Abstract
T-cell lymphomas are a heterogeneous group of rare malignancies with overlapping clinical, immunologic, and histologic features. Recent advances in our understanding of T-cell differentiation based on gene expression profiling, next-generation sequencing, and transgenic mouse modeling studies have better elucidated the pathogenetic mechanisms underlying the diverse biology of T-cell lymphomas. These studies show that although genetic alterations in epigenetic modifiers are implicated in all subtypes of T-cell lymphomas, specific subtypes demonstrate enrichment for particular recurrent alterations targeting specific genes. In this regard, RHOA and TET2 alterations are prevalent in nodal T-cell lymphomas, particularly angioimmunoblastic T-cell lymphomas, peripheral T-cell lymphomas (PTCLs) not otherwise specified, and nodal PTCLs with T-follicular helper phenotype. JAK-STAT signaling pathways are mutationally activated in many extranodal T-cell lymphomas, such as natural killer/T-cell and hepatosplenic T-cell lymphomas. The functional significance of many of these genetic alterations is becoming better understood. Altogether these advances will continue to refine diagnostic criteria, improve prognostication, and identify novel therapeutic targets, resulting in improved outcomes for patient with T-cell lymphomas.
Introduction
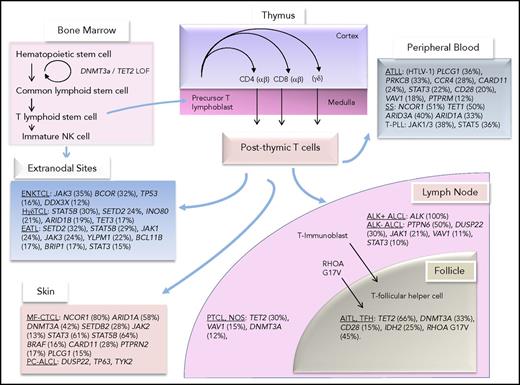
Peripheral T-cell lymphomas (PTCLs) represent 5% to 10% of non-Hodgkin lymphomas in the United States, although in Asian countries, the incidence may be higher (35%).1,2 T-cell lymphomas are highly heterogeneous in their clinical presentation, histologic features, pathogenesis, and prognosis. Importantly, with the exception of a minor subset of disease categories, such as patch stage mycosis fungoides, the morbidity and mortality rates of patients with T-cell lymphomas remain high.3 Postthymic T cells differentiate to function in either the innate or the adaptive immune response. Conceptually, T cells within these functional compartments acquire genetic aberrations that contribute to the biology of T-cell malignancies that are characteristic of lymphomas that arise from nodal and extranodal sites. Figure 1 highlights the common genetic aberrations that have been identified in subtypes of T-cell lymphomas and their postulated cellular counterparts in the innate and adaptive immune systems.
T-cell development and postulated normal counterparts of T-cell lymphomas along with their most common mutations.
T-cell development and postulated normal counterparts of T-cell lymphomas along with their most common mutations.
The NPM-ALK gene fusion was identified as the predominant genetic alteration in ALK+ anaplastic large-cell lymphoma (ALCL) in 19944 ; however, recurrent chimeric fusions are only recently emerging as important pathogenetic events in T-cell lymphomas. A vast majority of oncogenic alterations in mature T-cell lymphomas are represented by point mutations, indels, and copy number changes.5-8 T-cell lymphomas frequently carry mutations leading to constitutive activation of pathways downstream of the T-cell receptor (TCR), costimulatory proteins, and/or cytokine receptors. Additionally, highly recurrent mutations are identified in many classes of epigenetic modulators. The numerous alterations in the DNA methylation and histone posttranslational modification machinery that occur as a result of mutations in epigenetic regulators and histone modifiers are only beginning to be understood, but they bear discussion because of their high frequency and likely role as early events in oncogenesis. Not surprisingly, genetic subversion of mechanisms exploiting immune evasion is also observed in T-cell lymphomas. In this review, we summarize recently published data from large-scale genomic and transcriptomic analyses into mechanistically based thematic insights into mature T-cell lymphomas (Table 1). Because excellent reviews covering individual T-cell lymphoma entities and insights into their diagnosis and prognosis have been published recently, here we briefly emphasize novel shared concepts of T-cell pathogenesis with a summary of key insights for 3 common subtypes of nodal T-cell lymphoma: AITL, PTCL NOS, and ALK− ALCL.
Frequently mutated genes in TCR signaling and epigenetic pathways in T-cell lymphomas
| Gene, mutation . | Change in activity . | Frequency, % . |
|---|---|---|
| AITL | ||
| RHOA, G17V | LOF12,31 /unknown29 | 42-6712,23,28,52,82 |
| CD28, point mutants, translocations | GOF23,41 | 10-1523,41,43,44 |
| DNMT3A, point mutants | LOF12,52 | 3-3312,52,83 |
| TET2, nonsense/point mutants | LOF12 | 13-7312,52,82,83 |
| IDH2, point mutants | Neomorphic97 | 20-4012,52,97 |
| ALK− ALCL | ||
| PTPN6, aberrant methylation and inactivation | LOF15,17 | 5016,17 |
| DUSP22, translocations | LOF7 | 307 |
| VAV1, translocations, point mutants | GOF7 | 117 |
| JAK1, point mutants | GOF54 | 2154 |
| STAT3, point mutants | GOF54 | 1054 |
| ATLL | ||
| VAV1, point mutants | Likely GOF14 | 1814 |
| PLCG1, point mutants | GOF14 | 3614,75 |
| PRKCB, point mutants | GOF14 | 3314,110 |
| CARD11, point mutants, CNV (gain) | GOF14 | 2414,75 |
| CD28, point mutants, CNV (gain), translocations | GOF14 | 2014,75 |
| CCR4, nonsense/point mutants | GOF14 | 2814,75 |
| PTPRM, point mutants | LOF14 | 1214 |
| STAT3, point mutations | GOF14 | 2214,75 |
| RHOA, point mutants | GOF14 /unknown29 | 814 |
| CTCL | ||
| PLCG1, point mutants (often S345F), CNV (gain) | GOF24,35,36 | 3-2024,35-37 |
| CARD11, CNV (gain), point mutants | GOF24,36 | 2924,36 |
| ZEB1, CNV (loss) | GOF36 | 6036 |
| JAK2, CNV (gain) | GOF24 | 1336 |
| STAT3, CNV (gain), point mutants | GOF24,36 | 6124 |
| STAT5B, CNV (gain), point mutants | GOF24,36 | 10-6224,36 |
| DNMT3A, CNV (loss) | LOF24 | 4-37.524,36 |
| SETD2, CNV (loss) | GOF24 | 2824 |
| ARID1A, CNV (loss) | LOF24,36 | 59-6324,36 |
| NCOR1, CNV (loss) | LOF36 | 8324 |
| RLTPR, point mutant (Q575E) | GOF24 | 724 |
| EATL/NKTCL | ||
| STAT5B, point mutants, indels | GOF34 | 2834 |
| JAK1, point mutants | GOF34 | 2334 |
| JAK3, point mutants | GOF34 | 2334 |
| SETD2, point/nonsense mutants | LOF34 | 3234 |
| BCOR, CNV (loss), point mutants | LOF85 | 3285 |
| HSTCL | ||
| STAT5B, point mutants | GOF68 | 3068 |
| STAT3, point mutants, indels | GOF68 | 968 |
| SETD2, nonsense/point mutants | LOF68 | 2568 |
| TET3, point mutants | LOF68 | 1568 |
| PTCL NOS | ||
| RHOA, point mutants | LOF12 /unknown23,29 | 12.5-4012,14,23,28 |
| VAV1, translocation, point mutants | GOF7,28 | 11-157,28 |
| DNMT3A, point mutants | LOF12 | 1212 |
| TET2, nonsense/point mutants | LOF12,25 | 11-3012,25 |
| SS | ||
| PLCG1, point mutants (often S345F), indels | GOF38 | 1138 |
| ZEB1, point/nonsense mutants | LOF38,39 | 11-5538,39 |
| TET1, CNV (loss), point mutants | LOF38 | 5038 |
| DNMT3A, CNV (loss), point mutants | LOF38 | 1838 |
| KMT2C, nonsense mutants | LOF38 | 3238 |
| KMT2B, nonsense mutants, CNV (loss) | LOF38 | 2238 |
| ARID1a, CNV (loss), point mutants | LOF38,39 | 33-4138,39 |
| NCOR1, CNV (loss), nonsense/point mutants | LOF38 | 5138 |
| Gene, mutation . | Change in activity . | Frequency, % . |
|---|---|---|
| AITL | ||
| RHOA, G17V | LOF12,31 /unknown29 | 42-6712,23,28,52,82 |
| CD28, point mutants, translocations | GOF23,41 | 10-1523,41,43,44 |
| DNMT3A, point mutants | LOF12,52 | 3-3312,52,83 |
| TET2, nonsense/point mutants | LOF12 | 13-7312,52,82,83 |
| IDH2, point mutants | Neomorphic97 | 20-4012,52,97 |
| ALK− ALCL | ||
| PTPN6, aberrant methylation and inactivation | LOF15,17 | 5016,17 |
| DUSP22, translocations | LOF7 | 307 |
| VAV1, translocations, point mutants | GOF7 | 117 |
| JAK1, point mutants | GOF54 | 2154 |
| STAT3, point mutants | GOF54 | 1054 |
| ATLL | ||
| VAV1, point mutants | Likely GOF14 | 1814 |
| PLCG1, point mutants | GOF14 | 3614,75 |
| PRKCB, point mutants | GOF14 | 3314,110 |
| CARD11, point mutants, CNV (gain) | GOF14 | 2414,75 |
| CD28, point mutants, CNV (gain), translocations | GOF14 | 2014,75 |
| CCR4, nonsense/point mutants | GOF14 | 2814,75 |
| PTPRM, point mutants | LOF14 | 1214 |
| STAT3, point mutations | GOF14 | 2214,75 |
| RHOA, point mutants | GOF14 /unknown29 | 814 |
| CTCL | ||
| PLCG1, point mutants (often S345F), CNV (gain) | GOF24,35,36 | 3-2024,35-37 |
| CARD11, CNV (gain), point mutants | GOF24,36 | 2924,36 |
| ZEB1, CNV (loss) | GOF36 | 6036 |
| JAK2, CNV (gain) | GOF24 | 1336 |
| STAT3, CNV (gain), point mutants | GOF24,36 | 6124 |
| STAT5B, CNV (gain), point mutants | GOF24,36 | 10-6224,36 |
| DNMT3A, CNV (loss) | LOF24 | 4-37.524,36 |
| SETD2, CNV (loss) | GOF24 | 2824 |
| ARID1A, CNV (loss) | LOF24,36 | 59-6324,36 |
| NCOR1, CNV (loss) | LOF36 | 8324 |
| RLTPR, point mutant (Q575E) | GOF24 | 724 |
| EATL/NKTCL | ||
| STAT5B, point mutants, indels | GOF34 | 2834 |
| JAK1, point mutants | GOF34 | 2334 |
| JAK3, point mutants | GOF34 | 2334 |
| SETD2, point/nonsense mutants | LOF34 | 3234 |
| BCOR, CNV (loss), point mutants | LOF85 | 3285 |
| HSTCL | ||
| STAT5B, point mutants | GOF68 | 3068 |
| STAT3, point mutants, indels | GOF68 | 968 |
| SETD2, nonsense/point mutants | LOF68 | 2568 |
| TET3, point mutants | LOF68 | 1568 |
| PTCL NOS | ||
| RHOA, point mutants | LOF12 /unknown23,29 | 12.5-4012,14,23,28 |
| VAV1, translocation, point mutants | GOF7,28 | 11-157,28 |
| DNMT3A, point mutants | LOF12 | 1212 |
| TET2, nonsense/point mutants | LOF12,25 | 11-3012,25 |
| SS | ||
| PLCG1, point mutants (often S345F), indels | GOF38 | 1138 |
| ZEB1, point/nonsense mutants | LOF38,39 | 11-5538,39 |
| TET1, CNV (loss), point mutants | LOF38 | 5038 |
| DNMT3A, CNV (loss), point mutants | LOF38 | 1838 |
| KMT2C, nonsense mutants | LOF38 | 3238 |
| KMT2B, nonsense mutants, CNV (loss) | LOF38 | 2238 |
| ARID1a, CNV (loss), point mutants | LOF38,39 | 33-4138,39 |
| NCOR1, CNV (loss), nonsense/point mutants | LOF38 | 5138 |
AITL, angioimmunoblastic T-cell lymphoma; ATLL, adult T-cell leukemia/lymphoma; CNV, copy number variation; CTCL, cutaneous T-cell lymphoma; EATL, enteropathy-associated T-cell lymphoma; GOF, gain of function; HSTCL, hepatosplenic T-cell lymphoma; LOF, loss of function; NKTCL, natural killer/T-cell lymphoma; NOS, not otherwise specified; SS, Sézary syndrome.
Conceptual overview of pathogenetic events in mature T-cell lymphomas
TCR signaling pathways
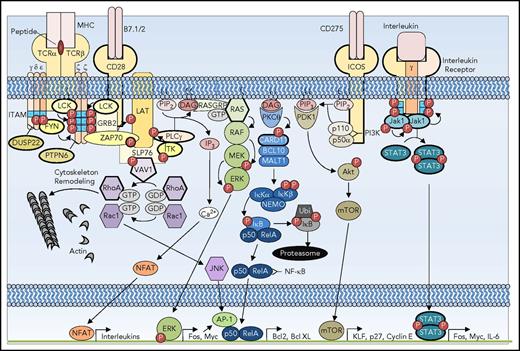
TCR signaling requires a sophisticated and highly coordinated interaction of membrane and cytosolic proteins to affect the responses of transcription programs in response to external stimuli. The combination of TCR, costimulatory, and cytokine signaling pathways is necessary for the survival, proliferation, and differentiation of T cells. Recent genomic analyses of T-cell malignancies have highlighted the emerging contribution of functional aberrations of numerous components of the TCR signaling pathways in the pathogenesis of T-cell lymphoma (Figure 2). Although certain mutations are restricted to specific subtypes of T-cell lymphoma, others are seen in several subtypes of T-cell neoplasms, underscoring the potential overlap in pathogenetic events.
Signaling pathways subverted in T-cell oncogenesis. TCR costimulatory receptors CD28 and inducible T-cell costimulator (ICOS) and interleukin receptor are depicted with a simplified representation of the downstream signaling mediators. The receptors and signaling pathway components that are recurrently mutated in T-cell lymphoma are depicted with bold outlines. DAG, diacylglycerol; GTP, guanine nucleotide triphosphate; IL-6, interleukin-6; ITAM, immunoreceptor tyrosine-based activation motif; IP3, inositol 3-phosphate; JAK, Janus kinase; MHC, major histocompatibility complex; mTOR, mammalian target of rapamycin; P, phosphorylation; PTPN, protein tyrosine phosphatase nonreceptor; STAT, signal transducer and activator of transcription.
Signaling pathways subverted in T-cell oncogenesis. TCR costimulatory receptors CD28 and inducible T-cell costimulator (ICOS) and interleukin receptor are depicted with a simplified representation of the downstream signaling mediators. The receptors and signaling pathway components that are recurrently mutated in T-cell lymphoma are depicted with bold outlines. DAG, diacylglycerol; GTP, guanine nucleotide triphosphate; IL-6, interleukin-6; ITAM, immunoreceptor tyrosine-based activation motif; IP3, inositol 3-phosphate; JAK, Janus kinase; MHC, major histocompatibility complex; mTOR, mammalian target of rapamycin; P, phosphorylation; PTPN, protein tyrosine phosphatase nonreceptor; STAT, signal transducer and activator of transcription.
TCR-associated kinases.
Binding of a major histocompatibility complex and peptide by the cognate TCR leads to recruitment of SRC family kinases (SFKs) such as LCK and FYN that phosphorylate tyrosine residues of immunoreceptor tyrosine-based activation motifs of the TCRζ chain and TCR-associated CD3 family. Immunoreceptor tyrosine-based activation motif phosphorylation leads to autophosphorylation of ZAP70 and recruitment of scaffolding proteins such as LAT. LCK also phosphorylates and activates ITK, which activates phospholipase Cγ (PLCG1), second messengers, and the RAS and NFAT pathways. Activation of SFKs in T-cell lymphomas occurs via numerous mechanisms, including somatic mutations, copy number alterations, and transcriptional deregulation. SYK protein is overexpressed in 94% of PTCLs relative to normal T cells.9 Chemical inhibition of SYK kinase in vitro induces apoptosis and decreases proliferation.10 Genomic gains of ITK are seen in 38% of PTCLs.11 Somatic activating mutations of FYN are present in 3% of PTCLs,12 as well as in CTCL13 and ATLL.14 A [t(5;9)(q33;q22)] translocation involving the pleckstrin, Tec, and SH3 homology domains of ITK with the SYK kinase domain functions as a constitutively active kinase in a subtype of PTCL,8 designated as follicular T-cell lymphoma. A similar mechanism of FER activation is postulated for the ITK-FER translocation in PTCL NOS.7
TCR pathway phosphatases.
PTPN family members PTPN6 (SHP1) and PTPN22 oppose the function of SFKs and serve as negative regulators of TCR signaling. Thus, loss of function of tyrosine phosphatases is a common event in T-cell lymphomas. The expression of PTPN6 is repressed via promoter methylation in NKTCLs15 and in up to 50% of ALK+ and ALK− ALCLs.16,17 Expression of PTPN6 diminishes proliferation of ALK+ ALCL cell lines.18 In ATLL, loss of PTPN6 expression is mediated by the HTLV-1–associated protein TAX via a transcriptional mechanism.19
Chromosomal rearrangements encompassing the DUSP22 locus reportedly lead to loss of DUSP22 function in up to 30% of ALK− ALCLs. DUSP22 encodes a dual-specificity phosphatase, which is expressed in a variety tissues and dephosphorylates both tyrosine and serine/threonine residues. It inactivates both LCK and ERK and is postulated to function as a tumor suppressor in ALK− ALCL.20 Notably, DUSP22-rearranged ALK− ALCLs exhibit a good prognosis.6,21 Importantly, DUSP22 rearrangement is mutually exclusive with other rearrangements observed in ALCL, including ALK and P63, suggesting distinct pathogenetic mechanisms in these categories of T-cell malignancies.6 The protein substrates and functional role of DUSP22 in T-cell lymphomas remain largely unknown at this time. Further credence for a pathogenetic role of DUSP22 in T-cell lymphomas is provided by the presence of inactivating splice site mutations of DUSP22 in PTCL NOS.20
TCR second messengers and GTPases.
Numerous RAS and RHO GTPases are deregulated in T-cell lymphomas. The RAS and RHO family members of GTPases transmit intracellular second messenger signals from the TCR to modulate proliferation, survival, and migration. TCR-mediated phosphorylation of phospholipase PLCG1 leads to the generation of DAG and inositol 3-phosphate, which activates RAS/MAPK signaling. These events are critical for activation of transcription factors JNK and AP-1, required for T-cell development and survival.22 The guanine exchange factor (GEF) VAV1 is activated by TCR-associated kinases and in turn activates RHO family members RAC1 and RHOA. These events lead to reorganization of the cytoskeleton and activation of the phosphatidylinositol 3-kinase (PI3K) pathway, whereas DAG leads to activation of protein kinase C, phosphorylation of the CBM complex (CARD11, BCL10, and MALT1), and NF-κB pathway activation.
Genetic alterations of VAV1 occur in diverse subtypes of T-cell lymphoma, including AITL,23 ATLL,14 CTCL,24 PTCL NOS,25 and ALCL (ALK+ or ALK−).7 VAV1 is activated by tyrosine phosphorylation at its inhibitory SH3 domain by SFKs, leading to a conformational change that exposes the Dbl homology domain with GEF function toward various GTPases.26 In addition, VAV1 has GEF-independent functions, perhaps as an adaptor to the actin cytoskeleton in a complex with PLCG1.27 The functional significance of VAV1 activation is highlighted by the various types of genetic alterations observed in T-cell lymphomas. Constitutive activation of VAV1 via point mutations is often seen in ATLL and PTCL NOS.14,23 Gene fusions involving VAV1 are also observed in ALK− ALCL and PTCL NOS.7 Recurrent in-frame deletions (VAV1 Δ778-786) generated by a focal deletion-driven alternative splicing event,28 as well as fusions to VAV1-THAP4, VAV1-MYO1F, VAV1-S100A7, and VAV1-STAP2, and VAV1-GSS, have been identified in PTCL and AITL.7,28,29 Notably, the breakpoints result in loss of the inhibitory SH3 domain, which leads to an increase in the activity of the Rac1 GTPase.30 Both of these mechanisms are thought to lead to increased activation of VAV1 catalytic and noncatalytic effector pathways.
RHOA encodes a ras-related GTP-binding protein that functions as a molecular switch regulating several cellular processes, including the cell cycle and actin-cytoskeleton dynamics and cell adhesion and migration. A recurrent mutation of the GTPase RHOA G17V is present in a majority of AITLs (53% to 71%) and follicular helper T (TFH)–class PTCLs NOS (17% to 18%) and is rare in other T-cell lymphomas.12,29,31,32 Using rhotekin-binding assays, it has been demonstrated that RHOA G17V is impaired in GTP binding and acts as a dominant negative in cell lines.31 Mouse models with RHOA G17V expression in CD4+ cells demonstrate increased numbers of TFH cells, and RHOA G17V expression in TET2-deficient mice leads to AITL-like tumors.33 Interestingly, both activating and inactivating mutations of RHOA at other codons have been reported in PTCL NOS23 and ATLL,14 whereas inactivating mutations are found predominantly in CTCL.24 In this regard, RHOA K18N, which exhibits a stronger GTP-binding capacity, has also been identified in some AITLs.23 Thus, the mutational type and positions seem to confer different biologic effects with regard to guanosine triphosphate/guanosine diphosphate (GTP/GDP)–binding kinetics and transcriptional regulation. In addition, the demonstration that mutant RHOA G17V but not WT RHOA is able to bind VAV1 and is associated with activation of PLCG1 suggests that the mechanism implicated by RHOA G17V may be distinct from its catalytic activity.29
Mutations affecting the MAPK pathway are present in a variety of T-cell lymphomas. Activating mutations of KRAS are present in TFH PTCL NOS and AITL,23 whereas activating mutations of MAP2K1 and inactivating mutations of NF1 are seen in CTCL,24 and KRAS and NRAS mutations are present in EATL.34 PLCG1 mutations are common in a diverse spectrum of T-cell malignancies14,23,35-38 and are associated with increased NFAT pathway activation. Although the functional consequences of PLCG1 mutations are not well characterized, it is notable that the cytotoxicity associated with stable expression of PLCG1 S345F in lymphoid cell lines was rescued by the coexpression of BCL2, suggesting that its pro-oncogenic effects may require compensatory antiapoptotic mechanisms.35
Activation of NF-κB is a key survival pathway for T cells and is also mediated by mutations in CBM complex member CARD11, which occurs in a variety of T-cell malignancies including Sézary syndrome,38 AITL and TFH PTCL NOS,12 ATLL,14 and CTCL.36 Activating mutations in both PLCG1 and CARD11 are generally mutually exclusive in Sézary syndrome and CTCL,36,38,39 although they co-occur in rare cases of AITL23 and ATLL.14
T-cell costimulatory signals.
Integrated signals from costimulatory and coinhibitory proteins determine the degree of T-cell activation and proliferation; thus, it is not surprising that mutations of costimulatory proteins (CD28) and coinhibitory proteins (CTLA-4) and downstream proteins are pathogenetic events in T-cell lymphoma. Interaction of CD28 (Figure 2) with its ligands B7.1/B7.2 expressed on antigen-presenting cells leads to downstream activation of NFAT and NF-κB and generation of IL-2.40 ICOS, a member of the CD28 family, is also critical for the formation and maintenance of the TFH phenotype. Mutations affecting both the extracellular and intracellular domains of CD28 are present in up to 11.3% of AITLs. CD28 D124V mutation in the extracellular domain increases its affinity for CD86. Similarly, CD28 T195P, located in the intracellular domain, increases its affinity for GRB2 and GRAP2 and leads to constitutive mammalian target of rapamycin signaling and cell proliferation.23,41,42 Notably, mutations in CD28 are associated with decreased overall survival in AITL41 ; however, its prognostic relevance in other types of T-cell lymphomas remain unknown. A recurrent RLTPR Q575E mutation has been described in CTCLs. RLTPR is a scaffolding protein in the TCR signaling pathway, and the pQ575E mutation is reported to upregulate NF-κB signaling and augment TCR-dependent production of IL-2 in activated T cells.24
Gene fusions involving costimulatory proteins have oncogenic functions in T-cell lymphoma. For example, a fusion protein comprising the extracellular and transmembrane domains of CTLA-4 and the intracytoplasmic domain of CD28 has been described in Sézary syndrome. CTLA-4–CD28 fusion transcripts also occur in AITL, PTCL NOS, and NKTCL, potentially expanding their clinical relevance.43,44 Although the mechanism by which the CTLA-4–CD28 fusion transcripts promote oncogenesis is not fully understood, it is predicted that the CTLA-4–CD28 fusion protein results in strong binding of CTLA-4 to B7.1. Furthermore, ectopic expression of CTLA-4–CD28 in Jurkat cells led to enhanced phosphorylated ERK signaling and increased proliferation.43 Additionally, ICOS-CD28 fusions where the CD28 coding sequence is placed under the influence of the ICOS promoter, leading to strong CD28 expression, have also been identified in AITL41 and ATLL.14 Opportunities for therapeutic intervention with regard to these gene fusions may exist, because treatment with anti–CTLA-4 antibody ipilimumab is associated with a therapeutic response.45
Cytokine signaling and aberrant JAK/STAT activation
T-cell differentiation, function, and proliferation are modulated by cytokine signaling. Cytokine receptors influential in T-cell function lack intrinsic tyrosine kinase activity (Figure 2). Instead, they function through the JAK family of cytoplasmic tyrosine kinases (JAK1, JAK2, JAK3, TYK2) and the second messenger STAT family proteins. Dimerization of STATs via their SH2 domains occurs as a result of JAK-mediated phosphorylation and permits the nuclear translocation critical for their function as transcription factors. Key transcriptional targets of STAT proteins regulate cell proliferation through p21 and p27, cyclin D1, and MYC, apoptosis through BCL2 and BCLXL, and immune cell differentiation and function.46
Constitutive activation of the JAK/STAT pathway is observed in nearly all T-cell lymphomas.14,36,38,47-53 Interestingly, various mechanisms of JAK/STAT activation contribute to disease pathogenesis in distinct subtypes of T-cell malignancies. Although a majority of ALCLs, NKTCLs, and CTCLs display nuclear localization of phosphorylated STAT3 and other features of JAK/STAT activation, only a minority display mutations in JAK/STAT, indicating the presence of alternate mechanisms of activation.49,53,54 Indeed, autocrine and paracrine mechanisms of cytokine/receptor-mediated mechanisms for JAK/STAT activation have been reported. A majority of ALK+ ALCLs express both IL-9 and IL-9 receptor55 and IL-22,56 which promote JAK3 activation and potentiation of NPM-ALK. Recent large-scale N-glycoproteome studies have highlighted the contributions of ALK-mediated cytokine/receptor-JAK/STAT signaling with novel therapeutic vulnerabilities within this axis in these neoplasms.57 In ATLL, the HTLV-1 TAX protein transactivates common γ chain interleukins IL-2 and IL-15 and their receptors, leading to activation of JAK1/3 and STAT5.58 Breast implant–associated ALCL cell lines also exhibit autocrine production of IL-6.59 Autocrine regulation of IL-13, IL-15, and IL-21 also contributes to proliferation and survival of tumor cells in CTCL and Sézary syndrome.60-62 Inactivating mutations of ZEB1, a transcription factor that represses IL-2 and IL-15 production, also contribute to proliferation and survival of CTCL, Sézary syndrome, and ATLL.36,38,63,64 Gene expression profiling (GEP) of PTCL NOS has revealed observable differences in survival that correlate with cytokine expression patterns. Specifically, PTCLs NOS with the GATA-3 expression signature are associated with TH2 interleukins, whereas those with the T-BET expression signature are associated with interferon-γ.65,66
Although cytokine receptor mutations are relatively uncommon in T-cell lymphomas relative to their leukemic counterparts, recent studies have revealed the presence of recurrent mutations in the chemokine receptor CCR4 in 25% of ATLLs. CCR4 mutations result in the deletion of the carboxy terminus, thereby impairing receptor internalization, and increase PI3K pathway signaling and enhance cell proliferation.67
Mutations of JAK family members have been identified in nearly all T-cell neoplasms, predominantly in JAK1 or JAK3.5,13,14,25,34,36,38,54,68-70 Mutations generally inactivate the pseudokinase domains, which function both as negative regulators of tyrosine kinase activity and as interaction sites with cytokine receptors and thus are activating in function.71 Alternatively, TYK2 and PCM1-JAK2 fusions in ALK− ALCL lead to strong activation of downstream STATs.5,54,72 STAT5B and STAT3 are also commonly mutated in T-cell lymphomas, particularly in γδ-PTCL and hepatosplenic T-cell lymphoma (HSTCL).13,25,36,38,52,54,68,69 A vast majority of STAT mutations are located in the SH2 domains,73 which mediate dimerization and thus activation through nuclear translocation.74 The loss of expression of PTPRK, which dephosphorylates STAT3, in up to 50% of extranodal NKTCLs, is consistent with its putative role as a tumor suppressor.75 Intriguingly, JAK/STAT pathway mutations are not mutually exclusive in some cases of T-cell lymphoma, providing additional complexity of genetic interactions and possible evidence for oncogenic cooperativity.54
Epigenetic modifiers
Modifications of DNA and histones, which alter accessibility to transcriptional machinery, contribute to epigenetic deregulation. Recurrent mutations in epigenetic regulators are present in all subtypes of T-cell lymphomas.
Deregulated DNA methylation.
Methylation of cytosine residues found in CpG islands is a potent mechanism of transcriptional repression. DNA methyltransferases (DNMTs) are enzymes that mediate DNA methylation. DNMT1 functions to maintain DNA methylation, whereas DNMT3A/B is important for de novo DNA methylation.76
Mutations in DNMT3A have been identified in nearly all subtypes of T-cell malignancies.12,25,32,36,38,39,52,77,78 Although a majority of mutations are located in the catalytic methyltransferase domain, the remainder are spread diffusely across the gene. The DNMT3A R882H mutation is the most common and acts as a dominant negative and results in decreased ability to form functional tetramers.79
Cytosine demethylation is a multistep process regulated by the TET family of DNA methylcytosine hydroxylases. Mutations of TET family genes are widespread in T-cell lymphoma. DNMT3A and TET2 mutations are present in nontumoral hematopoietic cells of patients with both myeloid and T-cell malignancies, raising the possibility that they may be early events in oncogenesis occurring in hematopoietic stem cells.32,80,81 Many T-cell lymphoma subsets harbor TET family mutations, with TET1 and TET2 mutations found at comparable frequencies in PTCL NOS,25 ATLL,14 and EATL,34 whereas both TET2 and TET3 mutations are seen in HSTCL.68 AITL and TFH PTCL NOS show marked enrichment for TET2 mutations relative to other TET family members,12,25,52,82,83 indicating the possibility of a differential effect of TET family members in distinct T-cell subsets. It is also intriguing to note the segregation of the TET2-mutant allele with DNMT3A and RHOA G17V mutations in AITL, suggesting the possibility of a functional interaction between these proteins, leading to a predilection for TFH phenotype.12,52 By contrast, CTCL and Sézary syndrome genomes are enriched for TET1 mutations.36,38,39
Deregulation of histone-modifying enzymes.
The histone methyltransferases and acetyltransferases and their corresponding methylases and acetylases control the posttranslational modifications of histones and thus accessibility to the transcriptional machinery. Histone methyltransferase activity promotes eu- or heterochromatin, depending on the site of modification. Lysine methyltransferases such as EZH1/2 promote transcriptional repression via trimethylation of lysine 27 of histone H3, whereas SETD1 promotes transcriptional activation via trimethylation of lysine 4 of histone H3.84 Furthermore, H3K36 methylation by SETD2 is context dependent and is associated with both transcriptional activation and repression. Mutations in a wide spectrum of histone-modifying enzymes occur in T-cell lymphoma. SETD2 methyltransferase mutations occur in ATLL,14 Sézary syndrome,38 HSTCL,68 EATL,34 and PTCL NOS,12 and several other SET family members are frequently mutated in CTCL/Sézary syndrome.13,38 Mutations in the MLL methyltransferase family, particularly KMT2B and KMT2C, are present in up to 56% of Sézary syndrome cases but only occasionally noted in mycosis fungoides or PTCL NOS.13,25,38 Other gene products that recruit histone deacetylases are also recurrently affected by mutations, particularly BCOR in Sézary syndrome38 and NKTCL85 and BCORL1 in PTCL NOS.25 The functional consequences of these mutations remain largely unexplored, although recent systematic analyses of the epigenomic landscape of CTCLs demonstrate that chromatin dynamics are associated with clinical response to histone deacetylase inhibitors.86
Immune evasion mechanisms
Similar to the hijacking of T-cell signaling pathways, oncogenic events in T-cell lymphomas subvert the costimulatory/coinhibitory checks and balances to prevent immune recognition and response. The interaction of B7 proteins with CTLA-4 expressed on antigen-presenting cells leads to coinhibitory signals, T-cell anergy, and expression of programmed cell death 1 receptor (PD-1). Expression of the PD-1 ligand (PD-L1) on malignant cells or tumor-associated macrophages provides a mechanism of immune evasion, which has been successfully targeted by therapeutic anti–PD-1 antibodies. PD-L1 is expressed at variable frequencies in T-cell lymphomas (15% to 64% of PTCLs,87,88 27% of CTCLs,87 22% of ATLLs,89 5% of AITLs,90 and 34% of NKTCLs91 ), although the mechanism of their regulation is largely unknown. In ALCL, oncogenic NPM-ALK regulates the expression of PD-L1 via STAT3 and promotes immune evasion.92 Activation of the NF-κB pathway by Epstein-Barr virus LMP1 leads to increased PD-L1 expression in NKTCL.91 The MEK/ERK pathway, which mediates extracellular signals in many subtypes of T-cell lymphoma, also regulates the expression of PD-L1, suggesting that growth factors and cytokines may regulate the local immune tumor microenvironment.93
Immunomodulatory therapy for T-cell neoplasms is in its early stages. A phase 2b study of anti–PD-L1 antibody demonstrated activity in relapsed/refractory mycosis fungoides and PTCLs, providing rationale for additional clinical trials.94 Treatment of relapsed NKTCL with the anti–PD-L1 antibody pembrolizumab led to striking benefit, with 5 of 7 patients achieving durable complete responses.95 The presence of recurrent mutations affecting costimulatory/coinhibitory proteins also lends support for clinical trials evaluating the efficacy of the anti–CTLA-4 antibody ipilimumab and CTLA-4–immunoglobulin fusions abetacept and belatacept (Bristol-Myers Squibb) for PTCL, which are under way.41,45
Deregulated cellular metabolism
Alteration in cellular control of energy metabolism represents a hallmark of cancer and has been recently shown to be a relevant pathogenetic mechanism in ALK+ ALCL and may also be implicated in other subtypes. The cancer-associated Warburg effect favors aerobic glycolysis, which permits both growth in hypoxic conditions and shunting of metabolic intermediates from energy production toward synthesis of nucleic acids, lipids, and amino acids needed for proliferation. Integrative mass spectrometry–based phosphoproteomic analysis and metabolomic profiling of ALK+ ALCL revealed widespread metabolic changes associated with the expression of NPM-ALK.96 Drastic changes in diverse metabolic pathways, including synthesis of nucleotides, glycerophospholipids, and amino acids, were generated by NPM-ALK. Notably, small molecular inhibitors of ALK led to decreased lactate production, indicating a reversal of the Warburg effect. Many of these changes were mediated by a critical phosphorylation-mediated switch in the activation of the pyruvate kinase isoform PKM2 (Y105), which was shown to be a novel substrate of NPM-ALK, and were reversed with small-molecule activators of PKM2. Interestingly, phosphorylated Y105 PKM2 expression was abundant in ALK+ ALCL, but it was not expressed in cell lines derived from ALK− ALCL, mycosis fungoides, or Sézary syndrome, suggesting that different T-cell neoplasms may alter cellular metabolism using alternate mechanisms.96
Gain-of-function mutations of isocitrate dehydrogenase (IDH2) lead to enhanced production and accumulation of the metabolite 2-hydroxyglutarate (2HG). IDH2 R172 mutations are prevalent in AITL and lead to increased 2HG in tumor tissues and serum of patients.12,52,77,97 The physiologic function of IDH enzymes is NADP+-dependent conversion of isocitric acid to α-ketoglutaric acid. The IDH mutants have been demonstrated to produce the oncometabolite 2HG. The mechanism by which elevated 2HG leads to lymphomagenesis of TFH-derived AITL is not entirely understood, although enhanced production of 2HG can inhibit dioxygyenases, including the TET enzymes, and alter DNA methylation.77 The highly selective nature of IDH2 R172 mutations for AITL and their co-occurrence with TET2 mutations in other T-cell lymphomas may suggest a functional relationship. Indeed, GEP studies have demonstrated a correlation between IDH2 R172 mutations and global DNA methylation changes in AITL, with downregulation of genes associated with TH1 differentiation (STAT1 and IFNγ) and enrichment of IL-12–induced gene signature.77
Highlights of nodal lymphoma pathogenesis and therapy
Having provided a general overview of the novel pathogenetic mechanisms that characterize T-cell lymphomas, the following section will succinctly highlight insights relevant for the understanding of 3 common subtypes of nodal T-cell lymphomas, the pathogeneses of which have become much better understood from recent research.
AITL.
AITL is an aggressive neoplasm of TFH cells, associated with widespread immune dysregulation. The malignant cells may frequently comprise a minor proportion of an involved lymph node, with vascular proliferations, granulocytes, histiocytes, and often clonal B-cell populations admixed, all of which contribute to the B symptoms and dysproteinemia of AITL. Nonetheless, AITL has become 1 of the best-characterized T-cell lymphomas, with unveiling of many of the mechanisms underlying its protean manifestations. GEP studies have revealed important insights into the pathogenesis of AITL. A comparison of AITL and TFH signatures revealed significant overlap and provided insights into the development of polyclonal gammopathy seen in AITL. In addition, GEP studies have demonstrated the existence of a B-cell signature associated with a favorable outcome, whereas a signature of monocytic, cytotoxic, or p53-induced genes portends a poor prognosis.66
Recurrent mutations have been identified in AITL, and the role of these mutations in disease pathogenesis is beginning to be better understood. Loss-of-function mutations in DNA demethylation enzyme TET2 are seen in a majority of AITL cases.12,52,82 The TET2 mutations in AITL are inactivating mutations, including nonsense and frameshift alterations, which are distributed throughout the length of the protein, as well as missense mutations, which cluster within the C-terminal catalytic domain in AITL, as observed in myeloid malignancies.33,81 TET2 mutations and DNMT3A mutations may occur together in the same patient with AITL. In addition, identical mutations in TET2 and in DNMT3a may be seen in both T- and B-cell subsets from the same patient, suggesting a common stem-cell origin and a possible contribution of alterations of these genes to the dysproteinemia and other disease pathogenetic features.32,80,81 Indeed, the DNMT3A R882H mutant has been demonstrated to cooperate with TET2 inactivation to induce lymphoid malignancies in mouse models.98 It is noteworthy that RHOA G17V mutation is also seen in a majority of AITL cases with TET2 mutations.12,52 Although varied roles for RHOA G17V have been proposed in AITL oncogenesis, it seems sufficient to drive a TFH phenotype in murine models and, in conjunction with TET2 loss, leads to TFH lymphomas with the often distinctive morphology of AITL.33,99 Also noteworthy is that DNMT3A is also inactivated by mutations distributed throughout the coding sequence. Intriguingly, in AITL, the IDH2 mutations are exclusively located at the p.R172 position.33 The coexistence of IDH2 and TET2 mutations in a significant proportion of AITL cases suggests a cooperative relationship. Indeed, DNA methylation and histone H3K27 methylation have been demonstrated to be elevated in AITL samples with TET2 and IDH2 mutations, as compared with those with only TET2 mutations and lacking IDH2 mutations100 Chemotherapy regimens for AITL have had historically little success, with frequent progression or early remission.101 Therapeutic trials for targeted therapies in AITL have achieved mixed success. The vascular proliferations eponymous to AITL suggested a role for vascular endothelial growth factor antagonist bevacizumab; however, considerable cardiac toxicity prevented further study.102 The hypomethylating agent azacytidine has been administered to some patients harboring TET2 mutations, with notable clinical response.103
ALK− ALCL
Although ALCLs share a common eponymous morphology, their disease course and mechanisms of oncogenesis are distinct. Recent research on ALK− ALCL and primary cutaneous ALCL has broadened the body of knowledge of the subtypes and even demonstrated similarities between the clinically heterogeneous diseases. Several gene families demonstrate recurrent mutations in ALK− ALCL. Similar to ALK+ ALCL, ALK− ALCLs exhibit constitutive STAT3 phosphorylation consistent with JAK/STAT pathway activation, which is mutationally dependent or independent.54 Both whole-exome sequencing and high-density single-nucleotide polymorphism array analyses demonstrate that ALK− ALCLs exhibit recurrent mutations in tumor suppressors TP53 and PRDM1 (BLIMP1).54,104 Monoallelic PRF1 germ line mutations have been identified in 27% of childhood ALCLs; the mechanism of predisposition is unclear.105 Several recurrent translocations have been identified in ALK− ALCL, although none with the prevalence of NPM1-ALK. Translocations involving DUSP22, TP63, and TYK2 are associated with distinct clinical behavior.5,6 ERBB4 expression is altered in one quarter of ALCLs, both by mutation and translocation, and early data suggest that ERBB4+ ALCLs and those with the aforementioned translocations are mutually exclusive.106
Although the gene expression profiles, mutations, and copy number variations differ between ALK+ and ALK− ALCLs, they display similar DNA methylation changes in genes involved in differentiation and immune response, which could suggest a role for demethylase or bromodomain inhibitor–based therapies.17
PTCL NOS
PTCL NOS represents a progressively shrinking diagnostic category comprising clinically, histologically, and molecularly heterogeneous neoplasms that are poorly understood. Refinement of the diagnostic criteria has facilitated redesignation of T-cell neoplasms that would have fallen into this category primarily as AITLs, ALK− ALCLs, or nodal PTCLs with TFH phenotype. GEP and next-generation sequencing have identified significant overlap between PTCL NOS and other diagnostic categories. GEP revealed that a significant portion of PTCLs NOS exhibit TFH features, although the cases lack the classic morphologic features of TFH cells.66 These cases are now considered TFH lymphomas in the 2016 World Health Organization classification. In addition, a significant portion of cases classified as AITLs are best considered PTCLs NOS.107 Both CD30+ PTCL NOS and ALK− ALCL have decreased expression of core TCR signaling kinases LCK, FYN, and ITK, costimulatory protein CD28, and transcription factor NFATC2.108 Despite these similarities, GEP did not demonstrate significant overlap between ALK− ALCL and PTCL NOS, but the distinction is important because CD30+ PTCL NOS likely has worse prognosis.107
Recent discoveries of novel recurrent pathogenic alterations underlying PTCL NOS have expanded our knowledge of the mechanisms of T-cell proliferation. In addition to the ITK-SYK fusion protein, several other oncogenic fusion proteins have been identified. Notably, rearrangements in GEF VAV1 have been identified in both ALK− ALCL7 and PTCL NOS.12,28
Although the clinical behavior of these neoplasms is typically aggressive, GEP has revealed considerable heterogeneity. The identification of a cytotoxic PTCL NOS signature by GEP65 characterized by high TBX21 expression66 demonstrates a poor prognosis,66 but this is better than the dismal prognosis of the GATA3 subtype with MYC and PI3K signatures.109
Overall, PTCL NOS remains a catchall/wastebasket category, but the mechanisms underlying its heterogeneity are being increasingly elucidated. Opportunities for biologically and/or therapeutically relevant paradigms for classification (eg, CD30 positivity and structural alterations) may reveal homogeneous categories that improve on current classifications based on traditional histopathologic assessment.
Conclusions and future perspectives
Recent unbiased analyses of the genome, transcriptome, and proteome of many subtypes of T-cell lymphomas have provided an unprecedented level of novel insights into the pathogenesis of these rare heterogeneous but aggressive neoplasms (Figure 2). T-cell lymphoma subtypes harbor genetic alterations that perturb T-cell signaling pathways, including tyrosine kinases, protein tyrosine phosphatases, and GTPases, to promote proliferation and survival. Deregulated cytokine/JAK/STAT signaling is a pervasive event in T-cell lymphomas and involves a diverse array of genetic and transcriptional alterations. Genomic aberrations targeting epigenetic modifiers and chromatin remodelers are also highly prevalent in T-cell lymphomas. Genetic and transcriptional mechanisms that promote evasion of immune surveillance by T-cell malignancies are also being increasingly recognized. Overall, continued efforts to understand the pathogenetic events in T-cell lymphomas will lead to better delineation of diagnostic categories and provide novel insights into both normal lymphocyte biology and lymphomagenesis that will result in more-refined methods for disease monitoring as well as development of novel therapies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: All authors wrote the paper.
Conflict-of-interest disclosure: The authors delcare no competing financial interests.
Correspondence: Kojo S. J. Elenitoba-Johnson, Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, 609 Stellar Chance, 422 Curie Blvd, Philadelphia, PA 19104; e-mail: kojo.elenitoba-johnson@uphs.upenn.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal