TO THE EDITOR:
Myeloid (or granulocytic) sarcoma (MS), that is, an extramedullary tumor consisting of myeloid blastic cells, is a unique clinical presentation of any type of acute myeloid leukemia (AML), according to the World Health Organization.1 It could present (1) de novo (isolated MS), (2) with concomitant AML, (3) as a relapse of AML, or (4) as a progression from myelodysplasia or myeloproliferative neoplasia.1 MS is reported in almost one fourth of patients with AML and is mostly localized in skin, lymph nodes, bone, testis, gut, and the central nervous system.2,3 However, isolated MS at presentation is rare, with an estimated incidence of 2 per million in adults.4 In the Swedish AML registry,5 less than 1% of AML cases present with extramedullary disease and less than 5% with marrow blasts.6 Most4 but not all7 of these patients subsequently develop overt AML. Outcome for patients with MS seems to be similar3,8 or worse9,10 than AML without MS; however, isolated MS may have a better outcome.11
The aberrant tropism of MS cells likely results from a combination of specific genetic changes, surface marker expression,3 and microenvironmental cues at the tumor site. Cytogenetic analysis of MS is rarely performed, but complex karyotypes and typical recurrent abnormalities are reported.7,8,11,12 Next-generation sequencing, using restricted panels of AML and myelodysplastic syndrome–associated genes, in isolated MS tissues from 613 and 514 cases, identified mutations in FLT3, NPM1, DNMT3A, RUNX1, TP53, IDH2, NRAS, EZH2, ASXL1, TET2, and WT1.
The concept of clonal hematopoiesis, with sequentially developing mutations in hematopoietic stem cells, most frequently DNMT3A, ASXL1, and TET2, predisposing to leukemia, is now established.15 Whether preleukemic clones also would predispose for isolated MS has not been addressed previously.
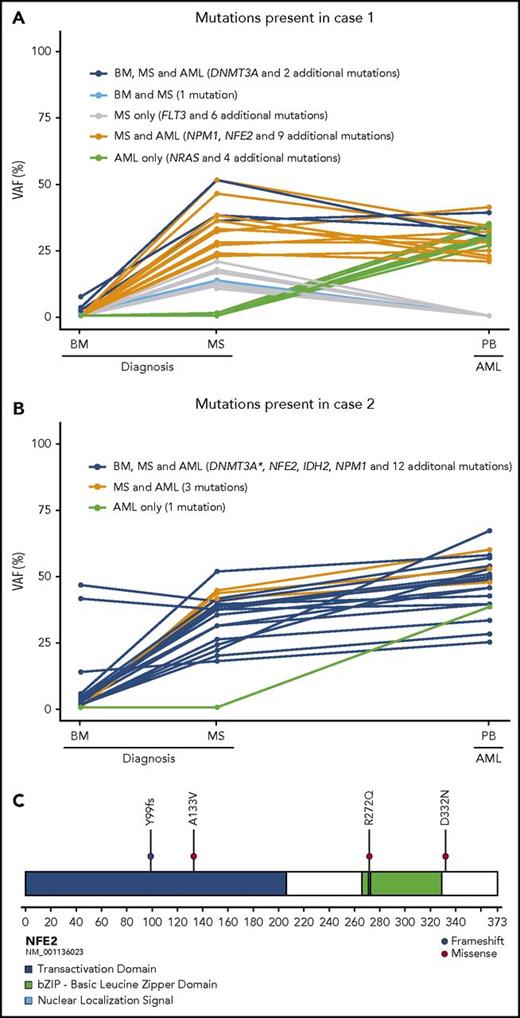
Here, we first performed whole-exome sequencing (WES) to gain insights into the molecular mechanisms underlying the development of isolated MS. Two females (cases 1 and 2) presented with vaginal bleeding, and diagnostic workup revealed isolated uterine myeloid sarcoma (Table 1). Following informed consent, we prospectively performed WES on MS biopsies, normal bone marrow aspirates at diagnosis, and peripheral blood at the development of AML. As reference, germline DNA was extracted from cultured fibroblast (supplemental Methods, available on the Blood Web site). WES of the MSs identified 21 and 20 somatic coding mutations, respectively, including mutations in recurrent AML driver genes: DNMT3A and NPM1 in both, a less common FLT3 point mutation in case 1, and IDH2 in case 216-18 (Figure 1A-B, Table 1, and supplemental Tables 1-2). Interestingly, both MS samples also harbored mutations in NFE2 (nuclear factor, erythroid 2), whereas the other variants found were different between the 2 cases (Figure 1C).
Clinical, cytogenetic, phenotypic, and molecular findings on 6 patients with isolated MS without AML at or before MS presentation
| Case . | Sex/age, y . | Medical history . | MS site and size . | MS . | BM at MS diagnosis . | Therapy . | Subsequent clinical history . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytogenetics and phenotype . | Variants detected by WES . | Cytogenetics and hematology . | Variants detected by WES . | |||||||
| 1 | F/59 | BMI 31 Concomitant serous adenocarcinoma of the ovary. | Vagina, cervix 5 × 4 cm | Karyotype: 46,XX IHC: CD34−, CD117+, CD56−, muramidase-, MPO−, CD61−, glycoA− | Variants detected in AML-associated genes: DNMT3A (p.V704fs) FLT3 (p.Y572C) NPM1 (p.W288fs) | Karyotype: 46,XX BM morphology: normal FCM: normal Normal CBC | Variants detected in AML-associated genes: DNMT3A (p.V704fs) | Daunorubicin + cytarabin. Surgery. Carboplatin + paclitaxel for ovarian cancer | Development of overt AML in the BM 28 mo after diagnosis. Dead | |
| Novel variants detected in: APOB CDH23 CMYA5 ETFDH FAM227B GIGYF2 LOC100128326 | MGA NFE2 (p.R272Q) PCDHGA1 PNPT1 PTEN RPL5 SETD2 STARD8 TTN | |||||||||
| 2 | F/59 | BMI 46. Subtotal hystero-oophorectomy with myoma and ovarian cysts. Ulcerative colitis | Cervix, uteri 9 × 7 cm | Karyotype: 46, XX IHC: CD34−, CD117+, muramidase−, MPO−, CD61−, glycoA− | Variants detected in AML-associated genes: DNMT3A (p.R320) IDH2 (p.R140Q) NPM1 (p.W288fs) | Karyotype: 46,XX BM morphology: normal FCM: normal Normal CBC | Variants detected in AML-associated genes: DNMT3A (p.R320*) Novel variants detected in: NFE2 (p.Y99fs) ODF1 TRAFD1 | Daunorubicin + cytarabin. | Development of overtAML in the BM 8 mo after diagnosis. Dead | |
| Novel variants detected in: BPIFB3 CKMT1 CLASP1 DLEC1 KCNB1 LPCAT2 MAMSTR | MASP2 NFE2 (p.Y99fs) ODF1 PCLO PRAMEF1 SEMA5A SOS1 TMEM44 TRAFD1 | |||||||||
| 4 | F/80 | MDS with 5q− since 2 y | Terminal ileum, 3 cm | Karyotype: NA IHC: CD34+, CD117+, muramidase+, MPO+ | Novel variants detected* in: NFE2 (p.D332N) | Karyotype: NA BM morphology: MDS, 4% blasts. Hb 111, WBC 2.3, ANC 0.4, Plt 129 | NA | Surgery due to ileus No further treatment | Died after 4 mo | |
| 5 | M/36 | Diabetes | Sub-mandibular tumor, 3 cm. | Karyotype: NA IHC: CD34−, CD117−, CD56+, muramidase−, MPO− | Novel variants detected* in: No variant detected in NFE2 | Karyotype: NA BM morphology: MDS EB1. Hb 137, WBC 10, ANC 9.4, Plt 344. | NA | AML-type chemotherapy | Allogeneic SCT. Alive | |
| 16 | F/69 | None | Vagina, cervix, 4 cm, with hydronephrosis | Karyotype: NA IHC: CD34−, CD117+, muramidase+, MPO− | Novel variants detected* in: No variant detected in NFE2 | Karyotype: NA BM morphology: MDS EB2. 11% blasts, abnormal monocytes. Hb 100, WBC 4.9, ANC 3.0, Plt 248 | NA | Nephrostomy. AML-type chemotherapy. CR | Local relapse at 10 mo. BM normal. Chemorefractory. Dead | |
| 17 | F/70 | PV since 12 y, P32 thrice, MF 1 y | Tonsil 3 × 2 cm + neck nodes | Karyotype: NA IHC: CD117+, MPO+ | Novel variants detected* in: NFE2 (p.A133V) | Karyotype: NA BM morphology: post PV-MF, cellularity 80%, no blast increase. Hb 120, WBC 3.2, ANC 2.0, Plt 81 | NA | Surgery | Two months later: WBC 29, ANC 9.1, monocytes 12.3, Plt 30. Dead | |
| Case . | Sex/age, y . | Medical history . | MS site and size . | MS . | BM at MS diagnosis . | Therapy . | Subsequent clinical history . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytogenetics and phenotype . | Variants detected by WES . | Cytogenetics and hematology . | Variants detected by WES . | |||||||
| 1 | F/59 | BMI 31 Concomitant serous adenocarcinoma of the ovary. | Vagina, cervix 5 × 4 cm | Karyotype: 46,XX IHC: CD34−, CD117+, CD56−, muramidase-, MPO−, CD61−, glycoA− | Variants detected in AML-associated genes: DNMT3A (p.V704fs) FLT3 (p.Y572C) NPM1 (p.W288fs) | Karyotype: 46,XX BM morphology: normal FCM: normal Normal CBC | Variants detected in AML-associated genes: DNMT3A (p.V704fs) | Daunorubicin + cytarabin. Surgery. Carboplatin + paclitaxel for ovarian cancer | Development of overt AML in the BM 28 mo after diagnosis. Dead | |
| Novel variants detected in: APOB CDH23 CMYA5 ETFDH FAM227B GIGYF2 LOC100128326 | MGA NFE2 (p.R272Q) PCDHGA1 PNPT1 PTEN RPL5 SETD2 STARD8 TTN | |||||||||
| 2 | F/59 | BMI 46. Subtotal hystero-oophorectomy with myoma and ovarian cysts. Ulcerative colitis | Cervix, uteri 9 × 7 cm | Karyotype: 46, XX IHC: CD34−, CD117+, muramidase−, MPO−, CD61−, glycoA− | Variants detected in AML-associated genes: DNMT3A (p.R320) IDH2 (p.R140Q) NPM1 (p.W288fs) | Karyotype: 46,XX BM morphology: normal FCM: normal Normal CBC | Variants detected in AML-associated genes: DNMT3A (p.R320*) Novel variants detected in: NFE2 (p.Y99fs) ODF1 TRAFD1 | Daunorubicin + cytarabin. | Development of overtAML in the BM 8 mo after diagnosis. Dead | |
| Novel variants detected in: BPIFB3 CKMT1 CLASP1 DLEC1 KCNB1 LPCAT2 MAMSTR | MASP2 NFE2 (p.Y99fs) ODF1 PCLO PRAMEF1 SEMA5A SOS1 TMEM44 TRAFD1 | |||||||||
| 4 | F/80 | MDS with 5q− since 2 y | Terminal ileum, 3 cm | Karyotype: NA IHC: CD34+, CD117+, muramidase+, MPO+ | Novel variants detected* in: NFE2 (p.D332N) | Karyotype: NA BM morphology: MDS, 4% blasts. Hb 111, WBC 2.3, ANC 0.4, Plt 129 | NA | Surgery due to ileus No further treatment | Died after 4 mo | |
| 5 | M/36 | Diabetes | Sub-mandibular tumor, 3 cm. | Karyotype: NA IHC: CD34−, CD117−, CD56+, muramidase−, MPO− | Novel variants detected* in: No variant detected in NFE2 | Karyotype: NA BM morphology: MDS EB1. Hb 137, WBC 10, ANC 9.4, Plt 344. | NA | AML-type chemotherapy | Allogeneic SCT. Alive | |
| 16 | F/69 | None | Vagina, cervix, 4 cm, with hydronephrosis | Karyotype: NA IHC: CD34−, CD117+, muramidase+, MPO− | Novel variants detected* in: No variant detected in NFE2 | Karyotype: NA BM morphology: MDS EB2. 11% blasts, abnormal monocytes. Hb 100, WBC 4.9, ANC 3.0, Plt 248 | NA | Nephrostomy. AML-type chemotherapy. CR | Local relapse at 10 mo. BM normal. Chemorefractory. Dead | |
| 17 | F/70 | PV since 12 y, P32 thrice, MF 1 y | Tonsil 3 × 2 cm + neck nodes | Karyotype: NA IHC: CD117+, MPO+ | Novel variants detected* in: NFE2 (p.A133V) | Karyotype: NA BM morphology: post PV-MF, cellularity 80%, no blast increase. Hb 120, WBC 3.2, ANC 2.0, Plt 81 | NA | Surgery | Two months later: WBC 29, ANC 9.1, monocytes 12.3, Plt 30. Dead | |
Variants with a variant allele frequency (VAF) >10% were first identified in the myelosarcoma, and subsequently their VAFs were determined in the corresponding bone marrow (BM); only variants with a VAF >5% are shown.
ANC, absolute neutrophil count 109/L; BM, bone marrow; BMI, body mass index; CBC, complete blood count; CD, cluster of differentiation; CR, complete remission; EB, excess blasts; F, female; FCM, flow cytometry; GlycoA, glycophorin A; Hb, hemoglobin, g/L; IHC, immunohistochemistry; M, male; MDS, myelodysplastic syndrome; MF, myelofibrosis; MPO, myeloperoxidase; NA, not available; Plt, platelet count 109/L; PV, polycythemia vera; P32, phosphorus-32; SCT, stem cell transplant; WBC, white blood cell count, 109/L.
No whole exome sequencing was performed, only targeted analysis of NFE2.
An overview of the mutations identified in the 2 MS patients. (A) In case 1 a small preleukemic clone was identified in the morphologically normal BM at MS diagnosis (shown in dark and light blue). In the MS, further mutations were gained (shown in gray and orange), and later in the developing AML, additional mutations had accumulated (shown in green), whereas some mutations were lost (shown in gray and light blue). (B) In case 2, the preleukemic clone comprised the majority of the BM cells at MS diagnosis (shown in dark blue). The accumulating mutations were identical in the MS and the subsequently developing AML (shown in orange), apart from 1 additional mutation (shown in green). (C) A schematic illustration of NF2E depicting mutations identified in the 4 patients with isolated MS. All variants were primarily identified as mutations (VAF above 10%) in the MS and in the developed AML and subsequently investigated in all corresponding samples. All variants less than 5% are regarded as uncertain because of the low number of reads supporting the variant allele. BM, bone marrow; PB, peripheral blood; VAF, variant allele frequency. *Two mutations were present in DNMT3A in case 2.
An overview of the mutations identified in the 2 MS patients. (A) In case 1 a small preleukemic clone was identified in the morphologically normal BM at MS diagnosis (shown in dark and light blue). In the MS, further mutations were gained (shown in gray and orange), and later in the developing AML, additional mutations had accumulated (shown in green), whereas some mutations were lost (shown in gray and light blue). (B) In case 2, the preleukemic clone comprised the majority of the BM cells at MS diagnosis (shown in dark blue). The accumulating mutations were identical in the MS and the subsequently developing AML (shown in orange), apart from 1 additional mutation (shown in green). (C) A schematic illustration of NF2E depicting mutations identified in the 4 patients with isolated MS. All variants were primarily identified as mutations (VAF above 10%) in the MS and in the developed AML and subsequently investigated in all corresponding samples. All variants less than 5% are regarded as uncertain because of the low number of reads supporting the variant allele. BM, bone marrow; PB, peripheral blood; VAF, variant allele frequency. *Two mutations were present in DNMT3A in case 2.
Mutations in NFE2 have previously only been described in 3 of 3294 investigated cases of hematopoietic malignancies, as are reported in the COSMIC database.16 However, rare insertion and deletion mutations in NFE2 were found in 8 of 456 (2%) myeloproliferative neoplasms (MPN),19 mainly resulting in premature stop codons. In case 1, we observed a somatically acquired missense mutation (p.R272Q) affecting amino acids (aa) in the nuclear localization signal, critical for transcriptional activation of the NFE2 protein.20 In case 2, a frame-shift mutation (p.Y99fs) resulted in a stop codon at aa99 (Table 1, Figure 1C). Because of the unexpected finding of recurrent NFE2 mutations, we next performed targeted NFE2-mutation analysis in a retrospective cohort of 16 additional formalin-fixed paraffin-embedded MS tissue samples (supplemental Methods). Twelve cases had concomitant or previous AML, but no variants in NFE2 were detected (supplemental Table 3). However, among 4 isolated MS samples, 2 displayed missense NFE2 mutations (case 4 a p.D332N and case 17 a p.A133V; Table 1, Figure 1C), none of them previously reported as normal variants, strongly suggesting that they were disease associated. Both of these cases had a previous history of myeloid neoplasia, without previous or concurrent AML transformation (Table 1). Hence, of the 6 investigated cases of MS without previous or concurrent AML in the bone marrow (BM), 4 (67%) harbored mutations in NFE2.
NFE2 is a transcription factor expressed in hematopoietic cells, mainly in erythroid, megakaryocytic, and mast cells.21 Mice deficient in Nfe2 display disturbed megakaryocyte formation and mild erythroid effects.21 NFE2 has been shown to be overexpressed in MPN patients, and acquired mutations result in enhanced activity of wild-type NFE2.19 Transgenic mice overexpressing wild-type NFE2 or retroviral overexpression of mutant NFE2 recapitulate features observed in MPN, including expansion of the progenitor and stem cell compartments and a propensity to develop AML.19,21 Mutated NFE2 could promote the development of MS, through altered homing of the leukemic cells or providing a selective advantage to cells growing as a tumor mass at distant sites, although this remains to be experimentally addressed.
Notably, WES of the morphologically normal BM in both cases 1 and 2 identified DNMT3A mutations present in the MS. In case 1, the DNMT3A mutation marked a small subclone (variant allele frequency [VAF] 7%) accompanied by 3 passenger mutations at even lower frequencies (Figure 1A, Table 1). In case 2, the DNMT3A mutation marked a larger clone (VAF 45%; Figure 1B, Table 1), and the detection of 14 additional low-frequency variants (VAF around 5%), including mutations in known AML-associated genes (DNMT3A, IDH2, NPM1), indicated subclonal evolution. Isolated MS may thus be accompanied by preleukemic BM clones that predispose to MS and later development of AML.
Both patients eventually developed overt AML, allowing us to infer clonal evolution. In case 1, all 4 variants that were present at a low frequency in the normal BM were also present in the isolated MS, and 3 of them remained in the subsequent AML. A total of 14 variants were shared between the isolated MS and the AML; 7 were only found in the isolated MS, whereas 5, including a NRAS mutation, were acquired in the AML phase (Figure 1A, supplemental Table 1). In case 2, the pattern was different, with all 17 variants present in the first normal BM shared with the MS, which in turn had 3 additional variants, and the subsequent AML sample acquiring 1 additional mutation (Figure 1B, supplemental Table 2). Case 2 is consistent with signs of a classical clonal evolution, in which the disease could have evolved through a preleukemic clone in the morphologically normal BM, spread to the distant site, followed by clonal evolution and MS growth, with subsequent tumor cell leakage from the MS into the BM and the development of full-blown AML. In case 1, however, this scenario is less likely, because a set of mutations in the isolated MS were not found in the subsequent AML, suggesting that although they had derived from a common preleukemic ancestor clone, they developed independently.
In summary, we conclude that DNMT3A-mutated preleukemia may predispose for the frequent bone marrow relapses following isolated MS and that recurrent NFE2 mutations may have a role in the development of isolated MS. Our results further shed light on the clonal evolution of isolated MS and suggest that WES of isolated MS and matched BM may help establish a firm diagnosis and initiate prompt treatment.
Acknowledgments
This work was supported by the Swedish Cancer Society, the Swedish Children’s Cancer Foundation, the Swedish Research Council, the IngaBritt and Arne Lundberg Foundation, and ALF, Region Skåne Research Grants.
The online version of this article contains a data supplement.
Authorship
Contribution: V.L., A.L., and G.J. collected patients, reported clinical outcomes, and provided samples; C.O.-P., H.L., M.R., L.P., and T.F. performed genetic analyses; L.P. and M.E. collected samples and evaluated morphology and immunohistochemistry; V.L., C.O.-P., T.F., and G.J. wrote the initial drafts of the manuscript; C.O.-P. and H.L. prepared graphs; all authors contributed to interpretation of data, provided comments on the manuscript, and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gunnar Juliusson, Stem Cell Center, Department of Laboratory Medicine BMC B10, Lund University, SE-22185 Lund, Sweden; e-mail: gunnar.juliusson@med.lu.se; and Thoas Fioretos, Department of Clinical Genetics, Akutgatan 22, Lund University, SE-22185 Lund, Sweden; e-mail: thoas.fioretos@med.lu.se.
References
Author notes
V.L., C.O.-P., H.L., G.J., and T.F. contributed equally to this study.