Key Points
Lyl1 compensates for loss of Scl in megakaryopoiesis.
Scl and Lyl1 share functional roles in platelet production by regulating expression of partner proteins including Gata1 and Fli1.
Abstract
The stem cell leukemia (Scl or Tal1) protein forms part of a multimeric transcription factor complex required for normal megakaryopoiesis. However, unlike other members of this complex such as Gata1, Fli1, and Runx1, mutations of Scl have not been observed as a cause of inherited thrombocytopenia. We postulated that functional redundancy with its closely related family member, lymphoblastic leukemia 1 (Lyl1) might explain this observation. To determine whether Lyl1 can substitute for Scl in megakaryopoiesis, we examined the platelet phenotype of mice lacking 1 or both factors in megakaryocytes. Conditional Scl knockout (KO) mice crossed with transgenic mice expressing Cre recombinase under the control of the mouse platelet factor 4 (Pf4) promoter generated megakaryocytes with markedly reduced but not absent Scl. These Pf4Sclc-KO mice had mild thrombocytopenia and subtle defects in platelet aggregation. However, Pf4Sclc-KO mice generated on an Lyl1-null background (double knockout [DKO] mice) had severe macrothrombocytopenia, abnormal megakaryocyte morphology, defective pro-platelet formation, and markedly impaired platelet aggregation. DKO megakaryocytes, but not single-knockout megakaryocytes, had reduced expression of Gata1, Fli1, Nfe2, and many other genes that cause inherited thrombocytopenia. These gene expression changes were significantly associated with shared Scl and Lyl1 E-box binding sites that were also enriched for Gata1, Ets, and Runx1 motifs. Thus, Scl and Lyl1 share functional roles in platelet production by regulating expression of partner proteins including Gata1. We propose that this functional redundancy provides one explanation for the absence of Scl and Lyl1 mutations in inherited thrombocytopenia.
Introduction
Megakaryopoiesis, a complex process involving lineage commitment, megakaryocyte maturation and platelet release, is regulated by a network of transcription factors including SCL (also known as TAL1), GATA1, FLI1, RUNX1, NFE2, and GFI1B.1 These factors work cooperatively to bind and activate platelet-specific genes throughout the genome while repressing genes of other lineages.2,3 Traditionally, our understanding of these transcription factors relied primarily on the use of experimental cell lines or small animal models. With the availability of next-generation sequencing, however, mutations in these factors have been increasingly recognized as a cause of hereditary platelet disorders, and they provide a natural experimental environment for better understanding their functions.4 Heterozygous mutations in GATA1, FLI1, RUNX1, and GFI1B have been reported to cause hereditary platelet disorders characterized by thrombocytopenia as well as platelet dysfunction.5 This is consistent with previous animal models showing critical roles in platelet production.6-9 The exception is SCL, which has never been reported to cause platelet disorders despite experimental evidence of its importance in platelet production.10,11
SCL is a basic-helix-loop helix (bHLH) transcription factor essential for hematopoietic development; constitutive deletion of murine Scl leads to in utero death at embryonic day 8.5 (E8.5) because of the complete absence of hematopoiesis and vascular defects.12,13 Once the hematopoietic system is established, however, Scl is no longer essential for maintaining normal hematopoietic stem cells.14,15 Specific roles for Scl in megakaryopoiesis were examined by Cre-mediated deletion of conditional Scl alleles.10,11 Overall, the effects were modest, with mild thrombocytopenia, defects in megakaryocyte growth, and a blunted response to thrombopoietin (TPO), the major regulatory cytokine for platelet production. Dysregulation of Cdkn1a and Nfe2 were proposed downstream mediators of these defects. Of note, heterozygous loss of Scl had no platelet phenotype.
One possible explanation for the relatively mild phenotype of Scl deletion in megakaryopoiesis and the absence of SCL mutations as a cause of hereditary thrombocytopenia is compensation by the highly related bHLH factor Lyl1. We have previously shown that Lyl1 can compensate for the loss of Scl in adult hematopoietic stem cells (HSCs) and primitive erythropoiesis.16,17 This functional redundancy is supported by genome-wide chromatin immunoprecipitation (ChIP) analyses of mouse and human stem and progenitor cells showing combinatorial binding of Scl and Lyl1 with other transcription factors, including those important for megakaryopoiesis.18,19 Whether this compensation between Scl and Lyl1 also occurs in megakaryocytes has not been examined because Scl-Lyl1 double knockout (DKO) adult HSCs die rapidly,16 which prevents the generation of DKO megakaryopoiesis. To address this question, we have used the platelet-specific Cre recombinase Pf4Cre to delete Scl on an Lyl1-null background to examine megakaryopoiesis in the absence of both Scl and Lyl1.
Materials and methods
Mice
All experiments were approved by the Alfred Medical Research & Education Precinct Animal Ethics Committee and performed in accordance with the institutional animal care guidelines. Mice with a loxP-targeted Scl allele (Sclf),20 a Lyl1 knockout (KO) allele (Lyl1LacZ),21 enhanced yellow fluorescent protein (EYFP) expressed in a Cre-dependent manner (Rosa26-EYFP),22 and transgenic mice expressing Cre recombinase exclusively in the megakaryocytic lineage (Pf4-CreT)23 have been described previously. Sclf/f mice were crossed with Pf4-CreT/+SclΔ/Δ mice to generate wild-type (WT) (Sclf/f) and megakaryocyte-specific deletion of Scl (Pf4-CreT/+SclΔ/Δ) mice. Sclf/fLyl1LacZ/LacZ mice were crossed with Pf4-CreT/+SclΔ/ΔLyl1LacZ/LacZ mice to generate Lyl1 KO (Sclf/fLyl1LacZ/LacZ) and DKO (Pf4-CreT/+SclΔ/ΔLyl1LacZ/LacZ) mice.
Stress tests for in vivo platelet production
Mice were bled for baseline platelet count, and then stress tests were performed by injecting mice with antiplatelet serum (Cedarlane) 2 μg intravenously (1 dose), 5-fluorouricil (5-FU) 150 mg/kg intraperitoneally (1 dose), or recombinant human TPO 2 μg intraperitoneally once per day for 5 days.
Platelet function analysis
Freshly isolated mouse platelets (1 × 106) were incubated with fluorescently conjugated antibodies against P-selectin (BD, 1 in 100 dilution) or he α-2b component of integrin αIIbβ3 (CD41) (Emfret, 1 in 6 dilution) for 15 minutes. Platelets were then treated with agonists (thrombin, collagen-reactive peptide, or calcium ionophore A23187) for 15 minutes before the addition of an equal volume of 4% paraformaldehyde and assessed by flow cytometry (FACSCalibur, BD).
Transmission electron microscopy of megakaryocytes and platelets
In situ bone marrow megakaryocytes were prepared by perfusion-fixation of anesthetized mice with 2% glutaraldehyde, 2.5% paraformaldehyde via the ascending aorta at a rate of 20.4 mL/h (Perfusor Compact S, Braun). The femurs were excised and immersion fixed for an additional 4 hours, rinsed in phosphate-buffered saline, and decalcified in 10% EDTA for 7 days at 4°C after fixation, dehydration, and embedding. Ultrathin sections of all samples were cut with a diamond knife (Diatome) on an ultramicrotome (Leica) and stained with methanolic uranyl acetate and lead citrate before examination with a transmission electron microscope (JEOL, Tokyo, Japan).
ChIP
ChIP was performed as previously described by using the human megakaryocytic cell line MEG-01 with 20 × 106 cells per condition and with antibodies that recognize human SCL (sc12984x, lots #A1613 and #B2312, Santa Cruz Biotechnologies) and human LYL1 (sc374164, lot #D2616, Santa Cruz Biotechnologies).19,21 Libraries were prepared and sequenced using the Illumina HiSeq2000 analyzer (BGI, Hong Kong).
ChIP-seq analysis
The raw ChIP sequencing (ChIP-seq) reads were filtered for adapter contamination and low quality scores, and we also excluded reads in which more than 10% of bases were unknown. The primary mouse ChIP-seq data set for Scl binding was publicly available from the NCBI Gene Expression Omnibus database through the GEO accession number GSE51337. The HOMER and MACS2 algorithms were used to call peaks with a stringent setting and statistical threshold of P < .01 compared with immunoglobulin G (IgG) control data. Peaks called by both algorithms were identified as high confidence binding sites for downstream analysis. Associated gene lists were generated using the Genomic Region Enrichment Annotations Tool (GREAT) to identify genes within 50 kb of the peak of interest. Cobinding of SCL and LYL was assessed by using a bootstrapping approach implemented in Matlab (v9.5.0.94).
Results
Loss of both Scl and Lyl1 in megakaryocytes leads to macrothrombocytopenia
The Pf4-Cre mouse strain used for our studies was recently shown to have megakaryocyte-restricted expression of Cre recombinase.24 To confirm these results, we crossed Pf4-CreT/+ mice with Rosa26-EYFPT/+ mice and harvested bone marrow for analysis of EYFP expression. Fluorescence-activated cell sorting analysis demonstrated EYFP expression in 15% of megakaryocyte progenitors (supplemental Figure 1A, available on the Blood Web site). Lineage-restricted erythroid progenitors were EYFP negative (supplemental Figure 1B), which together with previous analyses,24 suggested that Cre activity was limited to the megakaryocyte lineage. Expression of Pf4-Cre alone had no effect on platelet number or size (supplemental Figure 1C).
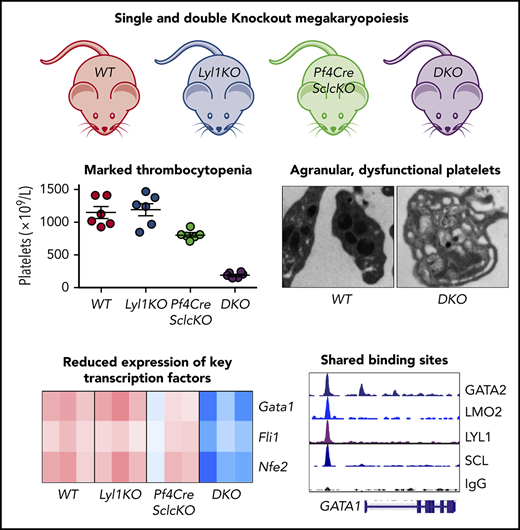
WT, Scl-deleted (Pf4Sclc-KO), Lyl1-KO, and DKO mice were generated at the expected Mendelian frequency (see “Materials and methods” for specific genotypes). To determine the efficiency of Scl gene deletion, we used RNA sequencing (RNA-seq) analyses, which are more quantitative than genomic polymerase chain reaction and can detect alternate transcripts (Figure 1A). Expression of Scl in cultured megakaryocytes from Pf4Sclc-KO demonstrated 95% loss of messenger RNA (mRNA) transcribed from the loxP-flanked exon 5 of Scl (Figure 1B). Importantly, DKO mice had similar levels of residual Scl expression. Expression of Lyl1 exon 4 was absent in Lyl1-KO mice but there was normal expression of the N-terminal exons as previously reported.25 Finally, in the single knockout (KO) mice, there was no compensatory increase in mRNA expression of the other transcription factor (Figure 1B). Overall, Pf4Cre-mediated excision led to low although not absent expression of Scl in both Pf4Sclc-KO and DKO megakaryocytes.
Scl and Lyl expression in cultured megakaryocytes. (A) Integrative genomics viewer visualization of Scl and Lyl1 messenger RNA (mRNA) in WT, Pf4Sclc-KO, Lyl1-KO, and DKO megakaryocytes. (B) Mean ± standard error of the mean (SEM). Counts per million (cpm) reads for the targeted exons of Scl and Lyl1 (n = 3 per genotype).
Scl and Lyl expression in cultured megakaryocytes. (A) Integrative genomics viewer visualization of Scl and Lyl1 messenger RNA (mRNA) in WT, Pf4Sclc-KO, Lyl1-KO, and DKO megakaryocytes. (B) Mean ± standard error of the mean (SEM). Counts per million (cpm) reads for the targeted exons of Scl and Lyl1 (n = 3 per genotype).
Full blood examination of adult Pf4Sclc-KO mice revealed mild thrombocytopenia (70% of WT levels) with large platelets (Figure 2A-B; Table 1). In contrast, Lyl1-KO mice displayed normal platelet numbers and size, as previously reported.21 Most striking was a severe thrombocytopenia (20% of WT levels) in DKO mice. Blood film examination revealed giant platelets resembling megakaryocyte fragments (Figure 2B). Hemoglobin and white cell counts did not differ among the 4 groups (Table 1).
Phenotype of single-KO and DKO mice. (A) Platelet count and volume for mice of each genotype with mean ± SEM shown. Results analyzed using two-way ANOVA with Tukey’s correction. *P < .01 and **P < .001 compared with WT mice. (B) Representative peripheral blood smear for each genotype showing enlarged platelets (arrowheads) in DKO mice. May-Grünwald-Giemsa stain. (C) Mean ± SEM number of bone marrow megakaryocytes (n = 3 per genotype). Results analyzed using two-way ANOVA with Tukey’s correction. ***P < .0001 compared with WT mice. (D) Representative bone marrow sections with abnormal megakaryocytes (arrowheads arrows) in DKO mice. Hematoxylin-eosin stain. (E) Mean ± SEM for ploidy of freshly isolated megakaryocytes (n = 3 per genotype). fl, femtoliter; hpf, high-power field; MPV, mean platelet volume.
Phenotype of single-KO and DKO mice. (A) Platelet count and volume for mice of each genotype with mean ± SEM shown. Results analyzed using two-way ANOVA with Tukey’s correction. *P < .01 and **P < .001 compared with WT mice. (B) Representative peripheral blood smear for each genotype showing enlarged platelets (arrowheads) in DKO mice. May-Grünwald-Giemsa stain. (C) Mean ± SEM number of bone marrow megakaryocytes (n = 3 per genotype). Results analyzed using two-way ANOVA with Tukey’s correction. ***P < .0001 compared with WT mice. (D) Representative bone marrow sections with abnormal megakaryocytes (arrowheads arrows) in DKO mice. Hematoxylin-eosin stain. (E) Mean ± SEM for ploidy of freshly isolated megakaryocytes (n = 3 per genotype). fl, femtoliter; hpf, high-power field; MPV, mean platelet volume.
Blood cell counts of mice lacking Scl, Lyl1, or both factors in megakaryocytes
| . | WT (n = 6) . | Pf4Sclc-KO (n = 6) . | Lyl1-KO (n = 6) . | DKO (n = 6) . |
|---|---|---|---|---|
| Hemoglobin, g/dL | 14.57 ± 0.37 | 15.47 ± 0.37 | 15.33 ± 0.79 | 14.42 ± 0.30 |
| White cells ×109/L | 9.91 ± 1.15 | 11.91 ± 0.95 | 12.72 ± 0.84 | 9.99 ± 0.75 |
| Platelets ×109/L | 1147 ± 88 | 807 ± 31* | 1194 ± 97 | 200 ± 15† |
| MPV, fL | 5.75 ± 0.18 | 6.58 ± 0.13† | 5.5 ± 0.13 | 6.83 ± 0.16† |
| . | WT (n = 6) . | Pf4Sclc-KO (n = 6) . | Lyl1-KO (n = 6) . | DKO (n = 6) . |
|---|---|---|---|---|
| Hemoglobin, g/dL | 14.57 ± 0.37 | 15.47 ± 0.37 | 15.33 ± 0.79 | 14.42 ± 0.30 |
| White cells ×109/L | 9.91 ± 1.15 | 11.91 ± 0.95 | 12.72 ± 0.84 | 9.99 ± 0.75 |
| Platelets ×109/L | 1147 ± 88 | 807 ± 31* | 1194 ± 97 | 200 ± 15† |
| MPV, fL | 5.75 ± 0.18 | 6.58 ± 0.13† | 5.5 ± 0.13 | 6.83 ± 0.16† |
Blood parameters obtained from 6- to 8-week-old mice. Values shown are the mean ± standard error of the mean. Analyzed by using ordinary one-way ANOVA with Tukey’s multiple comparison test.
fL, femtoliter; MPV, mean platelet volume.
P = .01 compared with WT mice.
P < .001 compared with WT mice.
Despite the marked thrombocytopenia, there was a twofold increase in the number of bone marrow megakaryocytes in DKO mice (Figure 2C). In addition to increased number, DKO megakaryocytes had abnormal morphology with an increased nuclear:cytoplasmic ratio resulting from loss of cytoplasm (Figure 2D; supplemental Figure 2). In contrast, loss of either Scl or Lyl1 had no effect on megakaryocyte number, morphology, or nuclear:cytoplasmic ratio (Figure 2C-D; supplemental Figure 2). Nuclear ploidy was not affected by loss of 1 or both genes (Figure 2E). To determine whether increased platelet clearance was a contributing factor to the thrombocytopenia in DKO mice, we performed platelet half-life studies using in vivo labeling with biotin. These studies demonstrated a shortened half-life of DKO platelets from 4.25 to 3.25 days (supplemental Figure 3), but this reduced half-life was unlikely to explain the severe thrombocytopenia in DKO mice.
Scl or Lyl1 are required for platelet production
Given the twofold increase in megakaryocyte numbers and mildly reduced platelet half-life, the marked thrombocytopenia in DKO mice was most likely explained by defects in platelet formation. In vivo platelet production was assessed by using 3 different strategies: chemotherapy (5-FU), anti-platelet serum, and recombinant TPO (Figure 3A). After 5-FU administration, platelet count in DKO mice fell to less than 10 × 109/L by 5 days, leading to catastrophic hemorrhage and death of animals. In contrast, single-KO mice had normal recovery with rebound thrombocytosis. After administration of anti-platelet serum, platelet counts in WT and single-KO mice dropped ∼70% within 24 hours before returning to normal by 10 days. Surprisingly, anti-platelet serum had no effect on platelet count in DKO mice. Finally, DKO mice were unable to mount any response to daily administration of TPO, whereas single-KO mice had normal response. This lack of platelet response in DKO mice may be explained in part by the absence of megakaryocyte expansion in response to TPO (Figure 3B). In contrast, reduction in nuclear size in response to TPO and ploidy were normal in DKO mice (supplemental Figure 4). To directly examine proplatelet formation, megakaryocytes were isolated from TPO-treated mice to visualize proplatelet formation. Using this assay, proplatelet formation was significantly reduced in Pf4Sclc-KO and DKO megakaryocytes isolated from TPO-treated mice (Figure 3C). Given that proplatelet formation is not reliant on TPO,24 this may be a consequence of an Scl-Lyl1-dependent factor that is also regulated by TPO. Taken together, these results indicate a major shared role for Scl and Lyl1 in platelet production.
Platelet production in single-KO and DKO mice. (A) Platelet count in response to 5-FU, anti-platelet serum (APS), or TPO (n = 3 per genotype). Baseline platelet count (day 0) was performed just before drug administration. (B) Mean ± SEM number of bone marrow megakaryocytes in mice treated with or without TPO for 5 days (n = 3 per genotype). Student t test compared with untreated mice of the same genotype. (C) Proplatelet production from in vitro cultured megakaryocytes obtained from untreated (left panel) and TPO-treated mice (right panel). Results analyzed using two-way ANOVA with Tukey’s correction. Megs, megakaryocyte; NS. not significant. **P < .001; ***P < .0001 compared with WT mice.
Platelet production in single-KO and DKO mice. (A) Platelet count in response to 5-FU, anti-platelet serum (APS), or TPO (n = 3 per genotype). Baseline platelet count (day 0) was performed just before drug administration. (B) Mean ± SEM number of bone marrow megakaryocytes in mice treated with or without TPO for 5 days (n = 3 per genotype). Student t test compared with untreated mice of the same genotype. (C) Proplatelet production from in vitro cultured megakaryocytes obtained from untreated (left panel) and TPO-treated mice (right panel). Results analyzed using two-way ANOVA with Tukey’s correction. Megs, megakaryocyte; NS. not significant. **P < .001; ***P < .0001 compared with WT mice.
Transmission electron microscopy was performed to further examine the morphologic abnormalities of DKO megakaryocytes and platelets. Single-KO megakaryocytes had subtle abnormalities: Pf4Sclc-KO megakaryocytes had increased emperipolesis whereas Lyl1-KO megakaryocytes had mild dilatation of the demarcated membrane system and small megakaryocyte cytoplasmic fragments (Figure 4A). In contrast, DKO megakaryocytes were markedly abnormal with a vesicular demarcated membrane system instead of the usual tubular phenotype, absent proplatelet regions, reduced granules, and prominent mitochondria (Figure 4A; supplemental Figure 5A). The lack of well-formed demarcation membrane system and proplatelet regions in mature DKO megakaryocytes was associated with the presence of giant platelets in the sinusoids (supplemental Figure 5B). The presence of platelets within sinusoids may be related to reduced expression of Vegfc, which has effects on blood vessel permeability (supplemental Figure 5C). Emperipolesis was pronounced in DKO megakaryocytes (supplemental Figure 5D). The giant platelets contained many small rounded vesicles and, in some cases, also contained parts of the demarcation membrane system (Figure 4B). DKO platelets also had significantly reduced α-granule content but normal numbers of dense granules (supplemental Figure 5E). Overall, these results show that Scl and Lyl1 play a common role in cytoplasmic maturation of megakaryocytes and platelets.
Transmission electron microscopy. (A) Representative images of megakaryocytes for each genotype. Arrows indicate emperipolesis (Pf4Sclc-KO) and prominent mitochondria (DKO). Upper panels (original magnification ×1000). Lower panels (original magnification ×8000). (B) Representative images of platelets for each genotype.
Transmission electron microscopy. (A) Representative images of megakaryocytes for each genotype. Arrows indicate emperipolesis (Pf4Sclc-KO) and prominent mitochondria (DKO). Upper panels (original magnification ×1000). Lower panels (original magnification ×8000). (B) Representative images of platelets for each genotype.
Platelets lacking both Scl and Lyl1 have broad-spectrum functional defects
We performed in vivo and in vitro assays to define the shared roles of Scl and Lyl1 in platelet function. Hemostatic function of platelets assessed by tail bleeding time demonstrated a significantly prolonged bleeding time in DKO mice compared with single-KO mice (Figure 5A). Bleeding times are difficult to interpret in the setting of marked thrombocytopenia, so we assessed platelet function by using flow cytometry assays of washed platelets in which the numbers of platelets could be equalized between all 4 genotypes. Platelet surface P-selectin expression was assessed because it normally resides within α granules but is released onto the surface upon platelet activation. Importantly, expression of P-selectin mRNA was not reduced in DKO megakaryocytes (data not shown). After exposure to thrombin, which activates platelets via the proteinase-activated receptors, P-selectin expression was markedly reduced in the DKO platelets despite maximal thrombin stimulation (Figure 5B). Pf4Sclc-KO platelets also had reduced expression of P-selectin in response to low doses of thrombin. Release of P-selectin by DKO platelets was also impaired in response to collagen-reactive peptide and the calcium ionophore (A23187), an artificial agonist that activates platelets by promoting influx of calcium from the extracellular compartment independent of platelet receptors (Figure 5C). Thus, these defects in platelet activation could not be solely attributed to the decreased expression of Gp1b and Gp2b (Figure 5D), receptors critical for platelet activation via von Willebrand factor and fibrinogen, respectively. In aggregate, these defects in DKO platelets show that Scl and Lyl1 share an important role for platelet function.
Platelet function tests of single-KO and DKO mice. (A) Tail bleeding times of mice. Mean ± SEM shown for 5 mice per genotype. Analyzed using two-way ANOVA with Tukey’s correction. (B) Surface platelet expression of P-selectin after stimulation with varying concentrations of thrombin. (C) Surface platelet expression of P-selectin after stimulation with thrombin, calcium ionophore (A23187), and collagen-reactive peptide (CRP). (D) Surface expression of glycoprotein 1b-α (Gp1ba) and glycoprotein 2b (Gp2b) on resting platelets. Mean ± SEM for 3 mice per genotype. **P < .001 compared with WT; ***P < .0001 compared with WT mice; ###P < .0001 compared with Pf4Sclc-KO. MCF, mean cell fluorescence.
Platelet function tests of single-KO and DKO mice. (A) Tail bleeding times of mice. Mean ± SEM shown for 5 mice per genotype. Analyzed using two-way ANOVA with Tukey’s correction. (B) Surface platelet expression of P-selectin after stimulation with varying concentrations of thrombin. (C) Surface platelet expression of P-selectin after stimulation with thrombin, calcium ionophore (A23187), and collagen-reactive peptide (CRP). (D) Surface expression of glycoprotein 1b-α (Gp1ba) and glycoprotein 2b (Gp2b) on resting platelets. Mean ± SEM for 3 mice per genotype. **P < .001 compared with WT; ***P < .0001 compared with WT mice; ###P < .0001 compared with Pf4Sclc-KO. MCF, mean cell fluorescence.
Scl or Lyl1 are required for expression of many genes important for megakaryopoiesis and platelet function
To identify gene targets coregulated by Scl and Lyl1 in megakaryopoiesis, we performed RNA-seq analysis on megakaryocytes isolated from lineage-depleted bone marrow cells cultured in TPO for 5 days. Multidimensional scaling using the 500 most differentially regulated genes confirmed clustering of samples according to genotype (supplemental Figure 6). Using a false discovery rate of 0.01, there were 384 genes differentially expressed more than twofold (180 up and 204 down) in Pf4Sclc-KO megakaryocytes and 40 genes (11 up and 29 down) in Lyl1-KO megakaryocytes (Figure 6A; supplemental Data Sets 1 and 2). Comparison of gene expression in Pf4Sclc-KO megakaryocytes data with a previous study (GSE24969)11 demonstrated significant correlation (supplemental Figure 7A). Loss of repression of the cell cycle inhibitor Cdkn1a (p21) was a proposed mechanism of defects in Scl-null megakaryocytes.11 Accordingly, we observed a fourfold increase in Cdkn1a expression in megakaryocytes lacking both Scl and Lyl1 (supplemental Figure 7B).
Gene expression profiling of megakaryocytes from single-KO and DKO mice. (A) Venn diagrams showing the numbers of genes up- and downregulated more than twofold in Pf4Sclc-KO, Lyl1-KO, and DKO megakaryocytes relative to WT mice (false discovery rate [FDR] <0.01). (B) Gene set enrichment analysis (GSEA) for platelet activation and aggregation genes between DKO and single-KO megakaryocytes. (C) Heat map showing expression of structural genes known to cause inherited macrothrombocytopenia. The FDR is shown for comparison of DKO with Pf4Sclc-KO cells. (D) Heat map showing expression of transcription factors that regulate megakaryopoiesis. FDR is shown for comparison of DKO and Pf4Sclc-KO cells. NES, normalized enrichment score; NS, not significant.
Gene expression profiling of megakaryocytes from single-KO and DKO mice. (A) Venn diagrams showing the numbers of genes up- and downregulated more than twofold in Pf4Sclc-KO, Lyl1-KO, and DKO megakaryocytes relative to WT mice (false discovery rate [FDR] <0.01). (B) Gene set enrichment analysis (GSEA) for platelet activation and aggregation genes between DKO and single-KO megakaryocytes. (C) Heat map showing expression of structural genes known to cause inherited macrothrombocytopenia. The FDR is shown for comparison of DKO with Pf4Sclc-KO cells. (D) Heat map showing expression of transcription factors that regulate megakaryopoiesis. FDR is shown for comparison of DKO and Pf4Sclc-KO cells. NES, normalized enrichment score; NS, not significant.
Megakaryocytes from DKO mice had the greatest expression changes with 1963 genes differentially expressed more than twofold (782 up and 1181 down), with the majority (631 up and 983 down) unique to DKO megakaryocytes (Figure 6A; supplemental Data Sets 1 and 2). Pathway analysis of the 631 genes upregulated only in DKO megakaryocytes revealed enrichment for cytokine receptor interactions including Il1r2, Il18rap, Il4ra, Il17ra, Cx3cl1, Cxcr3, Cxcl12, Tnfrsf13b, and Tnfrsf11b (supplemental Data Set 3). Pathway analysis of the genes downregulated only in DKO megakaryocytes revealed enrichment for genes involved in platelet activation, including Gp5, Gp6, Gp9, Gp1bb, P2ry1, P2ry12, Itga2, Col1a1, Col1a2, Col5a2, Vwf, Lyn, Fyn, and Mapk1 (supplemental Data Set 3). In contrast, pathway analysis of the 204 genes with reduced expression in both Pf4Sclc-KO and DKO megakaryocytes showed enrichment only for a small number of erythroid and platelet factors (Hba-a1, Hba-a2, Gypc, Gypa, Itgal, Pecam1, Icam1, Thbs1, and Thbs4) (supplemental Data Set 3). Consistent with the marked functional defects of DKO platelets compared with single-KO platelets, gene set enrichment analysis of DKO megakaryocytes compared with single-KO megakaryocytes also demonstrated reduced expression of genes regulating platelet activation, signaling, and aggregation (Figure 6B; supplemental Figure 7C). Furthermore, 10 of 16 nontranscription factor genes linked with inherited macrothrombocytopenia4 were significantly reduced in DKO megakaryocytes compared with single-KO megakaryocytes (Figure 6C; z-score, 8.5; P = 3 × 10−17). In addition, the transcriptional regulators of megakaryopoiesis (Gata1, Fli1, and Nfe2) were all significantly reduced (Figure 6D). In contrast, Mef2c, a proposed transcriptional target of Scl in megakaryopoiesis,26 was increased in DKO megakaryocytes. Gata2 expression was also reduced in megakaryocytes lacking Scl despite reduced Gata1 (supplemental Figure 7B). This contrasts with Gata1 KO mice in which Gata2 is significantly increased.27 Thus, Scl and Lyl1 coregulate many genes that are critical for megakaryopoiesis and platelet function causing inherited macrothrombocytopenia.
Given the reduced expression of many key transcription factors in DKO megakaryocytes (Gata1, Nfe2, and Fli1), we examined the gene expression profile of Gata1-deficient megakaryocytes (GSE2527)28 to determine whether Scl and Lyl1 expression were likewise dependent on Gata1. Consistent with the known Gata1-dependent transcriptional repression of Gata2,29 Gata2 was increased in the absence of Gata1 (supplemental Figure 7D). However, loss of Gata1 had no effect on the expression of Scl or Lyl1 in megakaryocytes. Importantly, many of the structural genes downregulated in DKO megakaryocytes (Flna, Actn1, and Tubb1) were also reduced in Gata1-deficient megakaryocytes (supplemental Figure 7D).
Many genes required for platelet production and function are direct targets of Scl and Lyl1
We used ChIP to identify the common binding targets of Scl and Lyl1 in megakaryopoiesis. The commercially available anti-Lyl1 antibodies were unable to pull down mouse Lyl1; therefore, we used the human megakaryocyte cell line MEG-01 for ChIP experiments to enable direct comparison of SCL and LYL1 DNA binding sites. Overall, 7420 SCL- and 2437 LYL1-binding peaks were identified in the MEG-01 genome (Figure 7A; supplemental Data Set 4). The majority of the LYL1 binding sites (1823; 75%) were also bound by SCL. In addition to E-BOX motifs, these common SCL- and LYL1-binding sites were enriched for GATA, ETS, and RUNX motifs (Figure 7B). In contrast, SCL-only binding sites were enriched for GATA and STAT motifs, whereas LYL1-only binding sites were enriched for SP1 motifs (supplemental Figure 8A).
Integration of ChIP and expression analyses. (A) Venn diagram showing numbers of common and unique binding peaks for SCL and LYL1 in the human megakaryocyte cell line MEG-01. (B) De novo motif discovery at common SCL/LYL1 binding sites in MEG-01 cells identifies significant enrichment for GATA, E-BOX, and ETS motifs and RUNX. (C) Venn diagram showing overlap of downregulated genes for Pf4Sclc-KO, Lyl1-KO, and DKO cells with mouse homologs of the common SCL/LYL1-binding sites identified in MEG-01 cells. (D) Read peak plot for ChIP-seq reads in MEG-01 cells and primary human megakaryocytes (GSE24674) displayed in the University of California Santa Cruz genome browser shows overlapping binding of SCL and LYL1 near the GATA1 gene.
Integration of ChIP and expression analyses. (A) Venn diagram showing numbers of common and unique binding peaks for SCL and LYL1 in the human megakaryocyte cell line MEG-01. (B) De novo motif discovery at common SCL/LYL1 binding sites in MEG-01 cells identifies significant enrichment for GATA, E-BOX, and ETS motifs and RUNX. (C) Venn diagram showing overlap of downregulated genes for Pf4Sclc-KO, Lyl1-KO, and DKO cells with mouse homologs of the common SCL/LYL1-binding sites identified in MEG-01 cells. (D) Read peak plot for ChIP-seq reads in MEG-01 cells and primary human megakaryocytes (GSE24674) displayed in the University of California Santa Cruz genome browser shows overlapping binding of SCL and LYL1 near the GATA1 gene.
To identify the functionally relevant binding targets shared by Scl and Lyl1, we integrated our ChIP-seq and RNA-seq data. We mapped the 1823 shared binding sites in MEG-01 cells to 1523 mouse genes (supplemental Data Set 4). Overall, 242 (16%) of these genes were differentially expressed in either single-KO or DKO mice (Figure 7C). Of these, the majority (177 genes; 73%) were differentially expressed in DKO megakaryocytes only (Figure 7C; supplemental Data Set 4). Pathway analysis of these 177 genes demonstrated the most enrichment for genes regulating platelet activation, including Gp6, Gp9, Lyn, and Fyn (supplemental Data Set 4). Furthermore, shared target genes for SCL and LYL1 included transcription factors that regulate megakaryopoiesis (GATA1, NFE2, ZEB2, and LMO2) and the platelet survival factor BCL2L1. In contrast, analysis of the 55 genes differentially expressed in Pf4Sclc-KO and DKO but not Lyl1-KO megakaryocytes (supplemental Data Set 4) showed no pathway enrichment.
We confirmed the Scl ChIP-seq results by using published data from primary mouse megakaryocytes.3 We mapped the 2043 Scl binding sites in primary mouse megakaryocytes to 2192 mouse genes (supplemental Data Set 4). Overall, 469 (21%) of these genes were differentially expressed in either single-KO or DKO megakaryocytes with the majority (368 [78%] of 469) only in DKO megakaryocytes (supplemental Figure 8B). Confirming the MEG-01 binding data, pathway analysis of these 368 target genes demonstrated greatest enrichment for platelet activation (supplemental Data Set 4).
Multifactor ChIP-seq analyses have suggested that SCL controls megakaryopoiesis in a multimeric complex that includes GATA1, GATA2, RUNX1, FLI1, and NFE2.2,30 Motif analysis of the 177 common target gene-binding sites demonstrated enrichment for GATA and E-BOX motifs (supplemental Figure 8C). This was directly confirmed by ChIP-seq analysis for the GATA1 locus, in which we saw coordinate binding of SCL, LYL1, LMO2, and GATA2 in MEG-01 cells. Although ChIP for LYL1 was not performed in primary human megakaryocytes, there was coordinate binding of SCL and GATA2 to the GATA1 locus (Figure 7D). A similar pattern of binding was observed for NFE2 (supplemental Figure 8D). Taken together, these binding and expression analyses suggest that Scl and Lyl1 share numerous target genes required for normal platelet production and function, including the transcription factors (Gata1 and Nfe2) known to form a multimeric complex with Scl.
Discussion
We used megakaryocyte-specific deletion of Scl together with constitutive KO of Lyl1 to examine the functional redundancy between these 2 related bHLH factors for platelet production and function. We showed that 5% residual expression of Scl in megakaryocytes was sufficient for relatively normal platelet number and function. However, in the absence of Lyl1, this low-level expression of Scl led to marked thrombocytopenia and severe defects in platelet structure and function. Integration of expression and ChIP analyses provided evidence that Scl and Lyl1 directly regulated many of the genes that cause inherited macrothrombocytopenia, including GATA1 and NFE2.
The platelet phenotype of Pf4Sclc-KO mice was less severe than previously reported by Chagraoui et al,11 who reported defects in megakaryocyte progenitor growth, polyploidization, and increased expression of the cell cycle inhibitor Cdkn1a. One explanation for this more severe phenotype is a different Scl KO allele. Alternatively, the more severe phenotype may be explained by deletion of Scl in more primitive progenitors. Although they used the same Pf4Cre transgenic mice, they observed deletion of Scl in erythroid and myeloid progenitors, indicating Cre activity before megakaryocyte lineage commitment. Supporting the idea that deletion of Scl in an earlier progenitor leads to a more severe phenotype, deletion of Scl in adult HSCs using the MxCre-1 transgene led to defects in stress megakaryopoiesis with loss of rebound thrombopoiesis after chemotherapy.10 By using the EYFP reporter mice, we demonstrated Cre-mediated deletion in phenotypic megakaryocyte progenitors but not the most closely related erythroid progenitors. Therefore, our results showing a less severe phenotype suggest that compensatory mechanisms at the committed megakaryocyte progenitor stage are relatively efficient. An example of this cell stage-dependent compensation is Cdkn1a, in which Scl is required for repression before megakaryocyte commitment, whereas we show that Lyl1 can compensate for loss of Scl in committed megakaryocytes (supplemental Figure 6B). However, this compensatory effect by Lyl1 may not be direct because we did not observe shared binding of SCL and LYL1 to the CDKN1A locus in the MEG-01 cell line (supplemental Data Set 4).
The striking platelet phenotype of DKO mice suggests that Lyl1 is the mechanism of compensation for loss of Scl in megakaryocytes and platelets. However, compensation by Lyl1 is incomplete as evidenced by the platelet phenotype of Pf4Sclc-KO mice: mild macrothrombocytopenia, defective activation response to low concentrations of thrombin, and the transcriptome profile that are more like DKO megakaryocytes. Functional redundancy for Scl and Lyl1 has been also reported in adult HSCs and embryonic erythropoiesis,16,17 although in the case of HSCs, Lyl1 may have a more important role than Scl because a single allele of Lyl1 can rescue the absence of Scl whereas a single allele of Scl cannot rescue the absence of Lyl1.16 The functional redundancy between Scl and Lyl1 is likely explained by their common binding targets, in which the majority (177 of 242) with altered expression are dysregulated only in DKO megakaryocytes (Figure 7C). Furthermore, this gene set is highly enriched for transcription factors known to be important for megakaryopoiesis and identified as causes of inherited macrothrombocytopenia. The ChIP analysis showing enrichment for Gata, Runx1, and Ets motifs at common Scl and Lyl1 binding sites supports the concept that Lyl1 can replace Scl in a multifactor complex analogous to the heptad reported in stem and progenitor cells.18
The reduced expression of Gata1 and Fli1 in DKO megakaryocytes only, together with the shared binding sites for Scl and Lyl1 at their promoters, indicates that either Scl or Lyl1 is sufficient for expression of these transcription factors. In contrast, Scl and Lyl1 expression are normal in megakaryocytes lacking Gata1. Furthermore, Gata2 is not increased despite loss of Gata1 expression. This suggests that Scl and Lyl1 may lie upstream of Gata1. The reduced expression of Gata1, Fli1, and Nfe2 likely explains the platelet phenotype of DKO mice. Gata1-KO mice display a similar phenotype to DKO mice with macrothrombocytopenia, increased megakaryocytes with disorganized demarcation system, and scant cytoplasm.6,31,32 Platelet function defects are also similar between DKO mice and individuals with GATA1 mutations, in which there is reduced expression of GPVI and GP1b-IX-V complexes.33 In addition, a number of structural genes (Flna, Actn1, and Tubb1) are reduced in both GATA1-KO and DKO megakaryocytes. Loss of the ETS factor Fli1 may also contribute to the platelet phenotype, which has reduced expression of GPVI, GPIX, and ITGA2B.34
In summary, by using megakaryocyte-specific deletion of Scl to overcome the embryonic lethality of germ line deletion, this study provides functional and genomic evidence that Scl and Lyl1 cooperatively regulate expression of numerous genes in which heterozygous mutations cause hereditary thrombocytopenia. Germ line homozygosity of Scl is lethal and therefore will not be identified as a cause of hereditary thrombocytopenia. In contrast, heterozygous loss of Scl or homozygous loss of Lyl1 have no platelet phenotype. This functional redundancy between Scl and Lyl1 provides 1 explanation for the absence of platelet phenotype in the Scl heterozygous state, and consequently, the failure to detect mutations of Scl in large sequencing studies performed for hereditary thrombocytopenia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Alfred Medical Research & Education Precinct (AMREP) animal service staff for animal husbandry and the AMREP flow cytometry core facility staff for technical assistance and advice; Kirill Tsyganov, Nick Wong, and the Monash Bioinformatics Platform for processing of RNA-seq data; and Stephen Cody and Chad Johnson of Monash Micro Imaging for measurement of nuclear and cytoplasmic areas.
This work was supported by a project grant (APP1052313) (D.J.C.) from the Australian National Health and Medical Research Council (NHMRC), a Leukaemia Foundation scholarship (S.K.C.), a Senior Medical Research Fellowship from the Sylvia and Charles Viertel Foundation (D.J.C.), a Cancer Institute University of New South Wales Fellowship (D.B.), and by funding from the NHMRC, Anthony Rothe Foundation (J.E.P.), Cancer Australia (D.B.), Gilead Sciences (D.B.) and an NHMRC Peter Doherty fellowship (D.B.).
Authorship
Contribution: D.J.C., C.S.T., J.E.P., and J.R.H. designed the study; S.K.C., S.L.O., M.J.M., J.S., S.E., Y.H., and D.B. performed the experiments; B.T.K. provided experimental animals and reagents; S.K.C., S.L.O., M.J.M., J.S., C.S.T., Y.H., and D.C. analyzed the data; and S.K.C., C.S.T., and D.J.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David J. Curtis, Australian Centre for Blood Diseases, 99 Commercial Rd, Melbourne, VIC, Australia 3004; e-mail: david.curtis@monash.edu.






![Figure 6. Gene expression profiling of megakaryocytes from single-KO and DKO mice. (A) Venn diagrams showing the numbers of genes up- and downregulated more than twofold in Pf4Sclc-KO, Lyl1-KO, and DKO megakaryocytes relative to WT mice (false discovery rate [FDR] <0.01). (B) Gene set enrichment analysis (GSEA) for platelet activation and aggregation genes between DKO and single-KO megakaryocytes. (C) Heat map showing expression of structural genes known to cause inherited macrothrombocytopenia. The FDR is shown for comparison of DKO with Pf4Sclc-KO cells. (D) Heat map showing expression of transcription factors that regulate megakaryopoiesis. FDR is shown for comparison of DKO and Pf4Sclc-KO cells. NES, normalized enrichment score; NS, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/134/10/10.1182_blood.2019896175/3/m_blood896175f6.png?Expires=1766022787&Signature=WSBesbpXW5uoGzoOmRvMUUFsazEANP1NOdY7IgWJez6ht5wa8NcgtZK~t0dcFzQoVaWQIw9sXf7lUD6mBQwjbZuU7Bubcpij8icM6UfnGVZatZ-bHd37618BAyvxhjarOQnS366Lh-M6W1b9z2KF8bWiaQsIr8zVIeza2COVFiCeCebQnHx-mx4issSuQGqke5z2cQuQLRJQthOw5AB287vO90gCpvsm3UM9TeMfmpowViAQrLgJHrXOhps~GFcuardcZedBsWR0GkNm24Ez9RYbXuwOSMqYsqqqMqkGlLQg7Yun4qHrvIn88u-J8vQqp-aFty2ePlw2GgJvqZJ~DQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal