Abstract
Chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL) are 2 well-defined entities that diverge in their basic pathogenic mechanisms and clinical evolution but they share epidemiological characteristics, cells of origin, molecular alterations, and clinical features that differ from other lymphoid neoplasms. CLL and MCL are classically considered indolent and aggressive neoplasms, respectively. However, the clinical evolution of both tumors is very heterogeneous, with subsets of patients having stable disease for a long time whereas others require immediate intervention. Both CLL and MCL include 2 major molecular subtypes that seem to derive from antigen-experienced CD5+ B cells that retain a naive or memory-like epigenetic signature and carry a variable load of immunoglobulin heavy-chain variable region somatic mutations from truly unmutated to highly mutated, respectively. These 2 subtypes of tumors differ in their molecular pathways, genomic alterations, and clinical behavior, being more aggressive in naive-like than memory-like–derived tumors in both CLL and MCL. The pathogenesis of the 2 entities integrates the relevant influence of B-cell receptor signaling, tumor cell microenvironment interactions, genomic alterations, and epigenome modifications that configure the evolution of the tumors and offer new possibilities for therapeutic intervention. This review will focus on the similarities and differences of these 2 tumors based on recent studies that are enhancing the understanding of their pathogenesis and creating solid bases for new management strategies.
Introduction
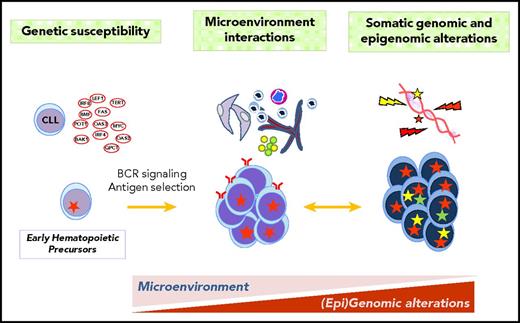
Chronic lymphocytic leukemia (CLL) and mantle cell lymphoma (MCL) are 2 lymphoid neoplasms characterized by the proliferation and accumulation of mature small CD5+ B cells that may involve bone marrow, blood, lymphoid tissues, and extranodal sites.1 They have different pathogenic mechanisms that translate into marked differences in the biological behavior, clinical evolution, and management of the patients.1-3 However, they also share epidemiological, biological, and clinical features that differ from other lymphoid neoplasms (Table 1).1,4-7 CLL and MCL are classically considered indolent and aggressive neoplasms, respectively. However, the clinical evolution of both tumors is very heterogeneous, with subsets of patients having an indolent behavior whereas others follow a very rapid evolution. Understanding the mechanisms underlying this diversity is becoming increasingly relevant due to novel therapeutic possibilities. The pathogenesis of the 2 entities encompasses complex interactions of 3 basic factors: genetic susceptibility, interactions between tumor cells and their microenvironment with the crucial role of the B-cell receptor (BCR), and acquired genomic/epigenomic alterations that will shape the evolution of the disease (Figure 1). This review is integrated in the context of the 2018 American Society of Hematology (ASH) Meeting on Lymphoma Biology and will focus on recent studies that are enhancing the understanding of the pathogenesis of these tumors.
Shared and divergent characteristics of CLL and MCL
| Characteristic . | CLL . | MCL . |
|---|---|---|
| Incidence*,4 | 5.1 | 0.8 |
| Hereditary susceptibility†,8 | 2.4- to 8.5-fold risk | 2-fold risk |
| Demographics1,4-7 | Elderly patients | |
| Males > Females | ||
| More common in Caucasians | ||
| Less frequent in Asians | ||
| Initial oncogenic events1 | del(13q), del(11q), tri(12), other (?) | t(11;14), SOX11 |
| Phenotype1 | ||
| SIg | Dim IgM, IgD | Strong IgM, IgD |
| CD5 | + | + |
| CD20 | Weak | Strong |
| CD23 | + | − |
| CD200 | + | − |
| Cytogenetic alterations1 | del(13q); del(11q); del(17p) | |
| t(11;14); del(9p); genomic instability | ||
| Somatic mutations66,67,116 | DNA damage response pathway (ATM, TP53) | |
| NF-κB pathway | ||
| NOTCH1 | NOTCH1/NOTCH2 | |
| SF3B1 and splicing machinery | Chromatin modifiers | |
| MYD88 | CDKN2A del | |
| MicroRNA dysregulation167 | miR-16-1, miR-29 a/b/c, miR-34a, miR-150, miR-155, miR-181 | |
| miR-15a, miR-21, miR-223, miR-650 | miR-17-92, miR-20b, miR-142-3p | |
| Characteristic . | CLL . | MCL . |
|---|---|---|
| Incidence*,4 | 5.1 | 0.8 |
| Hereditary susceptibility†,8 | 2.4- to 8.5-fold risk | 2-fold risk |
| Demographics1,4-7 | Elderly patients | |
| Males > Females | ||
| More common in Caucasians | ||
| Less frequent in Asians | ||
| Initial oncogenic events1 | del(13q), del(11q), tri(12), other (?) | t(11;14), SOX11 |
| Phenotype1 | ||
| SIg | Dim IgM, IgD | Strong IgM, IgD |
| CD5 | + | + |
| CD20 | Weak | Strong |
| CD23 | + | − |
| CD200 | + | − |
| Cytogenetic alterations1 | del(13q); del(11q); del(17p) | |
| t(11;14); del(9p); genomic instability | ||
| Somatic mutations66,67,116 | DNA damage response pathway (ATM, TP53) | |
| NF-κB pathway | ||
| NOTCH1 | NOTCH1/NOTCH2 | |
| SF3B1 and splicing machinery | Chromatin modifiers | |
| MYD88 | CDKN2A del | |
| MicroRNA dysregulation167 | miR-16-1, miR-29 a/b/c, miR-34a, miR-150, miR-155, miR-181 | |
| miR-15a, miR-21, miR-223, miR-650 | miR-17-92, miR-20b, miR-142-3p | |
SIg, surface immunoglobulin.
Rates are per 100 000 and age adjusted to the US standard population;
Risk of first-degree relatives of CLL and MCL of developing CLL or non-Hodgkin lymphomas, respectively.
CLL and MCL pathogenesis. The pathogenesis of the 2 entities integrates genetic susceptibility, interactions between tumor cells and their microenvironment, and acquired genetic and epigenetic alterations. Some genetic alterations may occur in progenitor cells. The influence of BCR activation and microenvironment interactions may be important from early stages of the disease whereas the acquisition of somatic mutations determines the progression and transformation.
CLL and MCL pathogenesis. The pathogenesis of the 2 entities integrates genetic susceptibility, interactions between tumor cells and their microenvironment, and acquired genetic and epigenetic alterations. Some genetic alterations may occur in progenitor cells. The influence of BCR activation and microenvironment interactions may be important from early stages of the disease whereas the acquisition of somatic mutations determines the progression and transformation.
Chronic lymphocytic leukemia
Genetic susceptibility
First-degree relatives of CLL patients have an increased risk (2.4- to 8.5-fold) of developing the disease.8 Approximately 40 susceptibility loci associated with CLL have been identifed.9-13 Most of them are in areas of active chromatin that may regulate genes influencing CLL development. Intriguingly, only a few of these candidate genes are altered in the overt disease (eg, POT1, BCL2, and IRF4).14 This difference suggests that susceptibility and driver CLL genes may influence different steps in the development of the disease or, although different, they may target similar pathways (eg, susceptibility variants in TERT and mutations in POT1 may interfere with telomeric function while the susceptibility BMF variant regulates BCL2 expression).9,11-13,15
Cells of origin and molecular subtypes
The cell(s) of origin of CLL is still controversial.16 CLL is a neoplasm of mature B lymphocytes but the earliest changes may already occur in hematopoietic stem cells primed for B-cell clonal differentiation.17 Common CLL genetic alterations such as tri(12), del(13q), and mutations of SF3B1, NOTCH1, and XPO1 have been found in hematopoietic progenitors of some patients.18,19 The mechanisms bridging these early changes to the clonal expansion of mature CLL cells are not well known but the immunogenetic analyses of the BCR strongly indicate that antigen selection may play a crucial role.20,21 One of the major factors determining CLL heterogeneity is the different cell of origin of the 2 molecular subtypes of the disease related to the immunoglobulin heavy-chain variable region (IGHV) mutational status (Table 2).22,23 CLL with unmutated IGHV (U-CLL) originates from B cells that have not passed through the germinal center and usually behaves more aggressively than CLL with mutated IGHV (M-CLL), which is derived from post–germinal center B cells. Gene expression profiling studies have suggested that M-CLL derives from a CD5+CD27+ post–germinal center memory B cell whereas U-CLL emerges from a pre–germinal center CD5+CD27− B cell that upregulates CD27 upon T-cell–independent antigen stimulation.24 Concordant with this idea, recent epigenetic studies have identified that M-CLL has a signature reflecting that its cell of origin has gone through the germinal center (ie, memory-like) whereas U-CLL originates from a cell that has maturated outside of the germinal center and still maintains a naive-like epigenetic signature.25,26 Intriguingly, these studies also identified a third epigenetic subtype with an intermediate methylation profile between the naive-like (U-CLL) and memory-like (M-CLL) CLLs, suggesting that it could originate in a not-yet-identified normal B cell. The 3 epigenetic CLL subtypes (naive-like, intermediate, and memory-like) differ in the profile of somatic mutations, usage of IGHV genes, and clinical outcome.25-28
Cells of origin and molecular subtypes of CLL and MCL
| . | CLL* . | MCL* . | |||
|---|---|---|---|---|---|
| IGHV-unmutated . | IGHV-mutated . | Conventional . | Leukemic nonnodal . | ||
| Gene expression24,109 | CD5+CD27−; pre–germinal center cell | CD5+CD27+; post–germinal center cell | Naive B-cell–like | Memory B-cell–like | |
| Germinal center relation, IGHV germ line identity | Unexperienced; IGHV ≥ 98% | Experienced; IGHV < 98% | Unexperienced† | Experienced† | |
| Epigenetic signature25-27 | Naive-like‡ | Intermediate‡ | Memory-like‡ | Naive-like | Memory-like |
| IGHV germ line identity, % (mean ± SD)25,27,109,111 | 99.7 (±1.0) | 96.4 (±2.4) | 92.9 (±3.2) | 98.7 (±2.6)† | 95.1 (±1.5)† |
| IGHV bias25,109,111 | IGHV4-34 | IGHV4-34 | |||
| IGHV1-69 | IGHV3-21 | IGHV4-34 | IGHV5-51 | IGHV5-51 | |
| IGHV4-39 | IGHV1-18 | IGHV3-7 | IGHV3-21 | IGHV1-8 | |
| IGHV3-23 | IGHV4-59 | ||||
| Stereotypes§,21,25,168 | Subsets 1, 6, 8 | Subset 2 | Subsets 4, 16 | ||
| Gene mutations66,116,168,169 | NOTCH1, ATM | SF3B1, ATM | MYD88 | ATM, CDKN2A del | CCND1, TLR2 |
| . | CLL* . | MCL* . | |||
|---|---|---|---|---|---|
| IGHV-unmutated . | IGHV-mutated . | Conventional . | Leukemic nonnodal . | ||
| Gene expression24,109 | CD5+CD27−; pre–germinal center cell | CD5+CD27+; post–germinal center cell | Naive B-cell–like | Memory B-cell–like | |
| Germinal center relation, IGHV germ line identity | Unexperienced; IGHV ≥ 98% | Experienced; IGHV < 98% | Unexperienced† | Experienced† | |
| Epigenetic signature25-27 | Naive-like‡ | Intermediate‡ | Memory-like‡ | Naive-like | Memory-like |
| IGHV germ line identity, % (mean ± SD)25,27,109,111 | 99.7 (±1.0) | 96.4 (±2.4) | 92.9 (±3.2) | 98.7 (±2.6)† | 95.1 (±1.5)† |
| IGHV bias25,109,111 | IGHV4-34 | IGHV4-34 | |||
| IGHV1-69 | IGHV3-21 | IGHV4-34 | IGHV5-51 | IGHV5-51 | |
| IGHV4-39 | IGHV1-18 | IGHV3-7 | IGHV3-21 | IGHV1-8 | |
| IGHV3-23 | IGHV4-59 | ||||
| Stereotypes§,21,25,168 | Subsets 1, 6, 8 | Subset 2 | Subsets 4, 16 | ||
| Gene mutations66,116,168,169 | NOTCH1, ATM | SF3B1, ATM | MYD88 | ATM, CDKN2A del | CCND1, TLR2 |
SD, standard deviation.
Immunogenetic analyses of the BCR in both entities strongly indicate that antigen selection plays an important role in the clonal expansion of all subtypes independently of the cell of origin.
Although the IGHV germ line identity differs significantly between conventional (SOX11+) and leukemic nonnodal (SOX11−) MCL, there is not a clear cutoff to distinguish both subtypes.109
Oakes et al26 identified the same 3 epigenetic CLL subgroups based on the methylation levels of the binding sites of the transcription factors AP-1, EBF1, and RUNX3. They named these subgroups low-, intermediate-, and high-programmed CLL based on the comparison of the changes in CLL with those occurring in normal naive B cells upon stimulation with CD40L and IgM.
BCR structure and signaling
The relevant role of the BCR in CLL and MCL is highlighted by the recent success of inhibitors of this pathway in their treatment.29,30 The BCR is composed of an immunoglobulin molecule, usually immunoglobulin M (IgM) and IgD, and the heterodimer CD79a/CD79b. BCR stimulation activates a molecular complex of different elements, including Bruton tyrosine kinase (BTK), phospholipase Cγ2 (PLCG2), and the phosphoinositide 3-kinase (PI3K), which propagate downstream signals promoting cell survival, proliferation, release of chemokines, and increased migration of the cells (Figure 2).31-33 The signaling response to BCR engagement is variable among cases ranging from intense to anergy, an unresponsive status of the B cell induced by chronic antigen stimulation in the absence of T-cell cooperation.34 This different response has a major impact on the heterogeneous behavior of the disease, being usually more effective in U-CLL than M-CLL due to different factors, including the higher expression of elements that enhance BCR activation such as IgM, CD38, ζ-chain associated protein kinase 70 (ZAP70), or miR-155 overexpression.35-37 Attenuating factors of the BCR response are less known and include CD5, CD22, CD72, and the phosphatase Src homology 2 containing inositol 5′ phosphatase 1 (SHIP1), whose expression is higher in ZAP70− CLL (Figure 2).31,38,39
BCR-signaling pathway. The initial steps in the response to the activation of the BCR are the phosphorylation of the CD79 ITAM motifs by the kinase LYN, followed by the docking of SYK and the subsequent activation of different components of the signalosome, a molecular complex organized at the cell membrane that will expand the downstream signals.31,34-39 This complex includes, among others, B-cell linker (BLNK), BTK, and PLCG2, together with PI3Kδ, recruited also by the adjacent activated CD19. The phosphorylation of the signalosome components propagates downstream messengers with the release of Ca2+ and diacylglycerol (DAG) and activation of protein kinase C (PKC), AKT, mitogen-activated protein kinase (MAPK) pathway with RAS/MEK/ERK, and the Rho-family GTPase among others. These signals lead to the activation of different transcriptions factors such as NF-κB, nuclear factor of activated T cells (NFAT), MYC, which result in the activation of cell survival and proliferation mechanisms, release of chemokines, and increase motility of the cells. The high expression of different elements such as IgM, ZAP70, and CD38 enhances the BCR response whereas other factors such as the phosphatase SHIP1, activated by LYN, act as a negative autoregulatory loop of the BCR stimulation. Other negative regulators are CD5, CD72, and CD22. Ag, antigen; PIP2, phosphatidylinositol (4,5)-bisphosphate; PIP3, phosphatidylinositol (3,4,5)-trisphosphate; PTEN, phosphatase and tensin homolog.
BCR-signaling pathway. The initial steps in the response to the activation of the BCR are the phosphorylation of the CD79 ITAM motifs by the kinase LYN, followed by the docking of SYK and the subsequent activation of different components of the signalosome, a molecular complex organized at the cell membrane that will expand the downstream signals.31,34-39 This complex includes, among others, B-cell linker (BLNK), BTK, and PLCG2, together with PI3Kδ, recruited also by the adjacent activated CD19. The phosphorylation of the signalosome components propagates downstream messengers with the release of Ca2+ and diacylglycerol (DAG) and activation of protein kinase C (PKC), AKT, mitogen-activated protein kinase (MAPK) pathway with RAS/MEK/ERK, and the Rho-family GTPase among others. These signals lead to the activation of different transcriptions factors such as NF-κB, nuclear factor of activated T cells (NFAT), MYC, which result in the activation of cell survival and proliferation mechanisms, release of chemokines, and increase motility of the cells. The high expression of different elements such as IgM, ZAP70, and CD38 enhances the BCR response whereas other factors such as the phosphatase SHIP1, activated by LYN, act as a negative autoregulatory loop of the BCR stimulation. Other negative regulators are CD5, CD72, and CD22. Ag, antigen; PIP2, phosphatidylinositol (4,5)-bisphosphate; PIP3, phosphatidylinositol (3,4,5)-trisphosphate; PTEN, phosphatase and tensin homolog.
Another remarkable feature of CLL cells, similar to MCL, is that they have a striking bias in the use of IG genes and in some cases have an identical or quasi-identical amino acid sequence of the complementary determining region 3 (CDR3), indicating a strong antigen-driven selection (Table 2). These BCR stereotypes are detected in ∼30% of cases, most U-CLL but also in M-CLL.20,21 Some of these stereotypes are associated with particular genomic alterations and clinical features suggesting that the immunogenetic origin may be an important determinant of the tumor biology.20,21,40
The search for antigens triggering CLL clonal selection has identified numerous autoantigens and environmental antigens exposed in apoptotic cells, microorganisms, and rheumatoid factors.41,42 Interestingly, recent studies have revealed that the IG of the CLL BCR may recognize homotypic epitopes generating interactions between IG molecules of the same or different cells able to trigger autonomous downstream signaling without the need of exogenous antigens.43,44 These homotypic interactions vary depending on the structure of the IG and generate responses of different intensity that may influence the behavior of the tumor.44 Whether this autonomous BCR signaling is sufficient to drive initial CLL leukemogenesis or requires additional factors needs to be further clarified.
CLL microenvironment
CLL cells recirculate between peripheral blood and tissues, mainly lymph nodes and bone marrow, where they receive survival and proliferation signals.45,46 These crucial interactions mainly occur in the proliferation centers of lymph nodes infiltrated by CLL that contain a loose meshwork of follicular dendritic cells (FDCs), a variable number of T cells, and other stromal cells (Figure 3).46-48 Bone marrow also provides a supportive microenvironment although less structurally organized. Tumor cells in proliferation centers become larger and have increased expression of downstream targets of the BCR signaling, NF-κB activation, cytokines, and antiapoptotic and proliferation-related markers.46,49-51 The expansion and high proliferation of these centers are associated with more aggressive behavior of the disease.52,53
CLL microenvironment. Tumor cells migrate to tissues attracted by the chemokines CCL19 and CCL21 produced by high endothelial venules, CXCL12 mainly secreted by nurse-like and stromal cells, and the CXCL13 of the FDCs, which interact with the tumor cell receptors CCR7, CXCR4, and CXCR5, respectively. Adhesion molecules (CD49d, CD44, CD62L) and their ligands (VCAM1, extracellular matrix proteins, CD34, among others) facilitate migration and homing of tumor cells. The presence of antigens in these niches may activate BCR signaling. TLRs recognize molecular patterns that activate the NF-κB pathway via interleukin-1 receptor activated kinase (IRAK)1/4 and MYD88, leading to increased secretion of inflammatory cytokines.163 This pathway may cooperate with the BCR stimulation, particularly in M-CLL.164 IL-4 expression by T cells enhances IgM and CD79b expression, particularly in U-CLL, amplifying the BCR signaling.165,166 Survival and proliferation stimuli are mainly provided by T cells, particularly through CD40L, and nurse-like and stromal cells through CD31, a proliferation-inducing ligand (APRIL), and B-cell activating factor (BAFF). All of these stimuli play an important role protecting tumor cells from drug-induced apoptosis. The downmodulation of CXCR4 on the CLL cells in tissue facilitates their return to the peripheral blood. CLL cells organize this favorable niche by secreting soluble factors, direct contact with surrounding cells, and release of extracellular vesicles that attract and activate stromal cells.
CLL microenvironment. Tumor cells migrate to tissues attracted by the chemokines CCL19 and CCL21 produced by high endothelial venules, CXCL12 mainly secreted by nurse-like and stromal cells, and the CXCL13 of the FDCs, which interact with the tumor cell receptors CCR7, CXCR4, and CXCR5, respectively. Adhesion molecules (CD49d, CD44, CD62L) and their ligands (VCAM1, extracellular matrix proteins, CD34, among others) facilitate migration and homing of tumor cells. The presence of antigens in these niches may activate BCR signaling. TLRs recognize molecular patterns that activate the NF-κB pathway via interleukin-1 receptor activated kinase (IRAK)1/4 and MYD88, leading to increased secretion of inflammatory cytokines.163 This pathway may cooperate with the BCR stimulation, particularly in M-CLL.164 IL-4 expression by T cells enhances IgM and CD79b expression, particularly in U-CLL, amplifying the BCR signaling.165,166 Survival and proliferation stimuli are mainly provided by T cells, particularly through CD40L, and nurse-like and stromal cells through CD31, a proliferation-inducing ligand (APRIL), and B-cell activating factor (BAFF). All of these stimuli play an important role protecting tumor cells from drug-induced apoptosis. The downmodulation of CXCR4 on the CLL cells in tissue facilitates their return to the peripheral blood. CLL cells organize this favorable niche by secreting soluble factors, direct contact with surrounding cells, and release of extracellular vesicles that attract and activate stromal cells.
The interactions between CLL cells, stromal cells, and the extracellular matrix are mediated by a complex network of adhesion molecules, cell surface ligands, chemokines, cytokines, and their respective receptors (Figure 3).54 Tumor cells actively organize their supportive “inflammatory” and “immune protective” microenvironment using different mechanisms that include secretion of soluble factors, direct contact with surrounding cells, and release of extracellular vesicles (Figure 3). Activated CLL cells by nurse-like cells or BCR stimulation secrete chemokines (CCL3, CCL4, and CCL22) and angiogenic factors that attract T and different stromal cells.32,54,55 Direct contact of CLL with T cells induce abnormal synapses impairing their antitumor response.56,57 The survival signals delivered by macrophages and stromal cells also require their direct contact with CLL cells that activate different pathways both in CLL and stromal cells including LYN, PKCβII, and NF-κB.58,59 These communication networks are also mediated by tumor-released extracellular vesicles carrying noncoding RNA and proteins that are internalized by distant T cells, monocytes, and stromal cells, enhancing their proinflammatory signals and promoting an immunoprotective environment.60-63
Genomic alterations
Genome-sequencing studies already performed in >1000 cases, including a subset of monoclonal B-cell lymphocytosis (MBL), have elucidated the tremendous mutational diversity of this disease.64-68 CLL has a low burden of somatic mutations (average 2500 per tumor).66 The number and type of mutations highly correlate with the IGHV mutational status. M-CLL accumulates more mutations than U-CLL (3000 vs 2000) despite their more favorable outcome. There are 3 main mutational signatures operational in CLL, corresponding to aging, activation-induced cytidine deaminase (AID), and a previously unknown signature named noncanonical AID (nc-AID) although it does not appear to be mediated by AID.66,68,69 This latter signature is responsible for the higher number of mutations in M-CLL, and is detected in other lymphoid neoplasms derived from germinal center–experienced cells.69
The relatively small number of genetic alterations that have shaped the prognostic models of CLL for almost 2 decades, such as del(13q)/miRs-15a/16-1, del(11q)/ATM, del(17p)/TP53, or tri12,70 has rapidly expanded to >60 driver71 genes and >12 recurrent structural variants (Figure 4).66-68 The profile of these mutations is very heterogeneous with few genes mutated at moderate frequency and a larger amount altered in <5% of the cases. Intriguingly, ∼15% of cases, mainly M-CLL, do not have any of the recurrent genetic lesions described, raising the possibility that uncommon mutations, epigenetic alterations, and/or specific BCR determinants may contribute to the growth and expansion of these apparently driver-less CLL.72,73
CLL mutational landscape. Comparison of CLL driver alterations in different cohorts of patients. (A) Mutational frequencies for a subset of 28 genes identified as frequent driver alterations in 2 whole genome/exome studies (International Cancer Genome Consortium [ICGC],66 Dana-Farber [DF]67 ) and 1 target sequencing at high coverage by Nadeu et al73 (Deep-Seq). Red, ICGC-CLL (n = 452); purple, ICGC-MBL (n = 54); blue, DF-pretreated CLL (DF-pre) (n = 123); green, DF-CLL8 cases enrolled in the clinical trial CLL8 (n = 278); and yellow, Deep-Seq (n = 406). The distribution of genomic alterations in MBL is similar to those of population-based CLL. Mutations in SF3B1, POT1, ATM, TP53, and RPS15 are more frequent in CLL8. MYD88 mutations are uncommon in clinical trial patients but are frequent in the DF-untreated, a cohort a decade younger that the ICGC. Detection of subclonal mutations by Deep-seq increases the global frequency of virtually all mutated genes. (B) Frequency of 11 CNA identified as putative driver alterations by Puente et al66 using single-nucleotide polymorphism (SNP) arrays (red, ICGC-CLL; purple, ICGC-MBL) and Landau et al67 using whole-exome sequencing data (blue, DF-pre; green, DF-CLL8). These CNAs were also analyzed by Nadeu et al73 by SNP arrays (yellow).
CLL mutational landscape. Comparison of CLL driver alterations in different cohorts of patients. (A) Mutational frequencies for a subset of 28 genes identified as frequent driver alterations in 2 whole genome/exome studies (International Cancer Genome Consortium [ICGC],66 Dana-Farber [DF]67 ) and 1 target sequencing at high coverage by Nadeu et al73 (Deep-Seq). Red, ICGC-CLL (n = 452); purple, ICGC-MBL (n = 54); blue, DF-pretreated CLL (DF-pre) (n = 123); green, DF-CLL8 cases enrolled in the clinical trial CLL8 (n = 278); and yellow, Deep-Seq (n = 406). The distribution of genomic alterations in MBL is similar to those of population-based CLL. Mutations in SF3B1, POT1, ATM, TP53, and RPS15 are more frequent in CLL8. MYD88 mutations are uncommon in clinical trial patients but are frequent in the DF-untreated, a cohort a decade younger that the ICGC. Detection of subclonal mutations by Deep-seq increases the global frequency of virtually all mutated genes. (B) Frequency of 11 CNA identified as putative driver alterations by Puente et al66 using single-nucleotide polymorphism (SNP) arrays (red, ICGC-CLL; purple, ICGC-MBL) and Landau et al67 using whole-exome sequencing data (blue, DF-pre; green, DF-CLL8). These CNAs were also analyzed by Nadeu et al73 by SNP arrays (yellow).
Genes commonly altered include NOTCH1 with mutations in the N-terminal and 3′ untranslated regions that truncate and stabilize the protein.66,74,75 NOTCH activation without mutations has been also found in ∼50% of CLL but the clinical significance of this finding is uncertain because it occurs at similar frequencies in U-CLL and M-CLL, whereas NOTCH1 mutations are more common in U-CLL than M-CLL (83% vs 17%, respectively).66,74,75 Another important player is the splicing regulator SF3B1 whose mutations impact in different downstream molecular processes such as DNA damage response, telomere maintenance, or NOTCH1 signaling.64,65,76-78 Other novel CLL genes are POT1, whose inactivation leads to telomere dysfunction, and MYD88, in which mutations tend to occur in younger patients, are associated with good prognosis, and lead to increased secretion of proinflammatory cytokines (CCL2, CCL3 and CCL4, interleukin 6 [IL6]) in response to Toll-like receptor (TLR) activation, reinforcing the role of genetic and microenvironment interactions.79-81
All of these mutations concentrate in a limited number of cellular processes, including DNA damage response (TP53, ATM, POT1), NOTCH1 signaling (NOTCH1, FBXW7), RNA maturation and export (SF3B1, XPO1), NF-κB (BIRC3, NFKBIE, EGR2, TRAF3, NFKB2), or B-cell signaling (MYD88, IKZF3, TLR2, BCOR, t(14;18)/BCL2, KRAS/NRAS), among others (Figures 4-5). By contrast, mutations in genes involved in chromatin remodeling or in cell cycle regulation, while relatively frequent in other lymphoid tumors including MCL, are scarce in CLL, with CHD2 as the most frequent alteration, almost exclusively present in M-CLL.82 Nonetheless, noncoding mutations in a PAX5 enhancer downregulate its expression, suggesting that mutations of regulatory elements may also contribute to CLL pathogenesis.66
CLL and MCL driver genes and molecular pathways. Main signaling pathways and mutated genes in CLL and MCL. Genes and pathways are divided in CLL-specific, MCL-specific, and those common to both entities.
CLL and MCL driver genes and molecular pathways. Main signaling pathways and mutated genes in CLL and MCL. Genes and pathways are divided in CLL-specific, MCL-specific, and those common to both entities.
Clonal and subclonal heterogeneity
The genomic landscape of CLL is highly dynamic. The variable frequency of mutations among studies reflects in part the heterogeneous clinical composition of the cohorts (Figure 4). The distribution of genomic alterations in MBL is similar to those of population-based CLL.66,83 However, mutations in SF3B1, POT1, ATM, RPS15, and TP53 are more frequent in patients entering clinical trials. The emergence of relapsed clones after chemotherapy, usually characterized by mutations in genes as TP53, IKZF3, or NRAS, is usually preceded by the presence of mutated subclones that can be detected prior to treatment.67,84,85 In contrast, mutations associated with resistance to ibrutinib observed in their target genes, BTK or PLCG2, are not usually detected before treatment or only at very low frequencies (below 2 in a million).86 However, they emerge progressively posttreatment and can be detected 3 to 15 months before clinical progression.87-89
Most CLL have a complex intratumor heterogeneity that influences the evolution of the disease. The earlier clonal alterations seem to correspond to the classical copy-number alterations (CNAs) del(13q), tri(12), and del(11q) that are followed by the acquisition of mutations in different genes.67,73,84,90 Sequencing at high coverage has expanded the detection of small mutated subclones in virtually all mutated genes (Figure 4).73,91-93 Small subclonal mutations in TP53, NOTCH1, NFKBIE, and POT1, among others, appear to have the same prognostic value as mutations in larger clones.73,91-93 Clonal tumors (ie, tumors in which all driver mutations are present in virtually all cells) have better outcome than cases with subclonal alterations.73,84 In addition, the increasing number of mutations worsens the evolution of tumors at diagnosis and relapse in both clonal and subclonal tumors.66,73,94 The influence of the number of mutations shortening the time to first treatment seems independent of specific drivers (TP53, SF3B1, ATM), clonal/subclonal composition of the tumor, and IGHV mutations.73 The increasing number of subclonal alterations confers shorter overall survival but it seems to be related to the IGHV status or TP53 mutations.66,67,73 These studies show the relevance of the subclonal architecture of CLL but need to be expanded in larger and homogeneously treated cohorts of patients.
The evolution of CLL is also associated with a marked alteration of the epigenome characterized by a global hypomethylation, particularly in the gene body and enhancers, which in part modulates the transcriptome.25,26 Intriguingly, increasing changes in the methylation of CLL correlate with the genomic complexity of the tumors, particularly in cases with subclonal genetic alterations whereas clonal cases maintain low methylation heterogeneity.95,96 The forces that govern the relationship between genomic and epigenomic alterations and the dynamic evolution of clonal and subclonal populations are poorly understood. These mechanisms are clinically relevant because chemotherapy selects high-risk fitter subclones, whereas new therapies promote the emergence of resistant clones.84,86,87,90
In spite of these advances, the use of this genomic information in the clinic is still limited. TP53 aberrations and IGHV mutational status already guide treatment decisions97-99 and NOTCH1 mutations may predict for impaired responses to rituximab.100,101 Up to 25 different genomic alterations appear to be associated with the outcome of the patients.66,67 Certain driver alterations, such as del(17p)/TP53, del(11q)/ATM, BIRC3, SF3B1, RPS15, or NOTCH1, are associated with worse outcome,66,85 whereas NOTCH1 mutations or loss of CDKN2A/B and 8p among others have been associated with Richter transformation.40,102-104 Unfortunately, the vast majority of these driver genes are mutated in very few patients (<2%) and are frequently found in combination with other adverse prognostic parameters preventing an easy evaluation of their clinical impact.66,73 The genomic information of the evolution of the disease is still limited.67,85 Novel agents may change the prognostic value of some alterations. Therefore, studies in large cohorts of patients encompassing the whole spectrum and treatments of the disease, using high-coverage sequencing to capture the subclonal composition are needed to evaluate the clinical implications of the CLL heterogeneous mutational landscape.105
Mantle cell lymphoma
Genetic susceptibility
The role of genetic susceptibility in MCL is less documented than in CLL. However, epidemiological studies have reported a twofold significant increase of hematological neoplasms among first-degree relatives of patients with MCL.7 Germ line mutations in ATM and CHK2 have been identified in occasional patients but, similar to CLL, these genes are not involved in the few studied families with lymphoid neoplasms and MCL.106 The low incidence of MCL is a challenge to identifying possible susceptibility loci.
Cells of origin and molecular subtypes
MCL is genetically characterized by the translocation t(11;14)(q13;q32) and CCND1 overexpression that is acquired in pre-B cells.107 This initial event is followed by 2 different molecular pathways that configure 2 distinct subtypes of the disease that parallel the 2 main CLL molecular subtypes (Tables 2-3; Figure 6).108,109 The most common form of MCL derives from mature B cells that have bypassed the germinal center and carry no or a limited number of IGHV mutations. The second, less common, subtype also derives from cells that have the t(11;14) but have experienced the germinal center and carry a higher number of IGHV mutations. These 2 subsets retain DNA methylation and expression signatures reminiscent of naive and memory B cells, respectively.109,110 Similarly to CLL, the BCR in MCL has a marked bias in the use of IGHV genes and 10% of cases carry IG-stereotyped sequences, that, although different from those used by CLL, highlight the influence of antigen selection in the clonal expansion of tumor cells (Tables 2-3).109,111,112
Clinical and biological characteristics of the 2 MCL subtypes
| . | Conventional MCL . | Leukemic nonnodal MCL . |
|---|---|---|
| Male/Female113-116,118 | 3-4 | 1 |
| Clinical presentation113-116,118 | Lymphadenopathy | Leukemic* |
| Extranodal | Splenomegaly* | |
| Cell of origin109,110 | Naive-like | Memory-like |
| Morphology113-115,118,170 | Classic*/blastoid | Small cell*/blastoid/plasma cell differentiation (37%) |
| Phenotype113-115,118,171 | ||
| SOX11 | + | − |
| CD5 | + (90%-100%) | − (25%-50%) |
| CD200 | − (90%) | + (40%-90%) |
| Chromosomal instability109,113-116,118 | High | Low |
| Somatic mutations, %116 | ||
| ATM | 55 | 0 |
| TP53 | 25 | 25 |
| CDKN2A del | 20 | 0 |
| CCND1 | 18 | 86 |
| KMTD2/MLL2 | 18 | 0 |
| NSD2/WHSC1 | 15 | 0 |
| UBR5 | 14 | 0 |
| BIRC3 | 7 | 7 |
| NOTCH2 | 6 | 4 |
| NOTCH1 | 4 | 0 |
| TLR2 | 0 | 3 |
| Clinical behavior113-115,118 | Aggressive | Stable/indolent |
| . | Conventional MCL . | Leukemic nonnodal MCL . |
|---|---|---|
| Male/Female113-116,118 | 3-4 | 1 |
| Clinical presentation113-116,118 | Lymphadenopathy | Leukemic* |
| Extranodal | Splenomegaly* | |
| Cell of origin109,110 | Naive-like | Memory-like |
| Morphology113-115,118,170 | Classic*/blastoid | Small cell*/blastoid/plasma cell differentiation (37%) |
| Phenotype113-115,118,171 | ||
| SOX11 | + | − |
| CD5 | + (90%-100%) | − (25%-50%) |
| CD200 | − (90%) | + (40%-90%) |
| Chromosomal instability109,113-116,118 | High | Low |
| Somatic mutations, %116 | ||
| ATM | 55 | 0 |
| TP53 | 25 | 25 |
| CDKN2A del | 20 | 0 |
| CCND1 | 18 | 86 |
| KMTD2/MLL2 | 18 | 0 |
| NSD2/WHSC1 | 15 | 0 |
| UBR5 | 14 | 0 |
| BIRC3 | 7 | 7 |
| NOTCH2 | 6 | 4 |
| NOTCH1 | 4 | 0 |
| TLR2 | 0 | 3 |
| Clinical behavior113-115,118 | Aggressive | Stable/indolent |
These parameters are more frequent in the subtype indicated but not exclusive.
MCL pathogenesis and molecular subtypes. MCL primary oncogenic event is the t(11;14) leading to CCND1 overexpression. The differential expression of SOX11 seems to be a relevant factor defining the 2 molecular subtypes. SOX11 expression represses BCL6 that may prevent cells from entering the germinal center. These cells carry unmutated IGHV and are genetically unstable. SOX11 may block the terminal B-cell differentiation of cells by forcing PAX5 expression and promote tumor cell growth, migration, and homing to lymph nodes via activation of the CXCR4/FAK/PI3K/AKT axis. SOX11− MCL cells enter the germinal center, carry mutated IGHV, and are genetically stable. The SOX11− tumor cells have very low invasive potential and remain in the blood as leukemic disease. Both MCL subtypes, conventional and leukemic nonnodal, may acquire additional genetic events such as TP53 mutations than promote progression to aggressive variants.
MCL pathogenesis and molecular subtypes. MCL primary oncogenic event is the t(11;14) leading to CCND1 overexpression. The differential expression of SOX11 seems to be a relevant factor defining the 2 molecular subtypes. SOX11 expression represses BCL6 that may prevent cells from entering the germinal center. These cells carry unmutated IGHV and are genetically unstable. SOX11 may block the terminal B-cell differentiation of cells by forcing PAX5 expression and promote tumor cell growth, migration, and homing to lymph nodes via activation of the CXCR4/FAK/PI3K/AKT axis. SOX11− MCL cells enter the germinal center, carry mutated IGHV, and are genetically stable. The SOX11− tumor cells have very low invasive potential and remain in the blood as leukemic disease. Both MCL subtypes, conventional and leukemic nonnodal, may acquire additional genetic events such as TP53 mutations than promote progression to aggressive variants.
The 2 molecular subtypes of MCL also diverge in their gene and microRNA expression profiles, genomic alterations, and clinical behavior (Table 3; Figure 6).108-111,113-118 Conventional MCL derives from naive-like B cells, expresses the oncogenic transcription factor SOX11, is genetically unstable, and tends to accumulate alterations in cell cycle regulatory genes, the DNA damage response pathway, chromatin modifiers, and cell survival mechanisms. Clinically, these cases usually have generalized lymphadenopathy and follow an aggressive clinical course. In contrast, MCL derived from memory-like cells is genetically stable, SOX11 is negative,119,120 and tumor cells mainly involve peripheral blood and spleen but not lymph nodes in early stages. This subtype has been recently recognized in the updated World Health Organization (WHO) classification as leukemic nonnodal MCL.121 The disease is stable and asymptomatic for long periods but some tumors may acquire additional genetic alterations such as TP53, facilitating the progression of the disease and transformation to more aggressive forms.108,113,115 Some studies have reported poor prognosis of SOX11− cases but this seems to be associated with TP53 alterations.122,123
Oncogenic role of cyclin D1 and SOX11
The relevance of cyclin D1 in MCL is reinforced by the selection of additional alterations, such as amplifications of the translocated allele and mutations/truncations of the 3′ untranslated region that increase its transcriptional levels.124-126 Interestingly, a subset of MCL does not carry the t(11;14), despite having similar pathological and clinical characteristics.127,128 These cases express SOX11 and ∼50% carry CCND2 translocations to IG genes, emphasizing the pathogenic role of both SOX11 and D-cyclins in these tumors.128 Cyclin D1 probably facilitates the transformation of B cells by dysregulating the G1/S-phase transition of the cell cycle. Cyclin D1 has been involved in other functions such as regulation of transcription,129-131 DNA damage response, or cell death among others.132-134 However, whether cyclin D1 plays any of these functions in MCL is not known. The observation that cyclin D1 inhibits the proapoptotic protein BAX in MCL expands its oncogenic mechanisms in these tumors.135
The other oncogenic element in MCL is SOX11, a transcription factor constitutively expressed in conventional MCL but absent in leukemic nonnodal MCL, all normal lymphoid cells, and virtually all mature B-cell neoplasms with the exception of 25% to 50% of Burkitt lymphomas.136,137 In contrast to CCND1, no genetic lesions are associated with SOX11 upregulation in MCL. However, a recent study has identified a distant enhancer whose activation seems to drive SOX11 expression through a 3-dimensional interaction with the promoter.110 SOX11 acts as an oncogene in MCL, promoting tumor cell growth in vivo and regulating a broad transcriptional program that includes B-cell differentiation, cell proliferation, and tumor cell–microenvironment interactions among others.138-140 One direct target of SOX11 is PAX5, a master regulator of B-cell differentiation whose downregulation is required for plasma cell development. The forced expression of PAX5 by SOX11 may contribute to lymphomagenesis by blocking this differentiation process.138 On the other hand, SOX11 represses BCL6 expression, which may prevent cells from entering the germinal center.139 Additionally, SOX11 regulates interactions of MCL cells with the microenvironment promoting angiogenesis through the induction of platelet-derived growth factor α (PDGFA),140 and MCL migration and stromal-mediated drug resistance through direct regulation of CXCR4 and FAK expression and focal adhesion kinase (FAK)/PI3K/AKT pathway activation.141 These mechanisms are consistent with the extensive nodal dissemination and more aggressive behavior of conventional SOX11+ MCL and provide new potential targets for intervention.
BCR signaling and microenvironment
Similarly to CLL, the tissue microenvironment provides an essential niche for MCL survival, proliferation, and drug resistance. These functions are activated through different mechanisms that include BCR activation of mucosa-associated lymphoid tissue 1 (MALT1)-driven MYC signaling, autocrine secretion of IL-1β, tumor necrosis factor α (TNFα), and CCL5, and stromal-induced FAK activation.142-144 BCR and NF-κB activation mainly occurs in lymph nodes, but some leukemic cases have this activation independent of the nodal microenvironment.145 Interactions between MCL cells and the microenvironment are mediated by different chemokines, adhesion molecules, and their respective receptors that are similar to those used by CLL (Figure 3).146-148 The immunoprotective role of the microenvironment in MCL is less known but high CD4:CD8 ratios seem related to more indolent behavior.149
Genomic alterations
MCL has increased genome instability compared with CLL, with a high number of structural alterations per case, frequently including chromothripsis/chromoplexy.116 However, recurrent translocations are relatively infrequent, with the exception of MYC, and the t(11;14) hallmark.150 By contrast, recurrent CNAs are common, with >90% of cases harboring at least 1. Some of them affect known target genes mainly related to cell cycle regulation, DNA damage response, and cell survival such as loss of 9p21/CDKN2A, 11q22/ATM, 13q14/RB1 or 17p13/TP53, and gain/amplification of 8q24/MYC, 10p12/BMI1, 12q13/CDK4 or 18q21/BCL2. Other recurrent alterations affect larger regions with no clear target genes, such as loss of 1p, 8p, or 9q, and gain of 3q, 7p, or 15q. All of these genetic alterations tend to accumulate in tumors with high proliferation and poor outcome. The high expression of a proliferation signature is considered a biological integrator of multiple genetic alterations that robustly discriminate the evolution of patients and can now be reliably detected in routine clinical samples.151,152
Genome studies have increased the number of known mutated genes in MCL.116,153-156 The number of somatic mutations is higher than in CLL (average 3700 per case), and most of them can be classified in the same 3 major mutational signatures described in CLL. The most common secondary alteration is the mutation of ATM (42%-55%), usually associated with 11q deletions, followed by mutation/deletion of TP53 (28%), reflecting the importance of DNA repair in the pathogenesis of MCL (Table 3; Figure 5). CCND1 mutations with the AID signature occur predominantly in IGHV-mutated MCL. Novel mutated pathways include NOTCH1/NOTCH2 (10%-14%) associated with poor outcome; chromatin modifiers MEF2B, KMTD2, NSD2, and TET2 (30%-50%), or the ubiquitin ligase UBR5 (18%), seldom mutated in CLL. Somatic mutations in the NF-κB pathway include BIRC3 (6%-10%), TRAF2 (15%), and less frequently TLR2, CARD11, MAP3K14, and IKBKB.116,153-156 Alterations in the alternative NF-κB pathway have been associated with resistance to inhibitors of the BCR pathway.157,158 BTK mutations have been identified in occasional MCL treated with ibrutinib,159 but the main mechanism of primary resistance seems to be related to the activation of FAK and the PI3K–AKT–mammalian target of rapamycin (mTOR) pathway that bypasses the BTK-inhibitory effect.143,159,160
Similar to IGHV subtypes in CLL, the distribution of MCL driver genes differs considerably between SOX11+ and SOX11− tumors.116 Thus, despite the limited number of cases analyzed, ATM, CDKN2A, and chromatin-remodeling genes are almost exclusively altered in SOX11+ cases (Table 3), whereas TLR2 mutations appeared only in SOX11− tumors. TP53, NOTCH2, and BIRC3 are mutated in both MCL subtypes.
The epigenome of MCL is variably altered compared with that of normal B cells.110 Similarly to CLL, the increasing number of somatic mutations seems to correlate with higher methylation changes and this epigenetic complexity is associated with higher proliferation and worse outcome, suggesting that interactions between genomic and epigenomic changes may influence the evolution of these tumors.110
Conclusions and perspectives
Recent biological and genomic studies have expanded our knowledge of the similarities and differences between CLL and MCL. They share common cells of origin in antigen-experienced cells that either have bypassed or experienced the germinal center, and dramatically influence the subsequent development of 2 different molecular and clinical subtypes in each disease. Genomic studies have virtually completed the profile of somatic mutations of pretreated CLL66,67,161 but the information in MCL and the genomic/epigenomic alterations in the evolution of both diseases is still very limited. The function of many new genes is also unknown. All of this information is essential to understanding the cross talk between tumor cells and the microenvironment and the interactions between genomic and epigenomic modifications that define the evolution of these tumors. One of the major challenges for implementing biology-based management strategies will be understanding of the mechanisms generating the clonal heterogeneity of the diseases and the dynamics of its evolution.162 The translation into the clinics of these new perspectives will require an integrated effort to better understand these basic mechanisms in the context of the complex evolution of the diseases.
Acknowledgments
The authors thank Carlos Lopez-Otín (Universidad de Oviedo); Virginia Amador, Silvia Beà, Dolors Colomer, Julio Delgado, Jose Ignacio Martín-Subero, and Armando Lopez-Guillermo (Hospital Clinic of Barcelona, IDIBAPS) for their helpful comments on the manuscript; and Silvia Xargay (University of Girona) and Ferran Nadeu (IDIBAPS) for their assistance in the design of the figures. The authors apologize to the authors whose work was not included due to space limitations.
X.S.P. was supported by Ministerio de Economía y Competitividad (MINECO) SAF2013-45836-R and Centro de Investigación Biomédica en Red de Cáncer (CIBERONC). P.J. was supported by grant FIS (PI14/00355) from the Instituto de Salud Carlos III. E.C. was supported by grants from Instituto de Salud Carlos III (PMP15/00007, CIBERONC, and ERA-NET TRANSCAN initiative [TRS-2015-00000143] AC15/00028, which is cofinanced by European Regional Development funds, “Una Manera de Hacer Europa”), MINECO SAF2015-64885-R y SAF2016-81860-REDT, Generalitat de Catalunya AGAUR 2014-SGR-795, and Gilead Spain (GLD15/00288).
E.C. is an Academia Researcher of the Institució Catalana de Recerca i Estudis Avançats (ICREA) of the Generalitat de Catalunya. P.J. and E.C. belong to Centres de Recerca de Catalunya (CERCA) Program/Generalitat de Catalunya.
Authorship
Contribution: X.S.P., P.J., and E.C. designed the review and wrote the manuscript.
Conflict-of-interest disclosure: E.C. has received research funding from Gilead Sciences, has provided expert testimony for Gilead Sciences, and is named inventor on 2 patents filed by the National Cancer Institute: “Methods for selecting and treating lymphoma types” licensed to NanoString Technologies and “Evaluation of mantle cell lymphoma and methods related thereof.” The remaining authors declare no competing financial interests.
Correspondence: Elías Campo, Unitat Hematopatologia, Hospital Clínic, Villarroel 170, 08036-Barcelona, Spain; e-mail: ecampo@clinic.cat.



![Figure 4. CLL mutational landscape. Comparison of CLL driver alterations in different cohorts of patients. (A) Mutational frequencies for a subset of 28 genes identified as frequent driver alterations in 2 whole genome/exome studies (International Cancer Genome Consortium [ICGC],66 Dana-Farber [DF]67) and 1 target sequencing at high coverage by Nadeu et al73 (Deep-Seq). Red, ICGC-CLL (n = 452); purple, ICGC-MBL (n = 54); blue, DF-pretreated CLL (DF-pre) (n = 123); green, DF-CLL8 cases enrolled in the clinical trial CLL8 (n = 278); and yellow, Deep-Seq (n = 406). The distribution of genomic alterations in MBL is similar to those of population-based CLL. Mutations in SF3B1, POT1, ATM, TP53, and RPS15 are more frequent in CLL8. MYD88 mutations are uncommon in clinical trial patients but are frequent in the DF-untreated, a cohort a decade younger that the ICGC. Detection of subclonal mutations by Deep-seq increases the global frequency of virtually all mutated genes. (B) Frequency of 11 CNA identified as putative driver alterations by Puente et al66 using single-nucleotide polymorphism (SNP) arrays (red, ICGC-CLL; purple, ICGC-MBL) and Landau et al67 using whole-exome sequencing data (blue, DF-pre; green, DF-CLL8). These CNAs were also analyzed by Nadeu et al73 by SNP arrays (yellow).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/21/10.1182_blood-2017-10-764373/4/m_blood764373f4.jpeg?Expires=1769098340&Signature=JFdCWZ2LJbN2aKyHVumW4a~0LHl2E83CCkr8I-ADOJa4j-rLY6KRXAxV6CxYtzc42VnDB-bBkeu1qz05TxEUXfm5NLRp78rwjePupCW2DHvcUUM3HnycTkkZTH7y1PB2Pm5veAgEw0XHZw4PfNUis2jieNazc9WAzX2RGnzOEXv0f7Qo4TAEvm3cw7q91P25YnQFFrTjA-6vM3mf8Zyd~TIByx9PlwT5Ef6rYtytOgKjbKP0EXBQzu0GzxlW5eWR9ylKw810JQuBjeVCxR90Ml9N7jBSYg94pTW7972MwMREFEPdCo9xGTVf2qgZ3~Xr4SdLR-gAu5j~t2XuyYqYcQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

