microscopy, and DLS, in reconstituted systems PF4 has the capacity to bind to vaccine constituents, leading to the formation of complexes that contain AV proteins (Figure 1; supplemental Figures 1 and 2). Although PF4/vaccine aggregates are exposed on platelet surfaces and recognized ex vivo by VITT patient anti-PF4 antibodies, the precise sequence of events occurring in vivo at injection sites or in blood requires additional studies. In humans, PF4 is enriched at the vessel wall and is locally released in high concentrations following platelet activation.32,33 Consistent with our imaging data, previous studies have shown that coronaviruses have the capacity to activate platelets34,35 and that AVs binding to platelets can lead to platelet-activation and release of PF4.36
Scheme of proposed procoagulant mechanisms in VITT. Early reactions: Initial VITT reactions are triggered by complexes formed by PF4 and vaccine constituents and are accompanied by an inflammation-induced “danger signal.” Both events occur early following vaccination (days 1-2). Late reactions: In some vaccine recipients, PF4/vaccine-induced activation of B cells produces high-titer anti-PF4 autoantibodies that bind to platelets and induce platelet activation. Anti-PF4 antibodies together with platelets activate granulocytes (neutrophils) to release procoagulant NETs (NETosis), culminating in VITT-associated thrombosis.
Scheme of proposed procoagulant mechanisms in VITT. Early reactions: Initial VITT reactions are triggered by complexes formed by PF4 and vaccine constituents and are accompanied by an inflammation-induced “danger signal.” Both events occur early following vaccination (days 1-2). Late reactions: In some vaccine recipients, PF4/vaccine-induced activation of B cells produces high-titer anti-PF4 autoantibodies that bind to platelets and induce platelet activation. Anti-PF4 antibodies together with platelets activate granulocytes (neutrophils) to release procoagulant NETs (NETosis), culminating in VITT-associated thrombosis.
ChAdOx1 nCoV-19 vaccine-derived AV aggregates bind to platelet surfaces and are transported via the bloodstream to the spleen, where they are phagocytosed by macrophages, ultimately inducing a pronounced B-cell activation in the marginal zone and follicles in mice.29 In line with the animal model findings, we visualized an interaction of AV-derived hexon proteins, PF4, and VITT patient-derived anti-PF4 antibodies on platelet surfaces by 3 independent techniques (Figure 1). Our observations suggest that AV binding to PF4 likely induces conformational changes in PF4 and creates potential neoantigen(s). Consistent with the hypothesis that VITT antibodies target neoantigen(s) in PF4, VITT patient anti-PF4 antibodies bind to PF4 following immobilization on plastic surfaces. PF4 binding to surfaces is known to induce conformational changes.37 Additionally, mutagenesis studies have shown that VITT anti-PF4 antibodies, similarly to polyanions (Figure 4A), induce PF4 clustering leading to platelet activation.38 Anti-PF4 antibody–stimulated platelet aggregation is not a new concept, as high-affinity anti-PF4 antibodies also cluster PF4, even in the absence of polyanions, and induce platelet activation in atypical HIT.13 In addition to AV proteins, we found substantial amounts (∼43% to 60% by content) of nonviral proteins originating from the T-REx HEK293 (RRID:CVCL_D585) human embryonic kidney-derived production cell line.29 HEK293 cells lack tissue-specific gene expression signature and express an array of markers of renal progenitor, neuronal, and adrenal gland cells with undefined immunogenicity.39 Potential other immunologic consequences triggered by intramuscular injection of ∼15-24 µg T-REx HEK293 proteins per vaccination dose remain to be established. In addition, vaccine protein contaminants have the potential to induce an acute inflammatory cosignal that enhances B-cell responses (immunologic “danger signal”).15,40,41 Synergistically, disseminated viral proteins potently activate innate immune reactions supporting early inflammatory reactions following vaccination.15,40,41 The inflammatory response provides a costimulus for anti-PF4 antibody production by preformed B cells in HIT.8,14 Consistently, western blotting revealed that vaccination increased titers of preexisting antibodies that bind to an array of vaccine components separated by SDS-PAGE. Additionally, the ChAdOx1 nCoV-19 vaccine contains EDTA, with the capacity for increasing capillary leakage at the inoculation site by a VE-cadherin-dependent pathway.27 Disruption of the endothelial barrier facilitates dissemination of vaccine constituents into the circulation (Figure 3). Alternatively, accidental intravenous injection may contribute to vaccine dissemination.39 Furthermore, viruses can reach tissues distant from their infection site by invading endothelial cells and gaining access into the extravascular space. Viruses can also be transported across the vascular wall or be cotransported in infected immune cells such as neutrophils, which have the capacity to transmigrate through the endothelial barrier.42 Systemic dissemination of vaccine components is not unique to ChAdOx1 nCoV-19; consistent with our murine model data (Figure 3B), another ChAdOx1 vector variant (with a hepatitis B vector insert) was detectable in multiple organs, including liver, heart, and lymph nodes, at days 2 and 29 after intramuscular injection in mice.43
Similar to HIT pathology,18 marginal zone B cells stimulated by PF4/vaccine neoantigens in the presence of inflammatory costimuli produce the pathogenic anti-PF4 antibodies underlying VITT. The detailed role of marginal zone B cells in anti-PF4 VITT antibody production and the mode of exposure of PF4/vaccine neoantigens (Figure 1A) remains to be established in patients39; however, in murine models an intravenous vaccine injection induces a pronounced B-cell immune response.29 At day 5 to 20 postvaccination, pathologic anti-PF4 antibodies reach high titers and are capable of clustering PF4 on platelet surfaces and activating platelets by binding FcγRIIA (Figure 4B). Anti-PF4 mediated platelet activation likely involves extracellular polyanions such as DNA and polyphosphate (Figures 1C and 4A). Clustering of PF4 by pathologic anti-PF4 autoantibodies is also a central mechanism for platelet activation in autoimmune HIT.13 Cross-talk of PF4, activated platelets, and VITT anti-PF4 antibodies activates neutrophils leading to NETs formation in VITT patient serum (Figure 5). NETs are degraded by DNases,44 and extracellular circulating DNA was increased in VITT patients (Figure 6A), which amplifies platelet activation in VITT (Figure 4). Furthermore, DNA within NETs binds PF4.45 The resulting PF4/DNA complexes create an additional target for anti-PF4 antibodies and increase the resistance of NETs to DNase-mediated degradation, further amplifying their procoagulant activity.46 This sequence of events31,46 culminates in massive Fcγ receptor-dependent activation of neutrophils, platelets, and, by analogy with autoimmune HIT, likely also monocytes and endothelial cells.47 Consistent with our data in VITT patients, activated neutrophils and NETs contribute to venous thrombosis in HIT mouse models, and HIT antibodies selectively bind PF4-NET complexes.46 The bimodal distribution of the neutrophil activation marker serum MPO observed in healthy controls (Figure 6C) may reflect the impact of smoking and hormonal contraceptive use, both of which increase MPO enzyme levels.48 Broadened reactivity of antibodies in a boosted immune response is the hallmark of other immune disorders besides VITT. Patients with severe COVID-19 often have anti-NET autoantibodies, which likely impair NET clearance and may potentiate SARS-CoV-2–mediated thromboinflammation.49,50 Autoimmune HIT often features initial heparin-dependent reactivity that extends to heparin-independent hyperreactivity.9 Similarly, posttransfusion purpura reflects a strong alloimmune response that progresses to include platelet-autoreactive properties.51 In this regard, the numerous cell culture-derived human proteins in ChAdOx1 nCoV-19 vaccine (Figure 2) we have identified raise concern. If such proteins express immunogenic structures (eg, a genetic polymorphism absent in the corresponding endogenous protein of the vaccinated individual), an alloimmune response with potential for autoreactivity in a susceptible vaccine recipient requires attention. Protein impurities and adverse immune reactions are not unique to ChAdOx1 nCoV-19. Xenogeneic antigens derived from the growth matrix used in the virus manufacturing process have been identified by mass spectrometry in swine influenza vaccines.52,53 Bioprocess impurities, originating from the production cell line of a bovine-virus diarrhea vaccine-induced alloreactive antibodies causing feto-maternal incompatibility with severe thrombocytopenia in calves.54 In humans, narcolepsy has been associated with Pandemrix influenza vaccine; however, the underlying immune mechanisms remain to be completely understood.55,56
Our study has limitations; detailed specifications of the ChAdOx1 nCoV-19 vaccine are not publicly available, and we focused on identification of protein and some small molecule content without claiming completeness. Additionally, we have not investigated the specific roles of B cells or T cells in the VITT immune response, nor a potential contribution of the complement system that is known to contribute to immunogenicity and downstream thrombosis in HIT.57-59 VITT is a rare adverse event with thrombosis occurring in 1 out of 30 000 to 50 000 vaccinated people. We currently can only speculate on the low incidence of VITT by drawing parallels to HIT, an adverse reaction that occurs only in a small subset of heparin-exposed individuals (∼1 to 100 to 1 in 1000 patients receiving UFH). VITT appears to share similarities with atypical HIT, as the latter is even more rare (<1 in 100 HIT cases), with similar approximate incidence (in the range of 1:100 000 cases) as VITT occurrence. Similar to current knowledge of HIT pathogenesis, definitive identification of risk factors for developing VITT remains challenging. Also in other adverse immune reactions, (eg, severe drug-dependent immune thrombocytopenia following treatment with antibiotics such as vancomycin), only a small fraction of treated individuals develop adverse effects, and underlying mechanisms have remained enigmatic. Furthermore, predominant localization of VITT-associated thrombi in cerebral sinus veins has remained puzzling. Preliminary findings based on systems biology and transcriptomics60 indicate low DNASE1 expression in central nervous system endothelial cells that can potentially lead to increased half-life and persistent procoagulant activity of NETs. However, the hypothesis requires additional human subjects and experimental confirmation.
Together, our data provide insight into the mechanisms by which the SARS-CoV-2 vaccine ChAdOx1 nCoV-19 initiates immune responses, leading to pathogenic anti-PF4 antibodies that trigger downstream prothrombotic reactions. Our findings have implications for the development of AV vector vaccines with improved patient safety.
Acknowledgments
The authors acknowledge all colleagues for helpful discussions, especially Sabine Eichinger-Hasenauer, Brigitte Keller-Stanislawski, Dirk Mentzer, Klaus Cichutek, and Axel Karger. They thank Elke Hammer, Manuela Gesell-Salazar, Sascha Blankenburg, and Katrin Schoknecht for supporting proteomics and immunoproteomics analyses, Martina Wurster and Simone Seefeldt for help with metabolomics analyses, Konrad for provided VITT patient material, and Frédéric Ebstein for the 25-mer DNA. They are grateful to Ulrike Strobel, Carmen Freyer, Katrin Stein, Ines Warnig, Ricarda Raschke, Julia Klauke, Jessica Fuhrmann, Petra Meyer, Mandy Jörn, and Anita Badbaran for excellent technical support.
The study was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) grants: 374031971; TRR240, 398967434; SFB/TR261, 25440785; SFB877, P6; KFO306, 80750187 - SFB841, DFG-FR4239/1-1, and INST 2026/13-1 FUGG, European Union (Marie Sklodowska-Curie grant agreement 840189), the German Heart Foundation (F/34/18), the Ministerium für Wirtschaft, Arbeit und Gesundheit Mecklenburg Vorpommern (project COVIDPROTECT), Leibniz WissenschaftsCampus; ComBioCat W10/2018, the Südmeyer fund for kidney and vascular research (“Südmeyer-Stiftung für Nieren- und Gefäßforschung”), and the Gerhard Büchtemann fund.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH, the US Department of Veterans Affairs, or the United States Government.
Authorship
Contribution: A.G. developed the concept, designed experiments, and wrote the first draft of the manuscript; J.W., S.H., R.P., K.A., and M.W. performed the platelet, granulocyte, DLS, ELISA, and some confocal microscopy experiments; M.L. and K.M. performed the NMR studies; U.V., C.H., S.M., L.S., and A.R. performed the proteome studies; L.S., K.S., and T.T. characterized patients, contributed to the concept, and helped write the manuscript; K.F. performed the electron microscopy studies; M.B. analyzed data, discussed the concept, and helped write the manuscript; S.K. and L.K. analyzed the vaccine and contributed to the study concept; F.S. and N.E. performed the 3D super-resolution microscopy; T.E.W. contributed to the concept, discussed results, and helped write the manuscript; R.K.M., C.R., and T.R. performed the DNase experiments, NETs analyses, mouse studies, discussed data, and wrote the revised manuscript; B.F. designed the digital PCR; M.F., H.E., and E.X.S. performed and analyzed the immunohistochemistry analyses; A.B. provided autopsy material; and all authors have critically revised and approved the final version of the manuscript. A.G., K.S., T.E.W., and T.R have accessed and verified the underlying data.
Conflict-of-interest disclosure: A.G. reports grants and nonfinancial support from Aspen, Boehringer Ingelheim, MSD, Bristol Myers Squibb, Paringenix, Bayer Healthcare, Gore Inc., Rovi, Sagent, and Biomarin/Prosensa; personal fees from Aspen, Boehringer Ingelheim, MSD, Macopharma, Bristol Myers Squibb, Chromatec, and Instrumentation Laboratory; and nonfinancial support from Boehringer Ingelheim, Portola, Ergomed, and GTH e.V. outside the submitted work. T.T. reports personal fees and other from Bristol Myers Squibb, personal fees and other from Pfizer, personal fees from Bayer, personal fees and other from Chugai Pharma, other from Novo Nordisk, personal fees from Novartis, and other from Daichii Sankyo outside the submitted work. M.B. reports grants from CureVac Company, Tübingen, Germany. The remaining authors declare no completing financial interests.
Correspondence: Andreas Greinacher, Institut für Immunologie und Transfusionsmedizin, Abteilung Transfusionsmedizin, Sauerbruchstr., D-17487 Greifswald, Germany; e-mail: andreas.greinacher@med.uni-greifswald.de; and Thomas Renné, Institute for Clinical Chemistry and Laboratory Medicine, University Medical Center Hamburg-Eppendorf (UKE), Martinistrasse 52, D-20246 Hamburg, Germany; e-mail: thomas@renne.net
For original data, please contact andreas.greinacher@med.uni-greifswald.de or thomas@renne.net.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![VITT patient anti-PF4 antibodies trigger NETosis. (A-B) Confocal laser scanning microscopy images of in vitro NETs generated by human neutrophils and platelets following incubation with VITT patient serum alone (A) or VITT patient serum in combination with PF4 (B). (C-D) NET formation induced in healthy control serum alone (C) or healthy serum in the presence of PF4 (D). (E-H) NET formation induced by affinity-purified (Affi.pur.) anti-PF4 IgG from VITT patients (E); combination of anti-PF4 IgG from VITT patients and PF4 (F); buffer; (G) or PF4 only (H). (I-J) Neutrophils (without [w/o] platelets) were incubated with VITT patient serum (I) or VITT patient serum and PF4 (J), and NET formation was measured. (K) Quantification of NETs (NETosis [%]) from confocal laser scanning microscopy images obtained from nuclear and extracellular DNA fluorescent channels was performed using at least 12 individual images for each condition, n = 3 experimental replicates. Statistical analysis was performed using the Welch and Brown-Forsythe ANOVA multiple comparisons test with post hoc 2-stage step-up method per Benjamini, Krieger, and Yekutieli, q = 0.05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/138/22/10.1182_blood.2021013231/5/m_bloodbld2021013231f5.png?Expires=1769083762&Signature=jwvfBZca~AkNVHF9TWRqUPx2YCo-GE7KYgAWKzGlVBnEzrfQiyXoqLOVRAhmzGs8kTuoSGw4GM2LEFnTd5M0InxeNWTeeaK-fUZip080KtdVVA1b2ybrTo8kBLE9XCunm80ypOMo68h2mhL3cn2a~ieEEUYzlpWbffF1DmKJzE-~IWfCIGkwJPQ98Yk750h1CPo0FOvxy71Cx1k9mXukEARez6j52JJppb2c8wzrU1u3lgCEE6sE~OH0AeEQM4BAqNDHUZk1wTOJ0v-0qAO9GXrwdKmbqdammDZP8TC7CTVXdPU83a6I2UPLEA-slDaij35Yp4Yz4ev9~1IqvaeFBw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal