Key Points
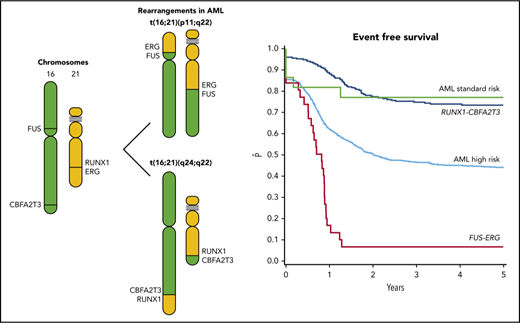
t(16;21) translocations in AML comprise t(16;21)(p11;q22) (FUS-ERG) as well as t(16;21)(q24;q22) (RUNX1-CBFA2T3).
Survival in pediatric AML with FUS-ERG is poor, whereas survival in RUNX1-CBFA2T3 is similar to other core-binding factor leukemias.
Abstract
To study the prognostic relevance of rare genetic aberrations in acute myeloid leukemia (AML), such as t(16;21), international collaboration is required. Two different types of t(16;21) translocations can be distinguished: t(16;21)(p11;q22), resulting in the FUS-ERG fusion gene; and t(16;21)(q24;q22), resulting in RUNX1-core binding factor (CBFA2T3). We collected data on clinical and biological characteristics of 54 pediatric AML cases with t(16;21) rearrangements from 14 international collaborative study groups participating in the international Berlin-Frankfurt-Münster (I-BFM) AML study group. The AML-BFM cohort diagnosed between 1997 and 2013 was used as a reference cohort. RUNX1-CBFA2T3 (n = 23) had significantly lower median white blood cell count (12.5 × 109/L, P = .03) compared with the reference cohort. FUS-ERG rearranged AML (n = 31) had no predominant French-American-British (FAB) type, whereas 76% of RUNX1-CBFA2T3 had an M1/M2 FAB type (M1, M2), significantly different from the reference cohort (P = .004). Four-year event-free survival (EFS) of patients with FUS-ERG was 7% (standard error [SE] = 5%), significantly lower compared with the reference cohort (51%, SE = 1%, P < .001). Four-year EFS of RUNX1-CBFA2T3 was 77% (SE = 8%, P = .06), significantly higher compared with the reference cohort. Cumulative incidence of relapse was 74% (SE = 8%) in FUS-ERG, 0% (SE = 0%) in RUNX1-CBFA2T3, compared with 32% (SE = 1%) in the reference cohort (P < .001). Multivariate analysis identified both FUS-ERG and RUNX1-CBFA2T3 as independent risk factors with hazard ratios of 1.9 (P < .0001) and 0.3 (P = .025), respectively. These results describe 2 clinically relevant distinct subtypes of pediatric AML. Similarly to other core-binding factor AMLs, patients with RUNX1-CBFA2T3 rearranged AML may benefit from stratification in the standard risk treatment, whereas patients with FUS-ERG rearranged AML should be considered high-risk.
Introduction
Despite intensive chemotherapy, current outcome of pediatric acute myeloid leukemia (AML) has reached a plateau,1 with 5-year event-free survival (EFS) rates ∼50% to 55% and 5-year overall survival (OS) rates reaching 70%.2-5 Apart from early clinical response, cytogenetic and molecular aberrations are the most reliable prognostic factors for survival.2,3,6 For future treatment stratification, identification of prognostic subgroups is important. Pediatric AML is a very heterogeneous disease; therefore, the prevalence of specific genetic subgroups can be too low to allow individual study groups to evaluate prognostic relevance and requires international collaboration.
Over the past few years, the international Berlin-Frankfurt-Münster (I-BFM) Study Group has described clinical and genetic characteristics of several rare pediatric AML subsets with the aim to provide clinicians with data for clinical decision making such as risk-group stratification.7-11 A pediatric AML group of interest is t(16;21), which, according to existing literature, is considered high risk. This is mainly based on case reports or small series in adult patients.12-16 Two different t(16;21) translocations resulting in different fusion transcripts can be distinguished. These include t(16;21)(p11;q22), resulting in the FUS-ERG fusion,12 and t(16;21)(q24;q22), resulting in the RUNX1-core binding factor (CBFA2T3) fusion.17
To date, 63 patients with FUS-ERG–rearranged AML have been described in patients from 1 to about 60 years of age,13 of which 19 were children.18 It has been reported that FUS-ERG AML presents with eosinophilia, micromegakaryocytes, and hemophagocytosis, and outcome has been described as poor.13,14,19 RUNX1-CBFA2T3 AML has been described in 24 patients, of which 5 were pediatric cases.18 This aberration is associated with the French-American-British (FAB) M2 phenotype and eosinophilia.16,20 In adults, RUNX1-CBFA2T3 has been associated with treatment-related AML and is reported to have a poor outcome.20 However, these data are mainly based on adult cases, and the prognostic impact of these rearrangements in pediatric AML is unknown.
To get more insight in the relevance of these somatic aberrations, we conducted a collaborative retrospective international study, gathering data from 14 study groups participating in the I-BFM Study Group. The aim of this study was to describe the biological and clinical characteristics and outcome of pediatric patients with t(16;21)-rearranged AML registered in I-BFM study group related data registries.
Patients and methods
Patients
To obtain the largest possible cohort of pediatric AML cases with t(16;21) aged 0 to 18 years of age, patient data were collected from 14 collaborative study groups and countries participating in the I-BFM Study Group (supplemental Table 1 , available on the Blood Web site). Patients diagnosed between 1 January 1995 and 1 January 2016 were included in the study. Patients were identified in the data registries of the study groups by reviewing karyotypes, fluorescence in situ hybridization (FISH) and/or polymerase chain reaction (PCR) analyses. Both t(16;21) translocations can be detected by conventional karyotyping. Associazione Italiana Ematologia Oncologia Pediatrica, BFM Austria, Japanese Children's Cancer Group, and the Belgian Society for Pediatric Hematology and Oncology confirmed the translocation with either FISH or PCR as standard of care. In 1 case from the Nordic Society of Paediatric Haematology and Oncology, the FUS-ERG fusion was detected through RNA sequencing.
For each case, a predefined set of data was collected and checked for consistency. This set included sex, age, date of diagnosis, white blood cell (WBC) count, extramedullary disease, relation with prior treatment or cancer, FAB morphology, eosinophilia and other morphological characteristics, presence of erythrophagocytosis, karyotype, treatment protocol, including data on allogeneic hematopoietic stem cell transplant (HSCT), response to therapy, including data on minimal residual disease detection (MRD) through flow cytometry and events, including relapse, resistant disease, occurrence of secondary malignancy, and death. Autologous HSCT was considered intensive chemotherapy.
Data of 1326 patients (excluding the t(16;21) cases) diagnosed between 1997 and 2013 were provided by the AML-BFM Study Group as a reference cohort. Patients with acute promyelocytic leukemia and Down syndrome were excluded. All patients in this cohort were classified as either standard risk (SR) patients, comprising inv(16) and t(16;21) or high-risk (HR) patients comprising other cytogenetic subtypes.
RNA sequencing data of 1035 patients with de novo AML with a median age of 9.9 (range, 0-29.6) from the Children’s Oncology Group (COG) AAML1031 trial (NCT01371981) were provided by COG for gene expression analysis21,22 and to identify the frequency of these aberrations. In this study, 93.9% of the patients were <18 years of age.
Central cytogenetic review
All karyotypes were centrally reviewed by 2 independent expert cytogeneticists, W.C. and M.P., following the International System for Human Cytogenetic Nomenclature (2016). Patients with inconclusive karyotypes were screened by reverse transcription (RT)-PCR (supplemental Methods). RNA was provided by the study groups.
Statistical analysis
Complete remission (CR) was defined as <5% blasts in the bone marrow, with regeneration of trilineage hematopoiesis and no leukemic cells in cerebrospinal fluid or elsewhere. If a patient did not obtain CR, treatment was considered a failure at day 0. MRD was measured by different study groups through flow cytometry after the first and second course of treatment. If >0.1% of the mononuclear cells were leukemic, MRD was considered positive. OS was calculated from the day of diagnosis until the date of last follow-up or death from any cause. EFS was measured from the day of diagnosis to the date of the first event or the date of last follow-up. Events considered in this analysis were resistant disease, relapse, occurrence of secondary malignancy, and death.
χ2 and Fisher's exact tests were used to compare clinical characteristics. OS and EFS analysis were estimated according to Kaplan-Meier and compared with log-rank test. Cumulative incidence of relapse (CIR) was calculated according to Kalbfleisch and Prentice and compared with the Gray test.23 The Mantel-Byar test was used to compare groups with and without allogeneic HSCT. The Cox proportional hazards model was used for multivariate analysis, considering age, WBC count at diagnosis, cytogenetic risk group (SR vs HR) as covariables, and HSCT as time-dependent variable. Analyses were performed with SPSS Statistics, version 21, and SAS 9.4. All tests were 2-tailed, and a P value < .05 was considered significant.
Gene expression analysis
Fusion transcripts from AML samples of patients included in the COG cohort were detected by RNA sequencing and validated by RT-PCR. Fractional counts were normalized to trimmed mean of m-values and counts per million mapped reads. The normalized counts were log2 transformed and filtered for genes with at least 1 count per million in 5% of samples. For hierarchical clustering, the relative level of expression per gene in each sample was determined by mean centering the expression values using the geometric mean. Pearson correlation coefficients were used as a measure of dissimilarity with the ward.D2 linkage algorithm implemented in the R statistical programming environment (R v.3.4.0). Sample correlations were derived from the expression of the 2412 differentially expressed genes, which are the union of those identified in FUS-ERG or RUNX1-CBFA2T3 vs other AML. Differential expression analysis was completed using Limma v3.32.5 R package. Genes with absolute log2 fold-change >1 and Benjamini-Hochberg adjusted P values < .05 were retained.
Results
Clinical features
A total of 55 patients with t(16;21) were identified, 32 patients with FUS-ERG–rearranged AML and 23 patients with RUNX1-CBFA2T3–rearranged AML. After central review of the karyotypes, there was 1 patient who did not meet our criteria because only 1 of 20 cells analyzed displayed a t(16;21)(p11;q22). It was not possible to confirm this fusion by RT-PCR; therefore, we excluded this patient from further analysis. The total cohort thus consisted of 54 patients with t(16;21), 31 with FUS-ERG–rearranged AML and 23 with RUNX1-CBFA2T3–rearranged AML. In the COG AAML1031 cohort, 5 FUS-ERG and 4 RUNX1-CBFA2T3 cases were identified, hence the frequency of FUS-ERG was 0.5% and 0.3% for RUNX1-CBFA2T3, compared with 0.3% and 0.1% in the BFM reference cohort (karyotype only), respectively. Clinical characteristics were compared with the AML-BFM Study Group reference cohort.
The patient characteristics of the t(16;21) subgroups are described in Table 1. No significant differences in sex and median age could be found when we compared patients in the FUS-ERG or RUNX1-CBFA2T3 groups with the reference cohort. No patients with FUS-ERG rearranged AML were <2 years of age and neither FUS-ERG nor RUNX1-CBFA2T3 rearrangements were found in infants <1 year of age (supplemental Figure 1). The median WBC of FUS-ERG (14.0 ×109/L) was not significantly different compared with the reference cohort (19.4 ×109/L, P = .66), whereas the WBC count of RUNX1-CBFA2T3 (12.5 ×109/L) was significantly lower (P = .030).
Clinical characteristics of pediatric cases with t(16;21)
| . | FUS-ERG . | RUNX1-CBFA2T3 . | I-BFM reference cohort . |
|---|---|---|---|
| N | 31 | 23 | 1326 |
| Median age (range), y | 8.5 (2.0-17.5) | 6.8 (1-17) | 8.7 (0-20.3) |
| Sex | |||
| % male | 61 | 46 | 51.6 |
| Median WBC (range), ×109/L | 14.0 (1-203) | 12.5 (0.01-185)* | 19.5 (0.01-190) |
| <20, n (%) | 17 (54.8) | 13 (61.9) | 670 (50.5) |
| 20-100, n (%) | 10 (32.3) | 7 (33.3) | 414 (31.2) |
| >100, n (%) | 5 (12.9) | 1 (4.8) | 242 (18.3) |
| FAB-type, n (%) | |||
| M0 | 3 (9.6) | 1 (4.3) | 46 (3.7) |
| M1 | 8 (25.8) | 3 (13.0) | 178 (14.5) |
| M2 | 8 (25.8) | 10 (43.5)* | 339 (27.6) |
| M4 | 6 (19.3) | 2 (8.7) | 293 (23.9) |
| M5 | 4 (12.9) | 1 (4.3) | 258 (21.6) |
| M6 | — | — | 29 (2.4) |
| M7 | 1 (3.2) | 1 (4.3) | 85 (6.9) |
| NOS | 1 (3.2) | 5 (21.7) | — |
| CNS involvement, n (%) | 6 (18.1) | 5 (22.7) | |
| Cytogenetics, n (%) | |||
| Sole abnormality | 12 (36.4) | 4 (21.1) | |
| Trisomy 8 | 6 (18.1) | 7 (36.8) | |
| Trisomy 10 | 4 (12.1) | — | |
| Complex karyotype | 10 (30.3) | 5 (26.3) | |
| Treatment, n (%) | |||
| CR obtained | 28 (87.5) | 22 (95.6) | |
| Refractory disease | 3 (9.4) | — | |
| HSCT in CR1 | 13 (40.6) | 8 (34.8) | |
| Survival (SE), % | |||
| 4-y EFS | 13 (9)† | 77 (9)† | 51 (1) |
| 4-y OS | 26 (8)† | 81 (8)† | 68 (1) |
| 4-y CIR | 69 (8)† | 0 (0)† | 32 (1) |
| . | FUS-ERG . | RUNX1-CBFA2T3 . | I-BFM reference cohort . |
|---|---|---|---|
| N | 31 | 23 | 1326 |
| Median age (range), y | 8.5 (2.0-17.5) | 6.8 (1-17) | 8.7 (0-20.3) |
| Sex | |||
| % male | 61 | 46 | 51.6 |
| Median WBC (range), ×109/L | 14.0 (1-203) | 12.5 (0.01-185)* | 19.5 (0.01-190) |
| <20, n (%) | 17 (54.8) | 13 (61.9) | 670 (50.5) |
| 20-100, n (%) | 10 (32.3) | 7 (33.3) | 414 (31.2) |
| >100, n (%) | 5 (12.9) | 1 (4.8) | 242 (18.3) |
| FAB-type, n (%) | |||
| M0 | 3 (9.6) | 1 (4.3) | 46 (3.7) |
| M1 | 8 (25.8) | 3 (13.0) | 178 (14.5) |
| M2 | 8 (25.8) | 10 (43.5)* | 339 (27.6) |
| M4 | 6 (19.3) | 2 (8.7) | 293 (23.9) |
| M5 | 4 (12.9) | 1 (4.3) | 258 (21.6) |
| M6 | — | — | 29 (2.4) |
| M7 | 1 (3.2) | 1 (4.3) | 85 (6.9) |
| NOS | 1 (3.2) | 5 (21.7) | — |
| CNS involvement, n (%) | 6 (18.1) | 5 (22.7) | |
| Cytogenetics, n (%) | |||
| Sole abnormality | 12 (36.4) | 4 (21.1) | |
| Trisomy 8 | 6 (18.1) | 7 (36.8) | |
| Trisomy 10 | 4 (12.1) | — | |
| Complex karyotype | 10 (30.3) | 5 (26.3) | |
| Treatment, n (%) | |||
| CR obtained | 28 (87.5) | 22 (95.6) | |
| Refractory disease | 3 (9.4) | — | |
| HSCT in CR1 | 13 (40.6) | 8 (34.8) | |
| Survival (SE), % | |||
| 4-y EFS | 13 (9)† | 77 (9)† | 51 (1) |
| 4-y OS | 26 (8)† | 81 (8)† | 68 (1) |
| 4-y CIR | 69 (8)† | 0 (0)† | 32 (1) |
CNS, central nervous system.
P < .05.
P < .001.
Patients with FUS-ERG had no predominant FAB type, whereas 76% of those with RUNX1-CBFA2T3 had an M1/M2 FAB type, compared with 42.1% in the reference cohort (P = .004). There was 1 patient with FUS-ERG with Auer rods. No other specific morphological features were reported.
Cytogenetics
At initial diagnosis, 9 of 31 (29.0%) patients with FUS-ERG had t(16;21)(p11;q22) as a sole aberration. Ten of 31 (32.3%) had a complex karyotype, defined as at least 3 chromosomal aberrations. In 3 cases, the t(16;21)(p11;q22) translocation was not detected by conventional karyotyping, but by PCR, FISH, or RNA sequencing (RNASeq). Recurrent additional cytogenetic aberrations were trisomy 8 (n = 6, 19.3%) and trisomy 10 (n = 4, 12.9%).
Complete karyotype data were available for 19 patients with RUNX1-CBFA2T3. In 3 (15.8%) of these patients, t(16;21)(q24;q22) was the sole abnormality. Five patients (26.3%) had a complex karyotype. Recurrent additional cytogenetic aberrations were trisomy 8 (n = 8, 42.1%) and deletion of the Y chromosome (n = 3, 15.7%). In 3 patients, cytogenetic analysis failed, but the RUNX1-CBFA2T3 translocation was detected by FISH or PCR. In 1 patient, the t(16;21)(q24;q22) translocation was not detected by conventional karyotyping, but was detected by PCR. A detailed list of the karyotypes is provided in supplemental Table 1.
Secondary AML
A total of 7 patients had secondary AML, 2 presenting with FUS-ERG rearrangements and 5 with RUNX1-CBFA2T3. Those with FUS-ERG–rearranged AML, presented with AML with myelodysplastic features and received chemotherapy before HSCT. One patient relapsed and died of disease; the second patient is still in remission after 7 years of follow-up.
Of the 5 patients diagnosed with RUNX1-CBFA2T3, 2 had Ewing sarcoma as primary malignancy. Four patients had been diagnosed with myelodysplastic syndrome (MDS) before AML development, 1 of whom also had previous Ewing sarcoma. The median time to development of AML after MDS diagnosis was 6.7 months (range, 4-28 months). None of the 4 patients with MDS was transplanted before AML diagnosis. In 1 patient, a 21q22 rearrangement was detected by FISH at time of MDS diagnosis. All patients received chemotherapy after being diagnosed with AML and 3 patients underwent HSCT in first CR.
Treatment and outcome
All patients in this cohort were treated with curative intent. CR was achieved in 87.1% of the patients with FUS-ERG and 82.6% of those with RUNX1-CBFA2T3. Two patients with RUNX1-CBFA2T3 AML suffered an early death before reaching CR.
In total, 23 patients had data available on MRD measured by flow cytometry, 12 with FUS-ERG and 11 with RUNX1-CBFA2T3 (supplemental Table 2). In the FUS-ERG group, 5 of 12 were MRD negative after the first course of chemotherapy (MRD1), and an additional 2 were MRD negative after the second course of treatment (MRD2). No difference in the incidence of relapse could be observed between the MRD positive and negative patients, as 10 of 12 patients experienced a relapse. One of the patients that did not suffer from relapse was MRD2 positive, but had a short follow-up time of only 2 months, the other patient was MRD1 negative and received an HSCT in first CR (CR1). In the RUNX1-CBFA2T3 group, 8/10 patients were MRD negative after the first course of chemotherapy. After the second course, all patients were MRD negative and none relapsed.
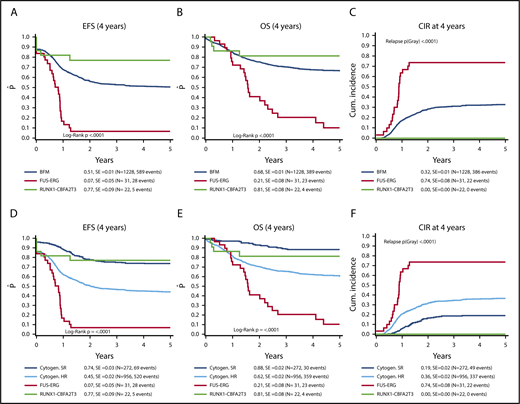
Four-year EFS, OS, and CIR for the reference cohort were 51% (standard error [SE] = 1%), 68% (SE = 1%), and 32% (SE = 1%), respectively. The SR group in the reference cohort had a 4-year EFS, OS, and CIR of 74% (SE = 3%), 88% (SE = 2%), and 19% (SE = 2%), respectively. For the HR group, EFS was 45% (SE = 2%), OS 62% (SE = 2%), and CIR 36% (SE = 2%).
Median follow-up for survivors in the t(16;21) cohort was 1.6 years for those with FUS-ERG and 5.0 years for RUNX1-CBFA2T3. Patients with FUS-ERG had a 4-year EFS of 7% (SE = 5%, P < .0001), an OS of 21% (SE = 8%, P < .0001), and a CIR of 74% (SE = 8%, P < .0001). The median time to relapse was 10.2 months. Almost all relapses occurred early within the first year after start of treatment (18/21).
For RUNX1-CBFA2T3, 4-year EFS was 77% (SE = 9%, P = .06), OS was 81% (SE = 8%, P = .34), and CIR was 0%. EFS rates of the SR patients was 74% (SE = 3%), Thus, the patients with RUNX1-CBFA2T3 had a similar outcome as BFM SR patients (Figure 1).
Survival of FUS–ERG and CBFA2T3/RUNX1 AML compared with a pediatric AML reference cohort. (A-C) Survival curves of patients with FUS-ERG–rearranged AML and CBFA2T3/RUNX1–rearranged AML compared with the BFM reference cohort. (D-F) The reference cohort is split up according to high and standard risk.
Survival of FUS–ERG and CBFA2T3/RUNX1 AML compared with a pediatric AML reference cohort. (A-C) Survival curves of patients with FUS-ERG–rearranged AML and CBFA2T3/RUNX1–rearranged AML compared with the BFM reference cohort. (D-F) The reference cohort is split up according to high and standard risk.
A total of 30 patients underwent an allogeneic HSCT: 22/31 (71.0%) patients with FUS-ERG and 8/23 (34.8%) with RUNX1-CBFA2T3. Of the 22 FUS-ERG patients who received an HSCT, 14 (42.2%) received the HSCT in CR1. The 4-year EFS for transplanted patients with FUS-ERG was 15% (SE = 15%) compared with 0% (SE = 0%) for patients receiving chemotherapy only (P = .50).
Multivariate analysis of EFS and OS revealed that FUS-ERG was an independent predictor of poor outcome for both EFS and OS (hazard ratio [HR], 2.9, P < .0001; and HR, 2.61, P < .0001, respectively), whereas RUNX1-CBFA2T3 was a predictor of favorable outcome for EFS but not OS (HR, 0.33, P = .02; and HR, 0.42, P = .14, respectively). The HR for RUNX1-CBFA2T3 were comparable to the SR group of the reference cohort, with HR for EFS of 0.36 (P < .001) and OS of 0.25 (P < .001). In addition, WBC >100 ×109/L was an independent predictor of poor outcome for both EFS and OS (HR, 1.4, P = .0005; and HR, 1.27, P = .046, respectively) (Table 2). All other covariates, including HSCT, were not significantly associated with outcome.
Multivariate analysis of survival parameters of t(16;21)-rearranged AML
| . | pEFS . | pOS . | ||||
|---|---|---|---|---|---|---|
| . | HR . | 95% CI . | P . | HR . | 95% CI . | P . |
| FUS-ERG | 2.85 | 1.93-4.21 | <.001 | 2.61 | 1.71-4.00 | <.001 |
| WBC >10 × 109/L | 1.40 | 1.15-1.70 | <.001 | 1.27 | 1.00-1.60 | .046 |
| Age >10 y | 1.14 | 0.98-1.34 | .087 | 1.38 | 1.14-1.67 | .001 |
| Time to HSCT | 0.84 | 0.63-1.12 | .23 | 0.97 | 0.70-1.33 | .834 |
| Cytogenetic SR group | 0.36 | 0.28-0.47 | <.001 | 0.25 | 0.17-0.36 | <.001 |
| RUNX1-CBFA2T3 | 0.32 | 0.12-0.87 | .025 | 0.42 | 0.14-1.33 | .140 |
| . | pEFS . | pOS . | ||||
|---|---|---|---|---|---|---|
| . | HR . | 95% CI . | P . | HR . | 95% CI . | P . |
| FUS-ERG | 2.85 | 1.93-4.21 | <.001 | 2.61 | 1.71-4.00 | <.001 |
| WBC >10 × 109/L | 1.40 | 1.15-1.70 | <.001 | 1.27 | 1.00-1.60 | .046 |
| Age >10 y | 1.14 | 0.98-1.34 | .087 | 1.38 | 1.14-1.67 | .001 |
| Time to HSCT | 0.84 | 0.63-1.12 | .23 | 0.97 | 0.70-1.33 | .834 |
| Cytogenetic SR group | 0.36 | 0.28-0.47 | <.001 | 0.25 | 0.17-0.36 | <.001 |
| RUNX1-CBFA2T3 | 0.32 | 0.12-0.87 | .025 | 0.42 | 0.14-1.33 | .140 |
CI, confidence interval; pEFS, probability of event free survival; pOS, probability of overall survival.
Gene expression profiling
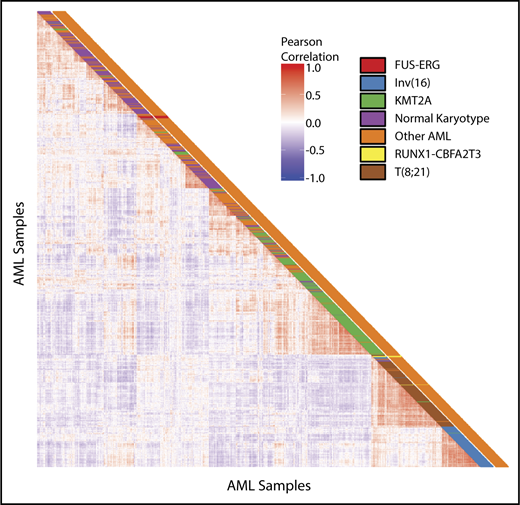
Unsupervised hierarchical clustering revealed that RUNX1-CBFA2T3 and FUS-ERG cluster separately (Figure 2). FUS-ERG also clusters separately from other cytogenetic subgroups, such as KMT2A rearrangements, t(16;21)(q22;q22) and inv(16)(p13;q22), whereas RUNX1-CBFA2T3 cases cluster in close proximity to t(16;21)(q22;q22) cases. Comparing gene expression of FUS-ERG with the remainder of the pediatric AML cohort revealed 1314 differentially expressed genes with an adjusted P value < .05. Among these, 428 genes were upregulated. In hematopoiesis, ERG is known to upregulate GATA2 and RUNX1; however, there was no differential expression of these genes in FUS-ERG–rearranged AML.24 The top 100 most differentially expressed genes of FUS-ERG are provided in supplemental Table 3.
Unsupervised clustering analysis. Pairwise sample correlations of 1037 samples of pediatric AML. The cells in the visualization are colored by Pearson’s correlation coefficient values. Cytogenetic subgroups are depicted in the first column. Presence of FUS-ERG or RUNX1-CBFA2T3 is depicted in the second column.
Unsupervised clustering analysis. Pairwise sample correlations of 1037 samples of pediatric AML. The cells in the visualization are colored by Pearson’s correlation coefficient values. Cytogenetic subgroups are depicted in the first column. Presence of FUS-ERG or RUNX1-CBFA2T3 is depicted in the second column.
Comparing RUNX1-CBFA2T3 gene expression to the remainder of the pediatric AML cohort revealed 119 differentially expressed genes in RUNX1-CBFA2T3, of which 76 genes were upregulated (supplemental Table 4). Because RUNX1-CBFA2T3 clustered in close proximity to t(16;21)(q22;q22) leading to the RUNX1-RUNX1T1 fusion, we analyzed whether these 2 groups share a gene expression profile. We detected differentially expressed genes in RUNX1-CBFA2T3 using the pediatric AML cohort, excluding the t(16;21) cases. Of the 2786 differentially expressed genes (2507 in t(16;21) and 279 in RUNX1-CBFA2T3), 187 were shared between the 2 groups. A total of 112 genes were upregulated in both groups, 70 were downregulated in both groups, and 5 genes were upregulated in RUNX1-CBFA2T3 and downregulated in t(16;21) (supplemental Table 5). Well-known targets of t(16;21) such as POU4F1, TRH, PSD3, MEIS1, and LAT2 were differentially expressed in RUNX1-CBFA2T3 AML.24,25
Discussion
Within the framework of this international collaboration, we studied the clinical and biological features of 2 translocations involving chromosome 16 and 21. We identified 2 clinically relevant, distinct subtypes of pediatric AML patients with different t16,21 rearrangements: FUS-ERG had poor outcome, whereas RUNX1-CBFA2T3 had favorable outcome. Our data suggest that patients with RUNX1-CBFA2T3–rearranged AML might benefit from treatment protocol for standard risk AML without stem cell transplantation, whereas those with FUS-ERG–rearranged AML seem to require high-risk therapy, including HSCT, or even experimental therapy.
Although 87.1% of the patients with FUS-ERG–rearranged AML reached morphological complete remission, the 4-year CIR was 74% and most relapses occurred within the first year after diagnosis. Currently, early response to therapy is increasingly used for risk-group stratification of therapy in AML.3,26,27 The detection of MRD through flow cytometry can provide a more accurate measure of therapy response; however, the additional benefit of MRD measurement is inconsistent between AML subtypes and studies and depends on the sensitivity of the applied technique.3,28-30 In our cohort, MRD data were reported in about one-half of the t(16;21) rearranged cases. Of those, almost 40% of the patients with FUS-ERG–rearranged AML, who had MRD data determined by flow cytometry, were MRD negative after the first course of treatment, and about one-half of the patients (7 of 12) after the second course of treatment. However, EFS was very low in both MRD-negative and MRD-positive patients and no significant difference in EFS between the 2 groups could be found. Despite numbers being small, this may suggest that MRD does not adequately predict relapse in this cytogenetic group. A reason for this might be that FUS-ERG–rearranged AML could be a leukemic stem cell–driven disease that is not successfully eradicated with current treatment protocols.
Currently, in high-risk AML subgroups with an EFS <30%, HSCT in CR1 is considered by some collaborative groups.31 Because EFS of FUS-ERG–rearranged AML is 7%, patients with this rearrangement should be considered for HSCT in CR1 despite the benefit from HSCT seeming limited in our analysis. Therefore patients with FUS-ERG–rearranged AML urgently require novel forms of therapy.
In contrast to FUS-ERG, no relapses were observed in RUNX1-CBFA2T3–rearranged pediatric AML. The only events that occurred were toxic events: 5 patients died from infections. Surprisingly, even patients with secondary AML did not suffer from relapse. To date, 24 RUNX1-CBFA2T3 cases have been reported, of whom only 5 were pediatric.15,16,20,32-45 In the literature, these patients were considered to be high risk. However, when we single out the pediatric cases, 2 died from an infection and 3 were in CR for at least 1 year. This seems to be consistent with our results, further supporting that RUNX1-CBFA2T3–rearranged AML should be stratified as SR. This suggests that outcome for RUNX1-CBFA2T3–rearranged AML differs between pediatric and adult patients. This may be related to the fact that this leukemia occurred often as a second malignancy in the adult cases, and perhaps was not treated with curative intent. Moreover, in general outcome in pediatric AML is better than in adults, which may reflect issues such as organ toxicity and tolerability for chemotherapy.
Of note, most of the patients in our pediatric cohort had de novo AML; however, 2 patients were diagnosed with Ewing sarcoma before AML development, and 3 additional patients had MDS before AML. The association between Ewing sarcoma and MDS/AML in pediatrics has been described previously, but not in combination with RUNX1-CBFA2T3.46 Surprisingly, even though secondary AML is known to be a poor prognostic risk factor, there was no difference in outcome between patients with de novo or secondary AML, with no relapses in either group. According to the World Health Organization classification of myeloid neoplasms and acute leukemia, patients with a t(16;21)(q22;q22) rearrangement and <20% blasts in the bone marrow should be classified as having AML and not MDS.47,48 This classification strategy could also be applied to RUNX1-CBFA2T3 because the “MDS cases” (<20% blasts in the diagnostic marrow) were cured without SCT.
This study showed that the 2 fusions give rise to different gene expression signatures. The gene expression profile of FUS-ERG–rearranged cases did not reveal any similarities with other cytogenetic subgroups. The t(16;21)(p11;q22) gives rise to a fusion of the N-terminal part of FUS, containing the transactivation domain of FUS and the C-terminal of ERG, containing the ETS DNA-binding site of ERG.49 FUS-ERG is known to bind at genomic regions that are also bound by other transcription factors associated with stem cell programs such as RUNX1, FLI1, and GATA2.50 However, we found no differential expression of these associated genes when we compared gene expression of FUS-ERG rearranged AML with the other AML cases. Furthermore, Sotoca et al found that the nuclear receptor heterodimer RARA:RXR binds to FUS-ERG–occupied genomic regions, suggesting possible modulation of the retinoic acid response in FUS-ERG–rearranged AML.50 This might make FUS-ERG–rearranged AML a potential target for treatment with all-trans retinoic acid.
On the other hand, RUNX1-CBFA2T3 can be classified as a core binding factor AML. The CBF is a heterodimer, consisting of RUNX1, RUNX2, or RUNX3 and CBFB. The CBF attaches to DNA and activates genes involved in hematopoietic development.51 In leukemia, recurrent fusions of these genes have been described.52 When CBFB or RUNX1 is part of a fusion gene, the function of the protein changes and, instead of activating the genes, it will repress them. In AML, 2 recurrent aberrations are currently classified as CBF AML: inv(16)(p11;q32)/t(16;16)(p11;q42) and t(8;21)(q22;q22), resulting in CBFB-MYH11 and RUNX1-RUNX1T1, respectively.11 There are striking similarities between RUNX1-CBFA2T3 and t(8;21) (also known as RUNX1-RUNX1T1) because both are mainly found in FAB M1/M2 AML and have a favorable outcome.6,53 Cytogenetically, both show recurrent loss of a sex chromosome, which is rare in other types of pediatric AML.11 Both fusions also have similarities in biology. They do not only share the RUNX1 gene, but CBFA2T3 and RUNX1T1 are paralogs and share 92% of their protein sequence.54 Furthermore, this study showed that RUNX1-CBFA2T3 and t(8;21) cluster in close proximity of each other and that RUNX1-CBFA2T3 and t(8;21) share 187 differentially expressed genes, among which target genes such as POU4F1, TRH, and MEIS1. These results are in line with the results Lavallee et al obtained when comparing gene expression profiles of t(8;21) and inv(16).55 These findings provide additional support that RUNX1–CBFA2T3 belongs to the CBF AML subgroup, similar to t(8;21).
In this study, we relied on cytogenetic analysis to detect t(16;21). Because cytogenetic analysis fails in about 10% of cases,56 there is risk of selection bias. Furthermore, as cytogenetic analysis can only detect large rearrangements; more subtle, complex rearrangements could be missed. More sensitive analysis such as FISH, RT-PCR, and RNASeq are more reliable to detect these rearrangements. RUNX1-CBFA2T3, for instance, can be detected through RUNX1-split FISH, which is usually performed as standard of care to detect RUNX1-RUNX1T1 rearrangements. The difference in incidence between the BFM and COG AAML1031 cohort seems to confirm that RNASeq might be slightly more reliable. However, because RNASeq needs high-quality RNA of samples with a high purity of blasts, not all patients can be analyzed by this method. We were not informed on whether these cohorts were truly population-based.
In conclusion, this international collaborative study describes 2 clinically relevant distinct subtypes of pediatric AML. Although numbers are small, reflecting the rarity of the diseases, FUS-ERG represents an extremely poor prognostic subgroup, whereas RUNX1-CBFA2T3 has a favorable outcome. Patients with RUNX1-CBFA2T3–rearranged AML might benefit from risk stratification to standard intensive therapy, as for CBF AML, whereas FUS-ERG–rearranged AML patients should be considered high risk and offered HSCT in CR1, even though the effect of HSCT in FUS-ERG–rearranged AML may be limited in this retrospective series. Although we unfortunately have no data on surface marker expression in these specific cases, more experimental therapy such as flotetuzumab or CAR T cells may offer opportunities to circumvent chemotherapy drug resistance and need to be explored in these high-risk FUS-ERG patients, certainly after relapse.57-60
Presented as an oral abstract at the 59th annual meeting of the American Society of Hematology, Atlanta, GA, 9 December 2017.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.N., M.Z., and M.M.v.d.H.-E. designed the study; M.Z., D.R., M.P., J.S., R.E.R., T.A.A., B.H., D.T., F.L., T.A.G., S.R., E.S., D.K.C., M.D., J.S., J.A., N.A.-C., M.C., B.D.M., H.H., and S.M. contributed materials and clinical data; M.Z., M.P., W.C., J.S., R.E.R., S.N., C.M.Z., and M.M.v.d.H.-E. analyzed data; S.N., M.Z., J.S., and R.E.R. performed statistical analysis; S.N., C.M.Z., and M.M.v.d.H.-E. wrote the paper; C.M.Z. and M.M.v.d.H.-E. supervised the study; and all coauthors performed critical review of the manuscript and gave their final approval.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the Berlin-Frankfurt-Münster Study Group appears in the online appendix.
Correspondence: C. Michel Zwaan, Princess Máxima Center, Heidelberglaan 25, 3584 CS Utrecht, The Netherlands; e-mail: c.m.zwaan@prinsesmaximacentrum.nl.