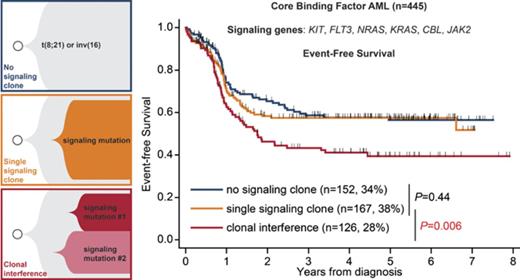
Key Points
Presence of ≥2 independent subclones in the receptor tyrosine kinase/RAS pathway, defining clonal interference, is found in 28% of CBF AMLs.
Clonal interference predicts shorter event-free survival independently of clinical variables and presence of specific signaling mutations.
Abstract
Mutations in receptor tyrosine kinase/RAS signaling pathway genes are frequent in core-binding factor (CBF) acute myeloid leukemias (AMLs), but their prognostic relevance is debated. A subset of CBF AML patients harbors several signaling gene mutations. Genotyping of colonies and of relapse samples indicates that these arise in independent clones, thus defining a process of clonal interference (or parallel evolution). Clonal interference is pervasive in cancers, but the mechanisms underlying this process remain unclear, and its prognostic impact remains unknown. We analyzed a cohort of 445 adult and pediatric patients with CBF AML treated with intensive chemotherapy and with deep sequencing of 6 signaling genes (KIT, NRAS, KRAS, FLT3, JAK2, CBL). A total of 152 (34%), 167 (38%), and 126 (28%) patients harbored no, a single, and multiple signaling clones (clonal interference), respectively. Clonal interference of signaling mutations was associated with older age (P = .004) and inv(16) subtype (P = .025) but not with white blood cell count or mutations in chromatin or cohesin genes. The median allele frequency of signaling mutations was 31% in patients with a single clone or clonal interference (P = .14). The repertoire of KIT, FLT3, and NRAS/KRAS variants differed between groups. Clonal interference did not affect complete remission rate or minimal residual disease after 1-2 courses, but it did convey inferior event-free survival (P < 10−4), whereas the presence of a single signaling clone did not (P = .44). This inferior outcome was independent of clinical parameters and of the presence of specific signaling clones. Our results suggest that specific clonal architectures can herald distinct prognoses in AML.
Introduction
Core-binding factor (CBF) acute myeloid leukemias (AMLs) account for 30% of pediatric AMLs and 15% of adult AMLs, and are characterized by chromosomal alterations resulting in gene fusions [RUNX1-RUNX1T1 fusion in t(8;21) or CBFB-MYH11 fusion in inv(16)/t(16;16)] involving subunits of the CBF transcription factor, a master regulator of hematopoiesis. CBF gene fusions lead to a preleukemic state, requiring secondary events to progress to AML.1
These secondary mutations notably target genes encoding receptor tyrosine kinases (RTKs; FLT3 and KIT) or involved in RTK signal transduction (CBL, JAK2), especially in the RAS pathway (NRAS, KRAS, PTPN11, and NF1). Such “signaling” mutations are found in two thirds of patients with t(8;21) and up to 90% of patients with inv(16), but their prognostic role is debated.2-5
Co-occurrence of signaling mutations has been reported in up to 25% of patients.6-8 Genotyping of individual colonies indicates that these signaling mutations are found in independent clones.9,10 Similar observations were made in other myeloid neoplasms.11-13 The coexistence of clones sharing a common ancestor and harboring independent lesions targeting the same pathway corresponds to a phenomenon of “clonal interference” (or parallel evolution), a well-characterized process in the dynamics of asexual populations under constrained evolution.14,15 Clonal interference is pervasive in cancers.16-18 In AML, it has notably been ascribed to FLT3 mutations, based on single-cell genotyping.19 The clinical relevance of clonal interference in cancers has yet to be reported. Here, we interrogate the clinical and molecular correlates of clonal interference of signaling mutations and its prognostic value in a large cohort of CBF AML patients treated with intensive chemotherapy.
Patients and methods
Patients and treatments
We retrospectively analyzed CBF AML patients with targeted sequencing data from the French AML Intergroup central laboratory (Lille University Hospital) and the Munich Leukemia Laboratory (MLL). The cohort included 142 adult patients from the French AML Intergroup CBF2006 trial20 (ClinicalTrials.gov NCT00428558) and 73 children enrolled in the pediatric ELAM02 trial (NCT00149162),21 whose mutational profiling was previously reported,4 as well as 230 patients referred from different German centers to the MLL, including 124 of 139 previously reported t(8;21) cases with adequate material8 and 106 unreported inv(16)/t(16;16) cases (supplemental Figure 1, available on the Blood Web site). French patients were diagnosed between October 2005 and November 2011, and German patients were diagnosed between August 2005 and January 2015. CBF AML was defined as the presence of t(8;21) translocation or inv(16)/t(16;16) by standard karyotyping or fluorescence in situ hybridization or the detection of RUNX1-RUNX1T1 or CBFB-MYH11 fusion transcripts.4,8,22
All patients (or parents) gave their informed consent for the use of laboratory results for scientific studies. Studies were approved by the ethics committees of Nimes University Hospital (CBF2006) and Saint-Antoine Paris University Hospital (ELAM02), as well as the Internal Review Board of MLL. All studies were conducted in accordance with the Declaration of Helsinki. Additional information on the patients’ treatments is reported in supplemental Methods.
Targeted high-throughput sequencing
Targeted high-throughput sequencing (HTS) of bone marrow samples collected at diagnosis was performed as reported previously by the CHU Lille Laboratory of Hematology for CBF2006 and ELAM02 patients.4 HTS of MLL samples is detailed in supplemental Methods. The detection limit for signaling genes (NRAS, KRAS, FLT3, JAK2, CBL, PTPN11, KIT) and for chromatin (ASXL1, ASXL2, BCOR, BCORL1, EZH2, KDM6A)/cohesin (RAD21, SMC1A, SMC3, STAG2) genes for both platforms was a variant allele frequency (VAF) of 1%. The overlap between regions sequenced in each platform for these genes is shown in supplemental Table 2. FLT3 internal tandem duplications (ITDs) and allele ratios (ITD/wild-type) were analyzed as previously described.4,8
Definitions
Each pathogenic variant identified within KIT, FLT3, NRAS, KRAS, JAK2, or CBL defined a signaling clone. Clonal interference was defined as the presence of ≥2 signaling clones in a single patient sample (eg, KRASG13D and NRASG12S). The total VAF of signaling clones was obtained by adding the VAF of each individual clone. For patients harboring FLT3-ITD clones determined using polymerase chain reaction (PCR), VAF was inferred from the allele ratio (ITD/wild-type) as follows: VAF = 1/(1+1/allele ratio).
Statistical analyses
Variables are reported as numbers and percentages or median and range (or interquartile range [IQR] when indicated). Group comparisons for continuous or categorical variables were performed with the Student t test or χ2 test, respectively. Multivariate analyses of dichotomic and continuous variables were carried out by logistic and linear regressions, respectively.
All time to events were computed from the date of diagnosis. Overall survival (OS) was determined until death or loss of follow-up, and event-free survival (EFS) was determined until remission induction failure, relapse, or death or until loss of follow-up, both with the Kaplan-Meier estimator. Univariate and multivariate analyses were performed with Cox models. Because indications of allogeneic stem cell transplantation (HSCT) were not uniform, survival analyses were censored at the time of HSCT in patients receiving HSCT in first complete remission (CR) but not later in disease evolution. A sensitivity analysis removing this censoring was also carried out. The linearity of continuous variables was verified by inspection of martingale residuals. The proportional hazard assumption was validated by graphical inspection of the Schönfeld residuals.23 Cumulative incidence of failure (CIF) was computed from diagnosis, considering primary induction failure and relapse as failures and death as a competing event. Univariate and multivariate analyses for CIF were done with Fine and Gray models.24 All analyses were stratified on the HTS platform (Lille vs MLL).
The relative prognostic importance of clonal interference and specific mutations was interrogated by bootstrapping.25 A total of 1000 bootstrap samples with replacement was created from the data set. Using the stepwise forward procedure, we determined the percentage of models including each of the initial variables. Validation of variable selection was based on lasso penalized regression, using the 1 SE rule with the glmnet package for R.26 Additional methods can be found in supplemental Methods.
Results
Baseline clinical and molecular characteristics
The study population included 445 CBF AML patients (251 males, 194 females) with a median age of 42 years (range 1-81), including 230 patients with t(8;21) AML and 215 patients with inv(16)/t(16;16) AML (Table 1). Median white blood cell count (WBC) was 9.6 × 109/L and 32.1 × 109/L in t(8;21) and inv(16) patients, respectively. Compared with patients from French trials, patients sequenced in the MLL were older (P < 10−4) but were otherwise comparable in terms of gender, WBC, and CBF subtype [t(8;21) vs inv(16); supplemental Table 1].
Baseline characteristics of patients
| . | t(8;21) . | inv(16) . | All . |
|---|---|---|---|
| Number of cases | 230 | 215 | 445 |
| HTS platform, n (%) | |||
| Lille | 106 (46) | 109 (51) | 215 (48) |
| MLL | 124 (54) | 106 (49) | 230 (52) |
| Age, median (range), y | 41 (2-80) | 43 (1-81) | 42 (1-81) |
| Sex, n (%) | |||
| Male | 124 (54) | 127 (59) | 251 (56) |
| Female | 106 (46) | 88 (41) | 194 (44) |
| WBC, median (range), × 109/L (n = 405) | 9.6 (1.3-170) | 32.1 (0.1-351) | 15.0 (0.1-351) |
| ACA, n (%) (n = 406) | |||
| Absent | 66 (29) | 110 (61) | 176 (43) |
| Present | 159 (71) | 71 (39) | 230 (57) |
| Induction outcome, n (%) | |||
| CR | 220 (96) | 204 (95) | 424 (95) |
| Early death | 6 (2) | 11 (5) | 17 (4) |
| Primary induction failure | 4 (2) | 0 (0) | 4 (1) |
| HSCT in first CR, n (%) | 24 (10) | 22 (10) | 46 (10) |
| Median follow-up, y | 5.0 | 4.5 | 4.7 |
| . | t(8;21) . | inv(16) . | All . |
|---|---|---|---|
| Number of cases | 230 | 215 | 445 |
| HTS platform, n (%) | |||
| Lille | 106 (46) | 109 (51) | 215 (48) |
| MLL | 124 (54) | 106 (49) | 230 (52) |
| Age, median (range), y | 41 (2-80) | 43 (1-81) | 42 (1-81) |
| Sex, n (%) | |||
| Male | 124 (54) | 127 (59) | 251 (56) |
| Female | 106 (46) | 88 (41) | 194 (44) |
| WBC, median (range), × 109/L (n = 405) | 9.6 (1.3-170) | 32.1 (0.1-351) | 15.0 (0.1-351) |
| ACA, n (%) (n = 406) | |||
| Absent | 66 (29) | 110 (61) | 176 (43) |
| Present | 159 (71) | 71 (39) | 230 (57) |
| Induction outcome, n (%) | |||
| CR | 220 (96) | 204 (95) | 424 (95) |
| Early death | 6 (2) | 11 (5) | 17 (4) |
| Primary induction failure | 4 (2) | 0 (0) | 4 (1) |
| HSCT in first CR, n (%) | 24 (10) | 22 (10) | 46 (10) |
| Median follow-up, y | 5.0 | 4.5 | 4.7 |
ACA, additional cytogenetic alteration.
All patients were treated frontline with intensive chemotherapy in the ELAM02 and CBF2006 trials for French patients and according to various regimens for German patients. Induction regimens were mostly 7+3 based, but 127 patients received intermediate doses (defined as ≥1 daily dose ≥ 1 g/m2) or high doses (≥1 daily dose ≥ 3 g/m2) of cytarabine during induction. Overall, 424 (95%) patients reached CR after induction therapy, 17 (4%) patients died during induction, and only 4 (1%) experienced primary induction failure. Most patients (335/363 with available information) received consolidation therapy based on 1 or several high-dose cytarabine courses, and 46 (10%) received HSCT in first CR. A total of 25 patients received a tyrosine kinase inhibitor (dasatinib, n = 23; sorafenib, n = 2) during induction (n = 4) and/or consolidation/maintenance (n = 25). The detailed distribution of treatments and early outcomes is reported in supplemental Figure 1.
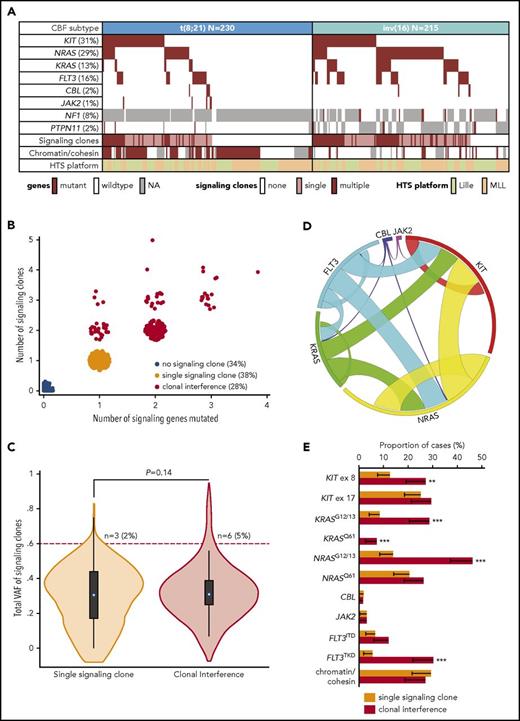
Molecular profiling of mutations in signaling genes was performed with a limit of detection of clones representing 1% of VAF, including FLT3 ITDs. Sequencing of KIT, NRAS, KRAS, FLT3 (including FLT3 ITD), CBL, and JAK2 was available for all patients, whereas sequencing of NF1 and PTPN11 was only available in 110 and 336 patients, respectively. The mutation pattern of t(8;21) and inv(16) patients is reported in Figure 1A, with the known enrichment of NRAS and KRAS mutations (both P < 10−4) and FLT3 (P = .014) in inv(16) compared with t(8;21) AML. Due to missing data on NF1 and PTPN11, we focused our analysis of signaling clones on 6 genes (KIT, NRAS, KRAS, FLT3, CBL, and JAK2). No significant difference between the 2 HTS platforms was noted in the mutation frequency of those 6 genes when adjusting for age and CBF subtype, with the exception of more frequent FLT3 variants in Lille patients (19%) compared with MLL patients (12%) (P= .02; supplemental Table 3). Mutations in the chromatin regulator genes ASXL1, ASXL2, EHZ2, KDM6A, BCOR, and BCORL1 or in the cohesin genes RAD21, STAG2, SMC1A, and SMC3, which have been shown to affect prognosis in CBF AML,4 were assessed in 339 patients (Lille, n = 215; MLL, n = 164). Chromatin/cohesin gene mutations were found in 57% and 7% of t(8;21) and inv(16) cases, respectively, with no interplatform difference (P = .15; supplemental Table 3).
Molecular pattern of clonal interference of signaling mutations in CBF AML. (A) Pattern of gene mutations in cases with t(8;21) and inv(16) AML, presence of clonal interference in the 6 comprehensively analyzed signaling genes (KIT, NRAS, KRAS, FLT3, CBL, JAK2), and presence of ≥1 mutation in chromatin (ASXL1, ASXL2, BCOR, BCORL1, EZH2, KDM6A) or cohesin (RAD21, SMC1A, SMC3, STAG2) genes. (B) Scatter plot indicating the number of signaling genes (KIT, NRAS, KRAS, FLT3, CBL, JAK2) and the total number of clones in these 6 signaling genes. Patients with ≥2 signaling clones are assigned to the “clonal interference” group. (C) Volcano plot of the total VAF of signaling clones in patients with a single or multiple signaling clones, together with the P value of a linear regression adjusted on HTS platform. The number of patients with a total VAF > 60% is reported. The difference in the proportion of patients with total VAF > 60% was not statistically different (logistic regression adjusted for HTS platform; P = .16). (D) Circos plot showing the pattern of comutations in signaling genes in patients with ≥1 signaling clone. (E) Proportion of patients with a single signaling clone (n = 167) or with clonal interference (n = 126) with ≥1 clone displaying the genotype indicated on the x-axis (the error bar indicates the 95% CI). P values for logistic regression adjusted on HTS platform: **P < .01, ***P < .001. Unspecified P values are >.05. ex, exon; NA, not available.
Molecular pattern of clonal interference of signaling mutations in CBF AML. (A) Pattern of gene mutations in cases with t(8;21) and inv(16) AML, presence of clonal interference in the 6 comprehensively analyzed signaling genes (KIT, NRAS, KRAS, FLT3, CBL, JAK2), and presence of ≥1 mutation in chromatin (ASXL1, ASXL2, BCOR, BCORL1, EZH2, KDM6A) or cohesin (RAD21, SMC1A, SMC3, STAG2) genes. (B) Scatter plot indicating the number of signaling genes (KIT, NRAS, KRAS, FLT3, CBL, JAK2) and the total number of clones in these 6 signaling genes. Patients with ≥2 signaling clones are assigned to the “clonal interference” group. (C) Volcano plot of the total VAF of signaling clones in patients with a single or multiple signaling clones, together with the P value of a linear regression adjusted on HTS platform. The number of patients with a total VAF > 60% is reported. The difference in the proportion of patients with total VAF > 60% was not statistically different (logistic regression adjusted for HTS platform; P = .16). (D) Circos plot showing the pattern of comutations in signaling genes in patients with ≥1 signaling clone. (E) Proportion of patients with a single signaling clone (n = 167) or with clonal interference (n = 126) with ≥1 clone displaying the genotype indicated on the x-axis (the error bar indicates the 95% CI). P values for logistic regression adjusted on HTS platform: **P < .01, ***P < .001. Unspecified P values are >.05. ex, exon; NA, not available.
Incidence of clonal interference
Of the 293 (66%) patients harboring ≥1 mutation in the 6 signaling genes (KIT, NRAS, KRAS, FLT3, CBL, and JAK2), 126 (28% of the total cohort) had ≥2 signaling variants, including 76 patients with mutations in different genes, 27 patients with several mutations in 1 gene, and 23 patients with both types of mutations (Figure 1B). The median VAF of signaling clones in patients having a single signaling mutation was 31% (IQR, 17-44%) compared with a median VAF of 31% (IQR, 25-39%) when adding VAFs of signaling mutations in the 126 patients with ≥2 mutations (P = .14; Figure 1C). Notably, only 6 (5%) patients with ≥2 signaling mutations had a cumulative VAF > 60% compared with 3 (2%) patients with a single signaling mutation (P = .16), in keeping with the presence of each of these lesions in mutually exclusive clones. Despite incomplete coverage, single-cell genotyping in 2 inv(16) patients harboring 2 and 4 signaling mutations was also compatible with this model (supplemental Figure 2; supplemental Table 4). Altogether, these findings suggest that, in most cases, each mutation in a signaling gene corresponds to an independent clone and that patients with ≥2 signaling clones harbor clonal interference.
Molecular pattern of clonal interference
Among CBF AML patients with ≥1 signaling clone, clonal interference was more frequent in inv(16) (83/173 cases, 48%) patients than in t(8;21) (43/120 cases, 36%) patients (P = .025). Clonal interference was notably associated with NRAS, KRAS, and FLT3 mutated clones (all P < 10−4) and, to a lesser extent, with KIT mutated clones (P = .03; Figure 1D), whereas CBF subtype did not have independent predictive value (P = .95). For a given gene, the mutation pattern of isolated clones and clones with interference differed. KIT mutations in exon 8 but not exon 17, NRAS mutations at the G12/G13 residue but not at the Q61 residue, and FLT3TKD but not FLT3ITD were significantly overrepresented in patients with clonal interference (Figure 1E). Conversely, chromatin/cohesin gene mutations did not influence the likelihood of clonal interference (P = .7). Individual inspection of nonsignaling genes sequenced in ≥90% of patients did not reveal any pattern associated with clonal interference (supplemental Figure 3), and the number of nonsignaling mutations was comparable between cases with a single signaling clone vs clonal interference (P = .38). The frequency and spectrum of additional cytogenetic alterations were comparable in patients with 1 or several signaling clones (Table 2; supplemental Figure 4). Altogether, these findings suggest that the occurrence of clonal interference is dictated by the incidence of signaling mutations themselves, but not by CBF subtype per se or other oncogenetic lesions, and that clonal interference is associated with a specific repertoire of signaling mutations.
Clinical characteristics of patients according to the presence of clonal interference
| . | Signaling clones . | P* . | ||
|---|---|---|---|---|
| No . | Single . | Interference . | ||
| Cases, n (%) | 152 (34) | 167 (38) | 126 (28) | |
| Age, median (range), y | 43 (4-79) | 39 (1-80) | 43 (2-81) | .004 |
| Sex, n (%) | .69 | |||
| Male | 79 (52) | 100 (60) | 72 (58) | |
| Female | 73 (48) | 67 (40) | 54 (42) | |
| CBF type, n (%) | .025 | |||
| t(8;21) | 110 (72) | 77 (46) | 43 (34) | |
| inv(16) | 42 (28) | 90 (54) | 83 (66) | |
| WBC, median (range), × 109/L | 7.1 (1-252) | 22.5 (0.1-231) | 24.8 (0.1-351) | .24 |
| ACA, n (%) (n = 406) | .62† | |||
| Absent | 42 (29) | 74 (49) | 60 (55) | |
| Present | 102 (71) | 78 (51) | 50 (45) | |
| Induction outcome, n (%) | .68 | |||
| CR | 147 (96) | 158 (94) | 119 (94) | |
| Early death | 2 (2) | 8 (5) | 7 (6) | |
| Primary induction failure | 3 (2) | 1 (1) | 0 (0) | |
| MRD log reduction, median (IQR) | n = 44 | n = 45 | n = 53 | |
| After 1 course | 3.1 (2.6-3.7) | 2.7 (2.2-3.3) | 2.6 (2.2-3.2) | .89‡ |
| After 2 courses | 3.6 (2.9-4.6) | 3.2 (2.5-4.3) | 3.1 (2.7-3.8) | .77‡ |
| . | Signaling clones . | P* . | ||
|---|---|---|---|---|
| No . | Single . | Interference . | ||
| Cases, n (%) | 152 (34) | 167 (38) | 126 (28) | |
| Age, median (range), y | 43 (4-79) | 39 (1-80) | 43 (2-81) | .004 |
| Sex, n (%) | .69 | |||
| Male | 79 (52) | 100 (60) | 72 (58) | |
| Female | 73 (48) | 67 (40) | 54 (42) | |
| CBF type, n (%) | .025 | |||
| t(8;21) | 110 (72) | 77 (46) | 43 (34) | |
| inv(16) | 42 (28) | 90 (54) | 83 (66) | |
| WBC, median (range), × 109/L | 7.1 (1-252) | 22.5 (0.1-231) | 24.8 (0.1-351) | .24 |
| ACA, n (%) (n = 406) | .62† | |||
| Absent | 42 (29) | 74 (49) | 60 (55) | |
| Present | 102 (71) | 78 (51) | 50 (45) | |
| Induction outcome, n (%) | .68 | |||
| CR | 147 (96) | 158 (94) | 119 (94) | |
| Early death | 2 (2) | 8 (5) | 7 (6) | |
| Primary induction failure | 3 (2) | 1 (1) | 0 (0) | |
| MRD log reduction, median (IQR) | n = 44 | n = 45 | n = 53 | |
| After 1 course | 3.1 (2.6-3.7) | 2.7 (2.2-3.3) | 2.6 (2.2-3.2) | .89‡ |
| After 2 courses | 3.6 (2.9-4.6) | 3.2 (2.5-4.3) | 3.1 (2.7-3.8) | .77‡ |
P values from logistic or linear regressions adjusted on HTS platform comparing patients with a single clone vs clonal interference.
P values from linear regressions adjusted on CBF type.
P values from linear regressions adjusted on CBF type and induction arm (7+3 or intensified), according to Jourdan et al.20
Clinical features and early outcome of clonal interference
Compared with patients with a single signaling clone, patients with clonal interference were older (median, 43 years [range 2-81] vs 39 years [range 1-80]; P = .004) but were comparable in terms of sex and WBC at diagnosis (Table 2). CR was obtained in 94% of patients with signaling mutations, regardless of the presence of clonal interference (P = .68). Adjusting on CBF subtype [t(8;21) vs inv(16)] did not change these results (data not shown). Minimal residual disease (MRD) was assessed by real-time quantitative PCR of RUNX1-RUNX1T1 or CBFB-MYH11 transcripts in the 140 CBF2006 trial patients in CR, as previously described.20 After adjusting on CBF subtype and induction arm (7+3 vs time sequential; supplemental Figure 1), there was no difference in MRD reduction after the first (P = .89) or the second (P = .77) course between patients with a single signaling clone (n = 45) and patients with clonal interference (n = 53; Table 2).
Prognostic impact of clonal interference
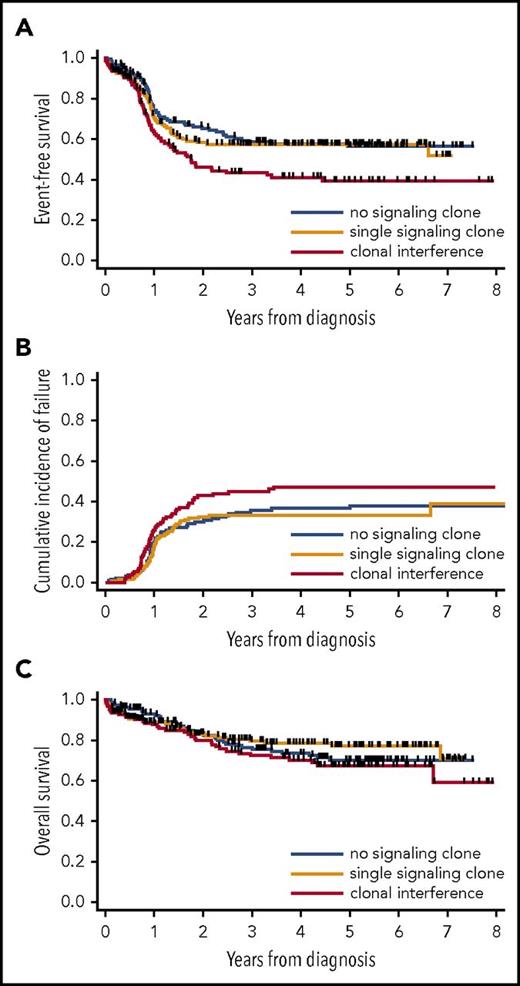
With a median follow-up of 4.7 years, 46 patients received an HSCT in first CR, and there were 142 relapses and 109 deaths. Censoring at HSCT in first CR, 3-year estimates for OS and EFS were 76.3% (95% confidence interval [CI], 71.6-80.3%) and 53.8% (95% CI, 48.6-58.6), respectively. In univariate analysis stratified by HTS platform, 3-year EFS was 58.6% (95% CI, 49.6-66.6) in patients without signaling mutation, 57.5% (95% CI, 49.1-65.1) in patients with a single signaling clone (P = .44 vs no signaling mutation), and 43.3% (95% CI, 33.9-52.5) in patients with clonal interference (P < 10−4 vs no signaling mutation; P = .006 vs single signaling clone; Figure 2A). The inferior EFS of patients with clonal interference could be attributed to a higher CIF, reaching 44.8% (95% CI, 35.4-54.3) at 3 years compared with 35.6% (95% CI, 27.2-43.9) and 33.2% (95% CI, 25.5-41.0) in patients with no or a single signaling clone, respectively (P = .04 for clonal interference vs single signaling clone; P = .04 for clonal interference vs no signaling clone; Figure 2B), whereas nonrelapse mortality was comparable between patients with a single or multiple signaling clones (P = .24). Three-year OS was 76.3% (95% CI, 67.7-82.8), 79.4% (95% CI, 71.7-85.2), and 72.4% (95% CI, 62.7-79.9) in patients with no, 1, and multiple signaling clones, respectively, with a trend toward a higher risk for death in patients with clonal interference compared with those with a single clone (hazard ratio [HR], 1.64; 95% CI, 1.01–2.66; P = .045) or with patients without a signaling clone (HR, 1.60; 95% CI, 0.99-2.57; P = .055; Figure 2C). With regard to EFS, the risk for reaching an event (induction failure, relapse, or death) increased in a linear way with each additional signaling clone (HR, 1.34; 95% CI, 1.16-1.55 [for each additional clone]; P < 10−4).
Outcome of patients according to clonal interference of signaling mutations in CBF AML. EFS (A), CIF (B), and OS (C) in patients with no, a single, or multiple signaling clones. EFS and OS are censored at HSCT in first CR.
Outcome of patients according to clonal interference of signaling mutations in CBF AML. EFS (A), CIF (B), and OS (C) in patients with no, a single, or multiple signaling clones. EFS and OS are censored at HSCT in first CR.
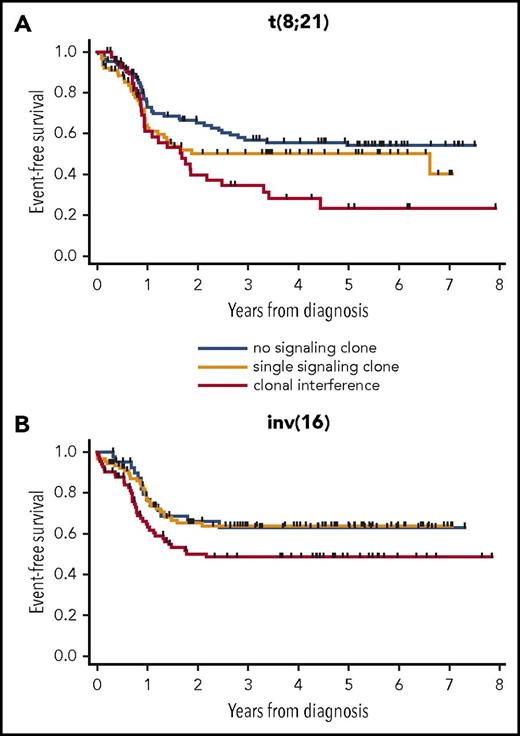
We next analyzed the prognostic impact of clonal interference in each subtype of CBF AML. In both CBF subtypes, EFS of patients with clonal interference was poorer than that of patients with a single or no signaling clone. In t(8;21) patients, 3-year EFS was 56.9% (95% CI, 46.0-66.3), 50.1% (95% CI, 37.6-61.4), and 34.5 (95% CI, 20.0-49.4) in patients with no, a single, or multiple signaling clones, respectively (P < 10−4 for multiple vs no clone; P = .10 for multiple vs single clone; Figure 3A). In inv(16) patients, 3-year EFS was 63.1% (95% CI, 45.7-76.3), 63.7% (95% CI, 52.0-73.3), and 48.6% (95% CI, 36.6-59.5) in patients with no, a single, or multiple signaling clones, respectively (P = .07 for multiple vs no clone; P = .01 for multiple vs single clone; Figure 3B).
EFS of patients according to clonal interference of signaling mutations in CBF AML subsets. t(8;21) patients (n = 230) (A) and inv(16) patients (n = 215) (B). EFS is censored at HSCT in first CR.
EFS of patients according to clonal interference of signaling mutations in CBF AML subsets. t(8;21) patients (n = 230) (A) and inv(16) patients (n = 215) (B). EFS is censored at HSCT in first CR.
Multivariate analyses of prognosis
We next used multivariate analyses to interrogate the prognostic impact of clonal interference stringently defined on the 6 genes with comprehensive data (KIT, FLT3, NRAS, KRAS, CBL, and JAK2). Presence of clonal interference conferred poorer EFS (P = .001) independently of older age (P = .001), higher WBC (P = .001), and t(8;21) subtype (P = .002), whereas total allele burden of signaling clones had no significant impact. Clonal interference was also an independent adverse prognostic factor for OS (P = .04; Table 3). In the 347 patients with available sequencing data, the presence of ≥1 mutation in chromatin/cohesin genes did not erase the poor impact of clonal interference on EFS (P = .003). There was no significant interaction between allocation to a more intensive induction regimen including ≥ 1 g/m2 cytarabine boluses (supplemental Figure 1) and clonal interference (P = .11). In particular, in the 127 patients receiving intensive induction, of whom 44 and 37 had 1 and multiple signaling clones, respectively, clonal interference retained its adverse impact on EFS (HR, 2.51; 95% CI, 1.24-5.10; P = .01). Among 362 patients with available information, only 35 (10%) received standard-dose cytarabine (<1 g/m2) instead of intermediate/high doses (≥1 g/m2; supplemental Methods) as consolidation therapy. Again, clonal interference retained its poor prognostic value on EFS independently of consolidation intensity (P < 10−4). Finally, in a bivariate analysis of the 142 CBF2006 patients with available MRD after 2 courses (ie, after the first consolidation course), the presence of clonal interference still conveyed poorer EFS (HR, 2.87; 95% CI, 1.35-6.99; P = .006) independently of the established adverse prognosis of MRD reduction < 3 logs (HR, 2.25; 95% CI, 1.32-3.84; P = .003).20 There was no significant interaction between clonal interference and MRD log reduction after 2 courses, suggesting that both variables independently worsen prognosis.
Multivariate analysis of the prognostic impact of clonal interference of signaling mutations on EFS and OS
| Risk factor . | EFS . | OS . | ||
|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | |
| Signaling clones | ||||
| None | 1 | 1 | ||
| Single | 1.38 (0.84-2.29) | .21 | 1.01 (0.5-2.02) | .98 |
| Clonal interference | 2.44 (1.44-4.14) | .001 | 2.07 (1.03-4.15) | .04 |
| Log(WBC) | 1.26 (1.09-1.44) | .001 | 1.11 (0.92-1.33) | .27 |
| CBF type | .002 | .008 | ||
| inv(16) | 1 | 1 | ||
| t(8;21) | 1.69 (1.2-2.37) | 1.88 (1.18-2.98) | ||
| Age, y | 1.02 (1.01-1.03) | .001 | 1.03 (1.02-1.04) | <.0001 |
| Total VAF of signaling clones, % | 0.5 (0.16-1.53) | .22 | 0.58 (0.12-2.73) | .49 |
| Risk factor . | EFS . | OS . | ||
|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | |
| Signaling clones | ||||
| None | 1 | 1 | ||
| Single | 1.38 (0.84-2.29) | .21 | 1.01 (0.5-2.02) | .98 |
| Clonal interference | 2.44 (1.44-4.14) | .001 | 2.07 (1.03-4.15) | .04 |
| Log(WBC) | 1.26 (1.09-1.44) | .001 | 1.11 (0.92-1.33) | .27 |
| CBF type | .002 | .008 | ||
| inv(16) | 1 | 1 | ||
| t(8;21) | 1.69 (1.2-2.37) | 1.88 (1.18-2.98) | ||
| Age, y | 1.02 (1.01-1.03) | .001 | 1.03 (1.02-1.04) | <.0001 |
| Total VAF of signaling clones, % | 0.5 (0.16-1.53) | .22 | 0.58 (0.12-2.73) | .49 |
All analyses are stratified on the HTS platform.
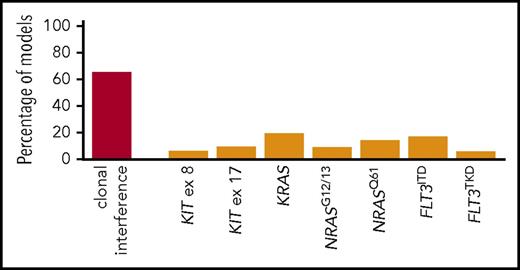
To determine whether the presence of a specific type of signaling mutation (grouped as per Figure 1E) underlies the poor prognosis of patients with clonal interference, we performed bootstrapping of multivariate Cox models of EFS in patients with ≥1 signaling clone. Only mutant clones present in ≥10 patients were retained for this analysis. CBL (n = 9) and JAK2 (n = 5) were not considered, and KRASQ61 (n = 9) was grouped with KRASG12/13. Only clonal interference was retained in >20% of models (Figure 4). Lasso penalized regression led to similar variable selection (data not shown). Of note, KIT exon 8 and 11 mutations were each retained in <10% of models, and, overall, KIT mutations, found in 138 (31.0%) patients, did not predict inferior EFS in univariate analysis (HR, 0.97; 95% CI, 0.71-1.32; P = .83). Thus, the number of signaling clones is a stronger predictor of outcome than the presence of specific signaling variants.
Bootstrapping analysis of the prognostic role of specific signaling mutant clones and clonal interference in CBF AML. Percentage of Cox multivariate models for EFS (y-axis) retaining the indicated variable (x-axis) as significant (P < .05) after forward selection of all variables indicated in the x-axis and stratified on the HTS platform. Results of 1000 bootstraps.
Bootstrapping analysis of the prognostic role of specific signaling mutant clones and clonal interference in CBF AML. Percentage of Cox multivariate models for EFS (y-axis) retaining the indicated variable (x-axis) as significant (P < .05) after forward selection of all variables indicated in the x-axis and stratified on the HTS platform. Results of 1000 bootstraps.
Discussion
In this large cohort of pediatric and adult CBF AML patients treated with intensive chemotherapy, we found that 28% of patients harbor multiple mutations in the RTK/RAS pathway. Our single-cell genotyping experiments are in keeping with previous observations that these mutations largely arise in independent subclones, thus resulting in clonal interference. We show that clonal interference is associated with a specific repertoire of signaling subclones but not with a greater proportion of RTK/RAS-mutated leukemic cells. We report for the first time that the presence of clonal interference, although not affecting early chemosensitivity, is associated with a higher incidence of relapse, resulting in inferior EFS independently of clinical and molecular confounders.
In the present study, we posit that each signaling mutation arises in an independent clone. This postulate, which is not mandatory for the clinical interpretation of our results, is supported by previous studies on single-colony genotyping of CBF AML samples9,10 or other myeloid malignancies.11-13 Our own single-cell genotyping data, although limited by patient numbers and PCR dropout rate, are also compatible with this hypothesis. The comparable VAF of signaling mutations in patients with 1 or several clones, <50% threshold in most cases, is also in keeping with this model,4,27 as the differential expansion or collapse of signaling variants at relapse in patients with several signaling clones.3,8,27-29 Our data cannot formally exclude the rare co-occurrence of 2 signaling mutations in a single clone, such as FLT3ITD and FLT3TKD,30 which, however, only accounted for a minority (16%) of signaling mutations in the present study. Such a co-occurrence has been reported infrequently in juvenile myelomonocytic leukemias in which RTK/RAS pathway mutations are initiating lesions,12,13 in contrast to CBF AML.31-33 The few double-mutated clones reported so far in juvenile myelomonocytic leukemias always involved rare, potentially weaker oncogenes (RRAS, SH2B3) than those considered in this study.12,13
Our present study focused on 6 of the most frequently mutated signaling genes, and it is possible that a more comprehensive characterization of signaling mutations, to include infrequently mutated signaling genes, would identify a slightly higher proportion of cases with clonal interference.34 Because we identify clones based on hotspot mutations, it is also possible that several similar genomic changes in these hotspot regions occur several times in leukemia evolution, leading to an underestimation of the number of signaling clones.19 Indeed, single-cell genotyping in UPN1 suggests that 2 independent FLT3TKD clones arose, including 1 from an ancestral BCOR mutated clone (supplemental Figure 2).
Our study population combined patients prospectively treated in clinical trials, together with real-life data from 2 countries. They were analyzed on 2 HTS platforms. Despite the almost perfect overlap of sequenced regions, similar mutation frequencies, and detection threshold, we acknowledge this potential source of heterogeneity by stratifying all of our analyses on the HTS platform. The relative homogeneity of intensive chemotherapy treatment schemes is exemplified by the high and comparable CR rates in both data sets, as well as by the high proportion of patients (90%) receiving intermediate/high-dose cytarabine as consolidation therapy. Because a higher proportion of German patients were transplanted in first CR, we censored all of our analysis at HSCT in first CR. Of note, removing this censoring did not affect our conclusions (data not shown). Although 29% of patients received intensified induction courses, we have previously demonstrated that intensification of induction increased early MRD log reduction without affecting the long-term outcome of CBF AML.20 Indeed, we found no significant interaction between induction intensity or MRD log reduction and clonal interference. Our results show that clonal interference does not abrogate the established prognostic value of MRD in CBF AML.20,35 However, we cannot exclude that different schedules or doses of cytarabine consolidation courses modulate the prognostic impact of clonal interference.
In our study, older age and the inv(16) subset of CBF AML were the only clinical parameters associated with a higher likelihood of developing clonal interference. The former could correspond to a longer disease latency, resulting in a higher likelihood of generating multiple competing signaling clones, whereas the latter reflects the known enrichment of signaling mutations in inv(16) compared with t(8;21).2-4,8,27,36,37 We found a biased repertoire of RAS, KIT, and FTL3 clones in patients with clonal interference. Molecular epidemiology and mouse models indicate that the oncogenic properties of specific RAS mutations differ in a context-dependent manner.6,38,39 KIT exon 8 mutants and FLT3TKD are often considered weaker oncogenes than KIT exon 17 mutants and FLT3ITD, respectively.40,41 Interestingly, the overrepresentation of weaker KIT and FLT3 hits in patients with clonal interference is in agreement with evolution theory, because strong oncogenes are more likely to reach fixation before the expansion of a second clone.42
Whether patients with clonal interference should be managed specifically remains debatable. The 15% drop in EFS only translated into a trend toward inferior OS. Only 11 patients with clonal interference received HSCT in first CR in our cohort, precluding a robust time-dependent investigation of the benefit of early HSCT in this context. Efforts to target the RTK/RAS pathway in AML are ongoing.43,44 The ongoing randomized study of dasatinib combination with chemotherapy AMLSG 21-13 (NCT02013648) will provide a unique opportunity to determine whether patients with clonal interference are specifically sensitive to such targeted interventions.
Our observations also raise important questions on the evolutionary dynamics of AML. Three nonmutually exclusive hypotheses could account for the increased risk for relapse associated with clonal interference. First, clonal interference may correlate with a higher proportion of leukemic cells harboring a RTK/RAS pathway alteration, regardless of its precise nature. Indeed, the prognostic relevance of clone size is well established for FLT3 in standard-risk AML,45 and clone size of signaling mutations in CBF AML has been suggested to carry prognostic information.37 However, we did not find a prognostic role for the total allele burden of signaling mutations. Furthermore, the average allele burden of signaling mutations (31%) was comparable in patients with 1 and multiple clones. A second, deterministic hypothesis posits that, within the repertoire of signaling clones, only a subset are chemoresistant. Thus, cases with clonal interference would have a higher likelihood of carrying such chemoresistant clones. Our bootstrapping analysis failed to identify such chemoresistant clones. We did not identify KIT mutations (all or specifically exon 17, including D816 variants) to predict adverse EFS, a notion that remains controversial.3-5,7,8,37 A third “stochastic” model would consider clonal interference as a proxy of a broader process, such as higher mutational instability, faster evolution, or clonal cooperation, that underlies the chemoresistance of these CBF AMLs. This model is in agreement with the instability of signaling clones at relapse3,8,27-29 and warrants further investigation.
Clonal interference is a recurrent phenomenon in many cancers types.16-19,46 Our findings expand on previous reports on the adverse prognostic value of intratumoral heterogeneity.47,48 They are the first to ascribe a prognostic value to a specific type of clonal architecture. Because of their relatively simple genomic landscape and consensual treatment, CBF AML may represent a unique model to understand the cellular mechanisms governing the emergence and chemoresistance of clonal interference and to uncover architecture-specific cancer vulnerabilities.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Christophe Roumier (Tumour Bank for the Acute Leukemia French Association certification NF 96900-2014/65453-1, Centre Hospitalier Regional Universitaire Lille) for handling, conditioning, and storing patient samples, as well as Sylvie Méléard and Alexandre Puissant for insightful discussions.
Authorship
Contribution: R.I. designed the study and conducted the analyses; R.I., N.D., A.F., N.B., T.H., and C.P. wrote the manuscript; N.D., A.F., A.M.-R., and M.M. performed targeted deep sequencing; G.D. and N.D. performed single-cell genotyping; E.J., A.P., H.L., J.-B.M., P.C.-L., N.I., G.L., H.D., and N.B. accrued patients and provided clinical annotations; T.H. and C.P. supervised the work; and all authors revised the manuscript and approved its final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Raphael Itzykson, INSERM/CNRS UMR 944/7212, Hematology Department, Hôpital Saint-Louis, Assistance Publique–Hôpitaux de Paris, Université Paris Diderot, 1, Ave Claude Vellefaux, 75010 Paris, France; e-mail: raphael.itzykson@aphp.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal