Abstract
Diffuse large B-cell lymphoma (DLBCL), the most frequent subtype of lymphoid malignancy, remains a significant clinical challenge, as ∼30% of patients are not cured. Over the past decade, remarkable progress has been made in the understanding of the pathogenesis of this disease, spurred by the implementation of powerful genomic technologies that enabled the definition of its genetic and epigenetic landscape. These studies have uncovered a multitude of genomic alterations that contribute to the initiation and maintenance of the tumor clone by disrupting biological functions known to be critical for the normal biology of its cells of origin, germinal center B cells. The identified alterations involve epigenetic remodeling, block of differentiation, escape from immune surveillance, and the constitutive activation of several signal transduction pathways. This wealth of new information offers unique opportunities for the development of improved diagnostic and prognostic tools that could help guide the clinical management of DLBCL patients. Furthermore, a number of the mutated genes identified are potentially actionable targets that are currently being explored for the development of novel therapeutic strategies. This review summarizes current knowledge of the most common genetic alterations associated with DLBCL in relation to their functional impact on the malignant transformation process, and discusses their clinical implications for mechanism-based therapeutics.
Introduction
Diffuse large B-cell lymphoma (DLBCL), the most common lymphoid malignancy in adulthood, is a heterogeneous disease that can arise de novo or from the histologic transformation of more indolent lymphomas, most commonly, follicular lymphoma (FL) and chronic lymphocytic leukemia (CLL).1 Although durable remissions can be achieved in >50% of cases, even at advanced stage, DLBCL remains a challenging clinical problem, with approximately one-third of patients not being cured by standard-of-care immunochemotherapeutic regimens.2,3 Current limits to effective treatment are related in part to the striking heterogeneity of this disease, which can be recognized at the morphologic, genetic, immunophenotypic, and clinical level. Indeed, modern genome-wide molecular analysis of DLBCL uncovered a multitude of altered cellular pathways that play key roles in tumor development and maintenance, as well as in the response to therapy. These discoveries are set to provide a molecular framework for the development of improved diagnostic and prognostic markers, allowing the design of more effective precision medicine approaches aimed at targeting oncogenic addictions specific to distinct lymphoma subtypes. This review focuses on the molecular pathogenesis of DLBCL not otherwise specified (NOS),1 with emphasis on the nature of recurrently involved genes/pathways that have been functionally characterized or clearly interpreted, and their implications for the development of novel targeted therapies. We refer the reader to other reviews for a more detailed survey on the expanding landscape of drugs targeting DLBCL,2,4 and a discussion on the increasingly important role of the tumor microenvironment, including its interplay with the lymphoma cells, in the pathogenesis of these tumors.5
Cell of origin and classification
DLBCL results from the malignant transformation of mature B cells that have experienced the germinal center (GC) reaction. GCs are dynamic microanatomical compartments that form when B cells are challenged by a foreign antigen, and represent the primary site for clonal expansion and antibody affinity maturation.6,7 These structures comprise two anatomically distinct areas where B cells constantly recycle bidirectionally: the dark zone (DZ), mostly composed of proliferating cells that mutate the variable region of their immunoglobulin (IG) genes through the process of somatic hypermutation (SHM); and the light zone (LZ), where B cells are selected to become either a plasma cell or a memory B cell based on their high affinity for the antigen, and also undergo class switch recombination (CSR) (Figure 1).6,7 The central role of the GC as the target structure of malignant transformation in lymphoma is highlighted by multiple observations, including evidence that DLBCLs carry somatically hypermutated IG genes,8 the occurrence of genetic lesions that are due to errors in GC-specific DNA remodeling events,9 and the similarity between the phenotype of the two major molecular subtypes of the disease (see next paragraph) and transcriptional programs that are associated with distinct functional phases of the GC.10,11
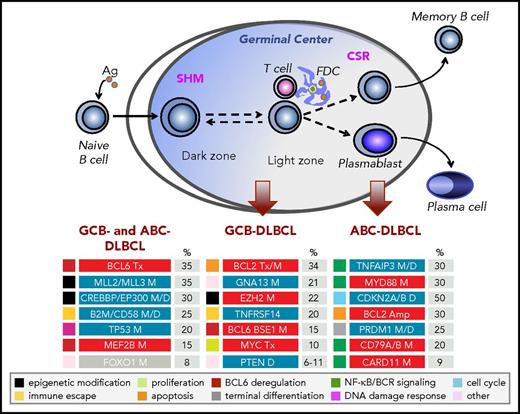
Cellular origin and genetic lesions associated with distinct DLBCL subtypes. Schematic representation of the GC reaction, and its relationship with the 2 molecular subtypes of DLBCL NOS, GCB-DLBCL, and ABC-DLBCL (unclassified DLBCL not shown). The most common, functionally characterized genetic alterations identified in this disease (including those shared across different subtypes and those subtype specific) are shown in the bottom panels, where blue indicates loss-of-function events and red indicates gain-of-function events; color codes on the left denote distinct categories, according to the subverted biological pathway. Ag, antigen; Amp, amplifications; D, deletions; FDC, follicular dendritic cells; M, mutations; Tx, chromosomal translocations. Note that, at lower frequencies, mutations affecting CARD11, TNFAIP3, and MYD88 residues other than the L265 hotspot can also be observed in GCB-DLBCL. CREBBP mutations can be found in all subtypes, although frequencies are significantly higher in GCB- (30%) than ABC- (15%) DLBCL. Modified from Pasqualucci and Dalla-Favera135 with permission.
Cellular origin and genetic lesions associated with distinct DLBCL subtypes. Schematic representation of the GC reaction, and its relationship with the 2 molecular subtypes of DLBCL NOS, GCB-DLBCL, and ABC-DLBCL (unclassified DLBCL not shown). The most common, functionally characterized genetic alterations identified in this disease (including those shared across different subtypes and those subtype specific) are shown in the bottom panels, where blue indicates loss-of-function events and red indicates gain-of-function events; color codes on the left denote distinct categories, according to the subverted biological pathway. Ag, antigen; Amp, amplifications; D, deletions; FDC, follicular dendritic cells; M, mutations; Tx, chromosomal translocations. Note that, at lower frequencies, mutations affecting CARD11, TNFAIP3, and MYD88 residues other than the L265 hotspot can also be observed in GCB-DLBCL. CREBBP mutations can be found in all subtypes, although frequencies are significantly higher in GCB- (30%) than ABC- (15%) DLBCL. Modified from Pasqualucci and Dalla-Favera135 with permission.
In 2001, the genome-wide analysis of gene expression profiles obtained from primary DLBCL biopsies led to the identification of at least 2 phenotypic subgroups of DLBCL NOS, with a subset of cases showing an intermediate, unclassifiable phenotype.10 Although both subtypes are more closely related to GC LZ B cells,11 the germinal center B-cell-like (GCB) DLBCL lacks the expression of early post-GC differentiation markers, whereas the activated B-cell-like (ABC) DLBCL displays a transcriptional signature analogous to that observed in mitogenically activated B cells or in a small subset of lymphocytes that are also located in the LZ of the GC and are poised to plasma cell differentiation (plasma blasts).10
DLBCL risk stratification according to cell of origin has prognostic value in the context of the current cyclophosphamide, doxorubicin, vincristine, and prednisone, plus rituximab–based therapeutic regimens, with GCB-DLBCL showing a more favorable course and ABC-DLBCL representing the most aggressive form of the disease.12-14 Application of this classification to the routine clinical practice has been facilitated by the development of novel technologies allowing the analysis of formalin-fixed paraffin-embedded tissue biopsies, such as the gene expression–based Nanostring platform.15 Accordingly, the distinction in GCB- and ABC-DLBCL has now been officially incorporated into the revised World Health Organization classification of hematologic malignancies.1
Other microarray-based gene expression profile studies captured an alternative taxonomy of DLBCL into groups that correlate with different aspects of DLBCL biology, including the tumor microenvironment, and are defined by the differential expression of genes implicated in oxidative phosphorylation, B-cell receptor (BCR) signaling, and host inflammatory responses.16
Mechanisms of genetic alteration
The pathogenesis of DLBCL represents a multistep process entailing the accumulation of multiple genetic lesions that alter the structure and/or the expression pattern of proto-oncogenes and tumor suppressor genes, as well as of other molecules of pathogenetic significance. Analogous to most human tumors, several mechanisms contribute to oncogenic dysregulation in DLBCL, including somatically acquired non–silent point mutations and gene copy number changes. In addition, the genome of DLBCL is altered by two mechanisms of genetic damage that are intimately connected to the physiologic IG DNA remodeling processes operating in B lymphocytes: (i) chromosomal translocations, due to errors occurring during VDJ recombination, SHM, and CSR9 ; and (ii) aberrant somatic hypermutation (ASHM), a byproduct of the activation-induced cytidine deaminase (AID)-mediated SHM process.17 The importance of the GC and its associated DNA remodeling events in the pathogenesis of lymphoma has been experimentally demonstrated in studies showing that deletion of Aicda, the enzyme required for CSR and SHM,18,19 in lymphoma-prone mouse models is sufficient to prevent the occurrence of MYC-IGH translocations and the development of GC-derived lymphomas.20,21
Chromosomal translocations
At variance with what is commonly observed in acute leukemias, DLBCL-associated chromosomal translocations do not typically generate fusion proteins. Rather, they juxtapose heterologous regulatory sequences (promoters or enhancers) derived from partner chromosomes, often involving the IG loci, in the proximity of the intact coding domains of proto-oncogenes, leading to deregulated expression of their normal proteins.22 As a consequence, proto-oncogenes that are tightly regulated in the preneoplastic counterpart may become constitutively expressed in the lymphoma cell (“homotopic deregulation”; eg, translocations of BCL6), whereas proto-oncogenes that are normally not expressed in the tumor precursor cell may become ectopically activated in the lymphoma cell (“heterotopic deregulation”; eg, translocations of MYC and BCL2, which encode for 2 proteins normally absent in most GC B cells23,24 ).
ASHM
The term ASHM defines a mechanism of genetic damage leading to the accumulation of multiple mutations around the 5′ sequences of genes that are otherwise found unmutated in normal GC B cells.17 The physiologic SHM activity relies on high-accuracy repair mechanisms25 that counteract the ability of AID to bind and mutate multiple DNA sequences across the genome.26,27 As a result, only the IG loci as well as a few additional loci (eg, BCL6) exhibit evidence of SHM in GC B cells. In contrast, over half of newly diagnosed DLBCL samples, irrespective of their molecular subtype (and, at lower frequencies, a few other B-cell non-Hodgkin lymphomas), show evidence of ASHM in >40 transcribed genes outside the IG loci, including the proto-oncogenes PIM1 and MYC.17,28 These mutations are biallelic, frequently multiple within the same allele, and display hallmarks of the SHM process, including the distribution within ∼2 kb from the transcription initiation site, the requirement for active transcription, a predominance of transversions over transitions, the preferential targeting of hotspot motifs, and frequencies that are several orders greater than the background mutation rate in mammalian cells.29 Depending on the genomic configuration of the target gene, both coding and noncoding regions can be affected. Thus, ASHM is considered a powerful mechanism of transformation that may cause perturbations in gene expression as well as changes in the structural and functional properties of numerous oncogenes or tumor suppressor genes. Because of the complexity of the genetic changes, a comprehensive assessment of the overall impact of ASHM on the pathogenesis of DLBCL is still lacking. Interestingly, recent work demonstrated that the sequences targeted by ASHM and chromosomal translocations are not randomly distributed across the genome, but are predominantly grouped within superenhancers or regulatory clusters in which sense and antisense transcription converge, indicating that these elements are key mediators of AID recruitment.30,31
The mutational landscape of DLBCL
The implementation of next-generation sequencing technologies, coupled with genome-wide functional shRNA or CRISPR/Cas9 screens, has revealed a remarkable structural and functional complexity in the DLBCL coding genome, compared with other hematologic malignancies. On average, each DLBCL biopsy displays 70 lesions affecting coding domains (including mutations and copy number aberrations), with great interindividual variability and a sizable fraction of genes found mutated in <10% of patients.32-36 Among these genes, ∼150 have been cataloged as significant drivers based on recurrence and mutation features in a recent most comprehensive analysis totaling ∼1000 DLBCL exomes.36 These figures may represent an underestimate of the functionally significant DLBCL mutation load, because the much greater noncoding portion of its genome remains largely unexplored, including sequences involved in ASHM, long noncoding RNAs, microRNAs, and other genomic regions with key regulatory functions. Indeed, initial whole genome sequencing analyses uncovered clusters of mutations at putative enhancers, and several mutation hotspots were found in untranslated regions.30,31 An additional layer of complexity that has only been marginally addressed in this neoplasm is represented by its significant intratumor subclonal diversity, with relapsed cases showing a pattern of early- and late-divergent evolution based on analysis of the rearranged IG variable region genes.37 Under the selective pressure imposed by the environment, including chemotherapy, this heterogeneity may give rise to the emergence of clones with improved fitness and possibly acquired resistance to therapy. Finally, the high number of genes that are mutated within a single case and across all DLBCL cases analyzed to date opens endless possibilities of cooperative interactions between genomic abnormalities, as well as between genetic lesions and cell signaling pathways, the significance of which remains currently unexplored. As we have only started to scratch the surface of this remarkably complex scenario, the next sections of this review cover the limited set of somatic genetic alterations that are most commonly observed in this malignancy and have been functionally interpreted. These include genomic abnormalities found in both COO subtypes and alterations specifically associated with GCB- or ABC-DLBCL. Of note, a study published while this review was in press and based on the analysis of combinatorial mutation patterns in ∼500 patients uncovered 4 genetic subgroups within the present COO classification, including one preferentially enriched in unclassified DLBCL.38 These groups display distinct transcriptional and clinical outcomes, providing a potential taxonomy for precision-medicine approaches. We note that a few germ line variants have also been found associated with DLBCL,39,40 but their contribution to heritable risk in the disease remains presently understudied.
Programs dysregulated in both GCB-DLBCL and ABC-DLBCL
Genetic lesions in histone/chromatin modifiers.
A consistent theme in DLBCL genomic analyses has been the discovery of recurrent mutations in genes that encode for histone/chromatin modifiers, including methyltransferases, acetyltransferases, and histones themselves32-34 (in particular, linker histone H1 family members,41 some of which are targets of ASHM), as well as, at low frequencies, ARID1A and TET2.36 Overall, alterations in at least 1 of these genes can be found in 85% of all DLBCL cases, and epigenetic modifiers comprised 11 of the 60 most highly recurrent targets identified in this lymphoma upon analysis of 1000 cases.36 Together, these findings point to a key role for epigenetic remodeling in lymphomagenesis. Of note, investigations aimed at reconstructing the history of clonal evolution during FL transformation (tFL) revealed that mutations of epigenetic modifiers represent early events introduced in a common ancestral clone before divergent evolution to FL or tFL.42-44 Because epigenetic changes, different from genetic events, are reversible, it is conceivable that drugs targeting the epigenome could provide therapeutic benefit to patients carrying these mutations by restoring physiologic acetylation or methylation levels, provided the tumor cells remain addicted to such modifications.
Inactivation of the methyltransferase KMT2D.
The KMT2D gene (also known as MLL2) encodes a member of the SET1 family of histone methyltransferases that induce an active chromatin conformation by predominantly mono- and dimethylating the lysine at position 4 of histone H3 (H3K4).45 In GC B cells, KMT2D occupies chromatin domains at enhancers, as well as at a smaller set of promoter regions, belonging to genes with key roles in B-cell physiology, including those involved in the positive regulation of apoptosis, in CD40 and BCR signaling, and in the control of cell migration.46,47 Monoallelic and, less commonly, biallelic somatic mutations of this gene are found in 30% of DLBCL cases, representing the single most frequent genetic aberration associated with this disease.32,33 KMT2D mutations comprise mostly truncating events that impair the protein enzymatic function by removing the C-terminal cluster of conserved domains, including the SET domain32,33 ; additionally, missense mutations in the same domains have been shown to suppress its ability to catalyze H3K4 methylation in vitro.47 Conditional deletion of Kmt2d in mouse pre-GC B cells, but not after antigen encounter (ie, during GC formation), leads to a significant expansion of GC B cells, supporting the notion derived from human tumors that KMTD2 inactivation is an early event, probably requiring multiple cell divisions to implement chromatin remodeling and gene expression changes.47 Mice with loss of Kmt2d coupled with BCL2 deregulation develop lymphoid malignancies that recapitulate the spectrum of phenotypes observed during human FL to DLBCL transition, demonstrating its tumor suppressor role in vivo.46,47 The demethylase involved in KMT2D-dependent modifications is not known and additional studies will be needed to identify vulnerabilities in KMT2D-mutated cells that could be therapeutically exploited.
Inactivation of the acetyltransferase genes CREBBP/EP300.
As many as one-third of DLBCL samples, with a significant prevalence in the GCB-DLBCL subtype, display somatic mutations and/or deletions affecting the acetyltransferase genes CREBBP (∼25%) and, less frequently, EP300 (5%).48 These enzymes are pleiotropic regulators of gene expression that catalyze the addition of acetyl groups to specific lysine residues in histone and nonhistone proteins, also including p53 (Figure 2).49,50 DLBCL-associated mutations and small indels disrupt the function of the CREBBP/EP300 proteins either by removing the histone acetyltransferase domain or by introducing amino acid changes within this domain, which cause diminished affinity for Acetyl-CoA, including a recurrent hotspot at R1446.48 In >80% of DLBCL cases, only 1 allele is affected in the tumor cells, and the residual wild-type allele is expressed, consistent with a haploinsufficient tumor suppressor function. In support of a dose-dependent pathogenic effect of these proteins, germ line loss of a single CREBBP (or EP300) allele is the causative genetic event in a rare congenital disorder known as Rubinstein-Taybi syndrome.51 Indeed, conditional deletion of CREBBP in B cells accelerated the development of FLs in several BCL2-driven mouse models.52-54
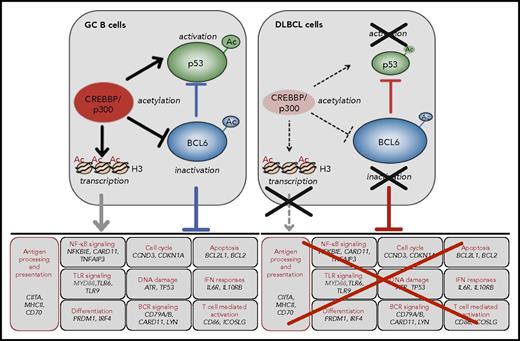
CREBBP modulated programs in the GC. CREBBP positively modulates multiple biological programs in the GC, through acetylation of histone and nonhistone proteins (eg, BCL6 and p53) (left). Prominent roles include its ability to regulate antigen presentation/processing via control of CIITA expression, and to counteract the activity of BCL6 by a dual mechanism entailing acetylation-mediated inactivation of its protein and the deposition of H3K27Ac marks on the promoter/enhancer regions of its target genes, which facilitates an active chromatin conformation (bottom panel, with representative cobound genes). This switch may allow the rapid induction of genes that are required for post-GC differentiation in the LZ. CREBBP-modulated programs are disrupted in tumors carrying inactivating mutations of its gene (right). Ac, acetylated lysines.
CREBBP modulated programs in the GC. CREBBP positively modulates multiple biological programs in the GC, through acetylation of histone and nonhistone proteins (eg, BCL6 and p53) (left). Prominent roles include its ability to regulate antigen presentation/processing via control of CIITA expression, and to counteract the activity of BCL6 by a dual mechanism entailing acetylation-mediated inactivation of its protein and the deposition of H3K27Ac marks on the promoter/enhancer regions of its target genes, which facilitates an active chromatin conformation (bottom panel, with representative cobound genes). This switch may allow the rapid induction of genes that are required for post-GC differentiation in the LZ. CREBBP-modulated programs are disrupted in tumors carrying inactivating mutations of its gene (right). Ac, acetylated lysines.
The mechanism by which CREBBP mutations contribute to lymphomagenesis involves the impairment of multiple biological programs that are critical to the normal GC reaction, and particularly to the LZ. Especially important is the ability of CREBBP to oppose the proto-oncogenic activity of BCL6 by a dual mechanism entailing (i) direct acetylation of the BCL6 protein, which prevents the recruitment of histone deacetylases (HDACs) and thus impairs its transrepressor function48,55 ; and (ii) H3K27 acetylation at the promoter/enhancer sequences of BCL6 target genes, which counteracts the repressive effect of BCL6 by facilitating transcription53,54 (Figure 2). Therefore, CREBBP may be involved in the switching between repressed and active chromatin states, allowing GC B cells to rapidly reprogram upon signals that are delivered at the exit of the GC. CREBBP-mediated acetylation is also required for the activation of the p53 tumor suppressor, which is itself a target of BCL6. Given that the amount of CREBBP in the cells is limited, haploinsufficiency may tip this balance by favoring the proto-oncogenic function of BCL6 over the tumor suppressor activity of p53. Finally, CREBBP binds and acetylates the regulatory sequences of several genes involved in antigen presentation/processing, including the CIITA transactivator and multiple major histocompatibility complex (MHC)-class II loci, thus contributing to tumor immune escape53,54,56 (Figure 2 and “Escape from immune surveillance”).
The identification of histone acetyltransferase mutations suggests that drugs targeting acetylation/deacetylation mechanisms (eg, HDAC inhibitors) could be effective in patients with CREBBP/EP300 mutant DLBCL. Although initial trials with HDAC inhibitors have yielded limited responses in unselected DLBCL patients,2 the efficacy of these drugs should be reevaluated in the context of proper patients stratification, and probably in combination with agents targeting cooperating oncogenic events. Also important will be the development of inhibitors that are specific for the involved deacetylases; among these, HDAC3 may represent an attractive therapeutic target in light of recent studies documenting its recruitment by BCL6 to repress transcription at shared chromatin targets.53
Deregulation of BCL6 activity.
BCL6 is a transcriptional repressor belonging to the BTB/Zinc finger family of transcription factors.57,58 In mature B cells, BCL6 is expressed exclusively in the GC, including all DZ and most LZ B cells, whereas it is downregulated in a subset of LZ B cells primed toward plasmablastic differentiation.59 BCL6 is an absolute requirement for the formation of GCs,60,61 where it negatively regulates multiple biological programs,62,63 including (i) cell cycle arrest (eg, by suppressing p21 expression,64 thus enforcing a highly proliferative program); (ii) the sensing and response to DNA damage (by regulating p53, ATR, and CHEK1,65-67 thus allowing to tolerate the physiologic DNA breaks involved in SHM and CSR); (iii) antiapoptosis programs (eg, by suppressing BCL224 ); (iv) terminal differentiation (by suppressing PRDM1/BLIMP168,69 ). These functions are restored once BCL6 is downregulated in the LZ, an event that is required for terminal differentiation. Approximately one-third of DLBCL cases, with a 2:1 ratio in ABC vs GCB-DLBCL,70 carry chromosomal translocations that prevent BCL6 downregulation by positioning the intact coding domain of the gene downstream to heterologous promoter sequences derived from >20 alternative chromosomal partners (most commonly, the IG heavy and light chain loci), leading to deregulation of BCL6 expression by a mechanism known as “promoter substitution.”71 The common feature of the promoters recruited by the translocations, which occur as byproducts of CSR or SHM, is their continued activity during post-GC differentiation. As a consequence, chromosomal translocations induce constitutive BCL6 expression and the pathologic maintenance of the GC phenotype, including tolerance to DNA damage and block of terminal differentiation.
The BCL6 gene can also be altered by somatically acquired point mutations that are distributed within a ∼2-kb region downstream to its transcription start site, encompassing the first noncoding exon (∼75% of DLBCL).72,73 Although some of these events reflect the physiologic activity of SHM in GC B cells, selected changes appear to be restricted to lymphoma; among these are mutations that abrogate a negative autoregulatory loop by which the BCL6 protein controls its own transcription,74,75 or prevent IRF4-mediated transcriptional repression induced by CD40 signaling in the GC LZ (Figure 3).76 Overall, as many as 35% of DLBCLs carry BCL6 genetic abnormalities of proven functional consequences, an estimate that may further expand as whole genome sequencing studies could reveal mutations in distant regulatory domains.
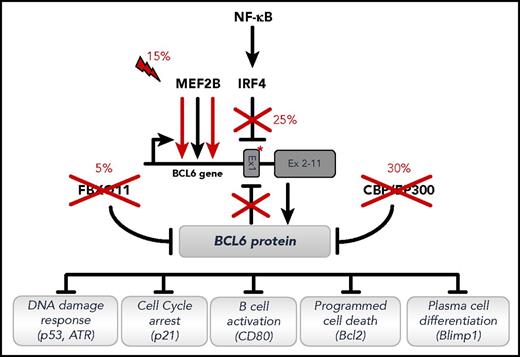
Deregulation of BCL6 activity by multiple mechanisms in DLBCL. Recurrent genetic alterations deregulating the function of BCL6 in DLBCL, either directly (by targeting the BCL6 gene) or indirectly (by targeting modulators of its activity). Only representative biological programs suppressed by BCL6 in the GC and disrupted as a consequence of these lesions are shown. Symbols depict loss-of-function (crosses) and gain-of-function (lightning bolt) genetic alterations. Asterisk represents point mutations in the BCL6 regulatory sequences, abrogating DNA binding sites used by the IRF4 transcription factor or by the BCL6 protein itself to negatively regulate BCL6 transcription. Reprinted from Pasqualucci and Dalla-Favera135 with permission.
Deregulation of BCL6 activity by multiple mechanisms in DLBCL. Recurrent genetic alterations deregulating the function of BCL6 in DLBCL, either directly (by targeting the BCL6 gene) or indirectly (by targeting modulators of its activity). Only representative biological programs suppressed by BCL6 in the GC and disrupted as a consequence of these lesions are shown. Symbols depict loss-of-function (crosses) and gain-of-function (lightning bolt) genetic alterations. Asterisk represents point mutations in the BCL6 regulatory sequences, abrogating DNA binding sites used by the IRF4 transcription factor or by the BCL6 protein itself to negatively regulate BCL6 transcription. Reprinted from Pasqualucci and Dalla-Favera135 with permission.
In addition to alterations that directly involve the BCL6 locus, several other genetic lesions deregulate the expression of BCL6 by alternative, indirect mechanisms (Figure 3). As mentioned, deleterious mutations of CREBBP/EP300 impair acetylation-mediated inactivation of the BCL6 transrepressive function.48,55 Approximately 15% of cases display somatic mutations in the MEF2B transcription factor,32,77 which is transcribed at high levels in GC B cells and occupies a large number of proximal and distal gene regulatory sequences (R.D.-F., unpublished data, 14 June 2017), among which is the BCL6 promoter/enhancer, upregulating its transcription.77 MEF2B mutations enhance its transactivator function and consequently BCL6 expression either by preventing physical interaction with the co-repressor CABIN1 (missense mutations in the N-terminal MEF/MAD domain), or by removing phosphorylation- and sumoylation-mediated negative regulatory motifs located in the C-terminal portion of the protein (truncating mutations).77 Finally, deregulation of BCL6 expression is achieved by a decrease in its catabolism in ∼5% of DLBCL cases displaying loss-of-function mutations and/or deletions of FBXO11, a ubiquitin adaptor protein that normally targets BCL6 for proteasomal degradation.78,79 Consistent with the functional interpretation of the human genetics data, mice engineered to recapitulate the chromosomal translocations involving BCL6, or any of the indirect mechanisms deregulating BCL6 expression, develop clonal lymphoproliferative disorders mimicking various stages of human pathology.54,79,80 Clearly, the contribution of MEF2B and FBXO11 mutations to the pathogenesis of DLBCL will likely entail the dysregulation of other pathways in addition to BCL6, given the multitude of substrates that have been (or have yet to be) recognized for these molecules.
Interestingly, transient deregulation of BCL6 expression in murine hematopoietic stem cells has been shown to induce the development of mature B-cell lymphomas lacking BCL6 expression, supporting a hit-and-run model for BCL6-mediated oncogenesis81 ; however, such model may not recapitulate the pathogenesis of the human tumors, because BCL6 translocations are mediated by errors occurring during CSR and SHM (ie, in a mature, antigen experienced B cell),9 and generally lead to variable levels of BCL6 expression.70,82 Overall, BCL6 continues to represent an attractive therapeutic target in DLBCL, and promising results have been obtained in vitro and in preclinical models with small-molecule inhibitors or HSP90 inhibitors, which also destabilize BCL6.83,84
Escape from immune surveillance.
Approximately 60% of DLBCL samples fail to express MHC class I and are therefore expected to evade cytotoxic T lymphocytes–mediated immune surveillance, due to a variety of genetic and epigenetic mechanisms. These include (i) biallelic inactivation and/or focal homozygous deletions of the B2M locus, encoding for the β2-microglobulin invariant subunit (29% of GCB- and 15% of ABC-DLBCL cases), which is necessary for the formation of the HLA-I complex on the cell surface; (ii) point mutations and genomic loss of the HLA loci; (iii) lack of expression or aberrant cytoplasmic localization of the B2M/HLA-I protein, in the absence of genetic lesions (30% to 45% of cases), consistent with epigenetic silencing or defects in the assembly and transport from the endoplasmic reticulum to the cell surface.85 Supporting its relevance in DLBCL pathogenesis, loss of HLA-I expression is infrequently observed in other B-cell non-Hodgkin lymphoma types (L.P., R.D.-F., unpublished observation, 2 December 2014), but is often associated with tFL.44,86 DLBCL also harbors recurrent genetic alterations inactivating the CD58 gene (23% of ABC- and 12% of GCB-DLBCL),85 a member of the immunoglobulin superfamily that functions as the ligand of the CD2 receptor expressed on T cells and natural killer cells.87 Of note, loss of HLA-I and CD58 expression are often concurrent in the same cases, suggesting that tumors may coselect these lesions in order to evade both cytotoxic T lymphocyte–mediated and natural killer cell–mediated immune surveillance mechanisms.85
Downregulation of MHC class-II expression has been reported in 40% to 50% of DLBCL, where this finding correlates with poor outcome.12,88 The mechanism underlying reduced MHC-II levels is thought to involve in part the genetic inactivation of CIITA, the gene encoding the MHC-II transactivator. CIITA represents a common target of ASHM (23% of cases)44,89 and can also be implicated in promiscuous chromosomal rearrangements leading to gene disruption or to dominant negative fusion proteins (3% of cases).90 Interestingly, CIITA is one of the functional targets of CREBBP in the GC, and reduced levels of MHC-II were observed in the B cells of Crebbp knockout mice, suggesting that genetic lesions of CREBBP may also contribute to this phenotype.53,54,56
Finally, gains, amplifications, and structural rearrangements of the genes encoding for programmed death ligands (PD-Ls), a common finding in primary mediastinal B-cell lymphoma and Hodgkin lymphoma, have been detected in a small subset of DLBCL-NOS (12%, 3%, and 4% of cases, respectively).91,92 These cytogenetic alterations correlate with increased expression of PD-L1, but not of PD-L2, and were more frequently observed in the non-GCB subtype of DLBCL. Interestingly, stable ectopic expression of wild-type PDCD1LG2 and the PDCD1LG2 fusions showed significantly reduced T-cell activation in coculture experiments. These findings suggest that treatments with PD:PD-L immune-checkpoint inhibitors might benefit the small group of patients carrying these lesions.
Mutations of FOXO1.
The transcription factor FOXO1 is a key player during B-cell differentiation, and its activity is negatively regulated by the PI3K-AKT and mechanistic target of rapamycin (mTOR) cascade. Within the GC, FOXO1 is expressed specifically in the DZ, consistent with the low level of PI3K signaling in this compartment, and is required for sustaining the DZ program, in part by cooperating with BCL6.93,94 FOXO1 mutations were identified in 8% to 10% of all DLBCL cases and comprise amino acid changes that cluster around a phosphorylation site required for AKT-mediated nuclear-cytoplasmic translocation.95 Thus, mutations have been suggested to prevent the inactivation of FOXO1 in response to PI3K signaling. However, a systematic examination of the functional consequences of these mutations in the context of B cells is still lacking. The occurrence of FOXO1 mutations appears to be associated with inferior prognosis and was enriched in GCB-type relapse/refractory DLBCL (36% of cases),96 although larger studies investigating sequential diagnostic/relapsed cases, and addressing the entire spectrum of FOXO1 aberrations, will be needed to conclusively determine the clinical impact of these events.
Genetic lesions associated with GCB-DLBCL
Chromosomal translocations of BCL2 and MYC.
BCL2 is a key antiapoptotic molecule expressed in most tissues but absent in the GC, due to BCL6-mediated suppression of its transcription,24,63 and consistent with the need of GC B cells to maintain a default proapoptotic program. This regulatory axis is disrupted in ∼30% of GCB-DLBCL by the t(14;18) translocation,12,97 which brings the BCL2 coding exons under the control of the IG locus, resulting in its ectopic expression (Figure 4). In line with its ability to confer survival advantage, deregulation of BCL2 has been associated with an inferior outcome, particularly when coupled with MYC deregulation.98 Small-molecule inhibitors targeting the BCL2 protein (eg, venetoclax) have demonstrated promising efficacy in lymphoid malignancies and are currently under investigation in combination with standard immunochemotherapy regimens.2,4
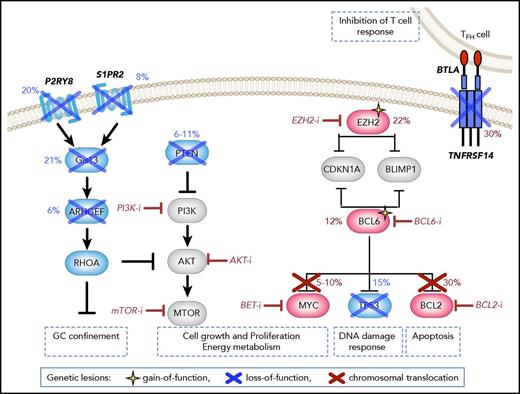
Disrupted signaling pathways in GCB-DLBCL. Genetic lesions preferentially associated with GCB-DLBCL include (i) chromosomal translocations of BCL2 (up to 35% of cases) and/or MYC (∼10% of cases), which lead to their ectopic expression in part by allowing them to bypass BCL6-mediated transcriptional repression; (ii) truncating mutations of the TNFRSF14 receptor, leading to weakened T-cell responses; (iii) gain-of-function mutations of EZH2 (∼20% of cases), which induce transcriptional silencing of various antiproliferative and tumor suppressor genes, including targets common to BCL6 (eg, CDKN1A and BLIMP1); (iv) point mutations in the BCL6 autoregulatory sequences (10% of cases). In addition, loss of PTEN expression is observed in as many as 55% of cases, as a consequence of genetic deletions (15%) and amplifications of miR17-92 (29%), resulting in activation of the PI3K/Akt/mTOR signaling pathway. Targeted agents currently in clinical trial (or, for BCL6, demonstrating activity in preclinical settings) are shown in red.
Disrupted signaling pathways in GCB-DLBCL. Genetic lesions preferentially associated with GCB-DLBCL include (i) chromosomal translocations of BCL2 (up to 35% of cases) and/or MYC (∼10% of cases), which lead to their ectopic expression in part by allowing them to bypass BCL6-mediated transcriptional repression; (ii) truncating mutations of the TNFRSF14 receptor, leading to weakened T-cell responses; (iii) gain-of-function mutations of EZH2 (∼20% of cases), which induce transcriptional silencing of various antiproliferative and tumor suppressor genes, including targets common to BCL6 (eg, CDKN1A and BLIMP1); (iv) point mutations in the BCL6 autoregulatory sequences (10% of cases). In addition, loss of PTEN expression is observed in as many as 55% of cases, as a consequence of genetic deletions (15%) and amplifications of miR17-92 (29%), resulting in activation of the PI3K/Akt/mTOR signaling pathway. Targeted agents currently in clinical trial (or, for BCL6, demonstrating activity in preclinical settings) are shown in red.
The MYC gene encodes for a transcription factor that controls numerous biological functions, including proliferation, cell growth, telomerase activity, energy metabolism, differentiation, and apoptosis,99 as well as DNA replication independently of its transcriptional activity.100 In most DZ and LZ B cells, MYC transcription is suppressed by BCL6, whereas its expression is reactivated in a subset of LZ B cells destined to recirculate into the DZ.23 MYC is ectopically and constitutively expressed in 10% to 14% of GCB-DLBCLs,101 often as the result of chromosomal translocations that join its intact coding domain to the IG heavy or light chains loci (Figure 4).22 Analogous to BCL2, the presence of MYC translocations has been linked to worse prognosis in DLBCL.102 Although the MYC oncogene has long been considered a particularly compelling target for cancer therapy, the MYC protein itself is not easily “druggable.” However, potent and selective inhibition of MYC has been obtained in preclinical studies by targeting BET bromodomain-promoter interactions, including the BRD4 transcriptional coactivator that epigenetically regulates MYC expression, and led to significant antiproliferative effects in lymphoma.2,4
In 5% to 10% of DLBCL, chromosomal translocations of MYC and BCL2 (or, less commonly, BCL6) coexist.103 These “double-hit lymphomas” (DHLs) have gained increasing attention due to their aggressive clinical course and particularly poor prognosis,103 although more recent prospective studies are starting to reveal comparable overall survival. Consistent with the highly preferential distribution of both translocations in GCB-like DLBCL, DHLs belong to this molecular subtype, even though the proposed revision of the World Health Organization classification recognizes them as distinct categories designated “high-grade B-cell lymphomas.”1 Owing to the rarity of the disease, a comprehensive characterization of the genetic makeup of DHL is still lacking.
DHL should be kept separate from DLBCL showing coexpression of MYC and BCL2 in the absence of chromosomal translocations, also a strong predictor of poor clinical outcome.103 The so-called double-expresser lymphomas presumably involve distinct underlying mechanisms, including BCL2 amplifications and constitutive activation by the NF-κB transcription complex, and appear to be primarily of the ABC subgroup, suggesting that they should be studied separately in a therapeutic perspective. Targeted intervention for DHL and “double expresser” may include anti-BCL2 and candidate anti-MYC compounds.2,4
Mutations of the EZH2 methyltransferase.
EZH2 encodes a SET-domain histone methyltransferase that is responsible for trimethylating the lysine 27 residue of histone H3 (H3K27me3), an epigenetic mark associated with transcriptional repression.104 As a component of the polycomb repressive complex-2, EZH2 is required for the formation of GCs, where it controls the expression of multiple genes, including those involved in cell cycle regulation (CDKN1A) and terminal differentiation (IRF4, BLIMP1) (Figure 4).105,106 Approximately 22% of GCB-DLBCL display heterozygous EZH2 gene mutations, which in most cases replace a single evolutionarily conserved residue (Y641) within the protein SET domain,107 enhancing its ability to catalyze the addition of the H3K27me3 mark.108 Expression of the mutant EZH2Y641 allele induces GC hyperplasia in mice, and when combined with BCL2 deregulation accelerates the development of mature B-cell lymphomas.105,106 Notably, small-molecule inhibitors of EZH2 have shown specific activity in preclinical models of GCB-DLBCL, independently of the presence of somatic mutations,105,109,110 supporting their early clinical testing for relapsed DLBCL.2,4
Mutations affecting B-cell migration.
The confinement of B cells within the B-cell follicle is modulated by the activity of two GC-specific G protein–coupled receptors: sphingosine-1-phosphate receptor 2 (S1PR2)111 and the orphan purinergic receptor P2RY8.112 In response to lipid ligands, these receptors recruit two closely related G proteins (Gα12 and Gα13) and stimulate RHOA activity through specific guanine nucleotide exchange factors, to ultimately suppress pAKT signaling and cell migration. GCB-DLBCL, but not ABC-DLBCL, displays recurrent inactivating mutations in several components of this pathway, including the S1PR2, GNA13, and, more rarely, ARHGEF1 and P2RY8 genes (overall, ∼30% of cases) (Figure 4).112,113 Accordingly, deletion of these genes in the mouse is associated with disruption of the GC architecture, followed by dissemination of GC B cells to the peripheral blood and bone marrow, eventually leading to the development of lymphomas that exhibit features of GCB-DLBCL.112,113
Mutations of TNFRSF14.
TNFRSF14 (also known as herpesvirus entry mediator [HVEM]) encodes for a member of the tumor necrosis factor–receptor superfamily that is expressed in both T and B cells and can deliver opposing signals based on its specificity for diverse ligands.114 Deletions and mutations of TNFRSF14, including missense (∼50%), nonsense (∼40%), and frameshift (2.5%) events confined to the exons encoding for its ectodomain, are recurrently found in DLBCL and exquisitely segregate with the GCB subtype (30% of cases) (Figure 4). Although the functional consequences of individual amino acid changes have not been studied, the observed mutation pattern (significant number of truncating events) and the existence of cases showing biallelic mutations/deletions indicate a strong selection against the function of this receptor during DLBCL development. The tumor suppressor role of TNFRSF14 has been documented in vivo in a FL-prone BCL2-driven mouse model, where silencing of this gene led to cell autonomous activation of B-cell proliferation and increased development of GC-derived lymphomas.115 One mechanism underlying the tumorigenic effect of TNFRSF14 loss is the inhibition of cell-cell interactions between this receptor and its ligand BTLA, which induces a tumor-supportive microenvironment marked by exacerbated lymphoid stroma activation and increased recruitment of T-follicular–helper cells.115 Consistent with this model, a mutually exclusive association has been reported in FL, but not yet in DLBCL, between TNFRSF14 mutations and BTLA downregulation, indicating that these two alterations may converge to create a favorable environment. These findings provide the rationale for therapeutically restoring this circuit by administration of the HVEM ectodomain protein. Indeed, soluble HVEM was able to bind BTLA and cause significant growth inhibition in BTLA-expressing lymphoma cells in vitro; moreover, it restored tumor suppression in established lymphomas in vivo, when produced locally and continuously by modified chimeric antigen receptor–T cells.115
Genetic lesions associated with ABC-DLBCL
The core biology of ABC-DLBCL is defined by alterations in genes encoding multiple signal transducers and adaptor molecules downstream of the BCR and Toll-like receptor (TLR). While these receptors can trigger a variety of signaling pathways, all of the lesions identified converge onto the constitutive activation of the NF-κB transcription complex, the expression of which is required for ABC-DLBCL survival. Alterations in NF-κB pathway components are complemented by lesions blocking terminal B-cell differentiation (Figure 5). In addition, ABC-DLBCL is characterized by recurrent amplifications of the BCL2 locus (∼30% of cases),12,116 homozygous loss of CDKN2A/B (∼50% of cases),33,116 and gains or amplifications of a genomic region spanning the SPIB locus on chromosome 19q (27% of cases).116
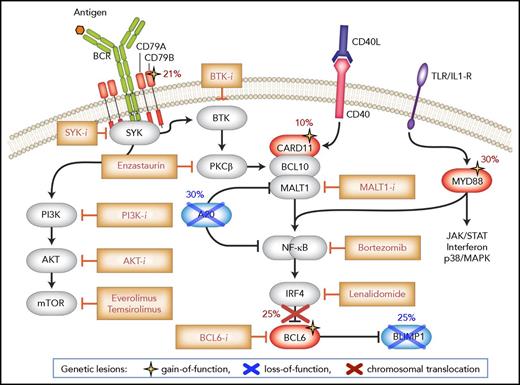
Disrupted signaling pathways in ABC-DLBCL. ABC-DLBCL is defined by multiple genetic alterations that fuel malignant transformation by sustaining constitutive NF-κB activity downstream of the BCR and TLR, while blocking terminal B-cell differentiation. Genes directly targeted by these lesions are shown in blue (inactivation) and red (deregulated expression/activity), and symbols at the bottom denote gain-of-function and loss-of-function events. Upstream inhibitors of the BCR, PI3K, and NF-κB signal transduction pathway have shown promising effects in early clinical trials involving ABC-DLBCL patients. Modified from Pasqualucci and Dalla-Favera135 with permission.
Disrupted signaling pathways in ABC-DLBCL. ABC-DLBCL is defined by multiple genetic alterations that fuel malignant transformation by sustaining constitutive NF-κB activity downstream of the BCR and TLR, while blocking terminal B-cell differentiation. Genes directly targeted by these lesions are shown in blue (inactivation) and red (deregulated expression/activity), and symbols at the bottom denote gain-of-function and loss-of-function events. Upstream inhibitors of the BCR, PI3K, and NF-κB signal transduction pathway have shown promising effects in early clinical trials involving ABC-DLBCL patients. Modified from Pasqualucci and Dalla-Favera135 with permission.
Alterations leading to constitutive activation of NF-κB.
Mutations in the BCR signaling pathway.
ABC-DLBCL displays a “chronic, active” form of BCR signaling that is sustained by genetic alterations affecting proximal members of the pathway.117 In 21% of cases, these are gain-of-function mutations in the immunoreceptor tyrosine-based activation motifs of the IG superfamily member CD79B (or, rarely, CD79A),117 which maintain BCR signaling by attenuating a negative feedback involving the phosphorylation-mediated activation of the Lyn kinase. In ∼9% of cases, mutations involve the gene encoding CARD11,118 a component of the “signalosome” complex that needs to be assembled for the proper transduction of BCR signaling.119 These two types of lesions enhance the amplitude of BCR signaling, but do not initiate this cascade de novo; thus, they cannot explain alone the dependency of ABC-DLBCL on chronic BCR signaling, which is also observed in a fraction of wild-type tumors following the knockdown of several proximal and distal subunits.117 Indeed, recent analyses revealed a role for self-antigens in the survival of ABC-DLBCL cells, consistent with the expression of a restricted IG variable region repertoire and with the demonstration that ABC-DLBCL cell lines rely on the ability of their BCR to interact with autoantigens.120 The dependence of ABC-DLBCL from the BCR signaling pathway is underscored by recent clinical studies reporting successful results with the use of agents that inhibit Bruton’s tyrosine kinase (BTK), a molecule linking signaling from the BCR to NF-κB selectively in patients with this molecular subtype and even in the absence of mutations (30% of those showing response).121 Of note, BCR emanates signals to multiple downstream effectors besides canonical NF-κB, including PI3K, MAPK/ERK, and NF-AT, all of which may play important roles in the neoplastic transformation of ABC-DLBCL cells. These vulnerabilities offer a unique opportunity for the development of combinatorial therapeutic strategies, as suggested by the cooperative toxicity observed upon combined inhibition of NF-κB and PI3K118 (Figure 5).
Mutations of MYD88.
Approximately 30% of ABC-DLBCLs harbor mutations leading to a hotspot L265P substitution in the hydrophobic core of the MYD88 TIR domain. This adaptor molecule is critical for relaying signals from the TLR (particularly, TLR9) (Louis M. Staudt, American Association for Cancer Research annual meeting, 3 April 2017) to the NF-κB transcription complex, as well as the interferon and JAK/STAT3 signaling cascade, another phenotypic trait of ABC-DLBCL cells required for their survival.122,123 The oncogenic potential of MYD88 L265P was documented in mice, where its B-cell–specific expression leads to the development of lymphoproliferative diseases, including a fraction of clonal lymphomas.124 Interestingly, although DLBCLs carrying the MYD88 mutant isoform did not respond to BTK inhibition in a recent clinical trial, exceptional responses were observed in tumors with concurrent MYD88 and CD79A/B mutations, suggesting that these pathways may be functionally coupled.121 A cooperative activity between MYD88 and CD79B mutations has indeed been shown in the mouse, where enforced expression of these proteins enabled the development of self-reactive B cells.125
Mutations of TNFAIP3.
Nearly 30% of ABC-DLBCL cases display biallelic truncating mutations and/or focal deletions inactivating the TNFAIP3 gene,126,127 which encodes a dual function ubiquitin-modification enzyme (also called A20) involved in the negative regulation of NF-κB responses triggered by TLR and BCR signaling.128 Inactivation of TNFAIP3/A20 may thus contribute to lymphomagenesis by inducing inappropriately prolonged NF-κB responses.126,127 Reexpression of functional TNFAIP3/A20 in DLBCL cell lines leads to cytoplasmic relocalization of NF-κB and apoptosis,126,127 supporting a role for this gene as a tumor suppressor. Consistent with its activity downstream of the BCR proximal signalling, patients with TNFAIP3-mutated DLBCL do not respond to BTK-inhibitors.121
Genetic lesions preventing terminal differentiation.
Genetic-driven constitutive activation of NF-κB in ABC-DLBCL is frequently complemented by lesions blocking terminal B-cell differentiation (Figure 5). In particular, 25% of cases display biallelic loss-of-function mutations/deletions of PRDM1/BLIMP1,129 a transcriptional repressor induced in a subset of LZ B cells committed to plasmacytic differentiation and required for plasma cell development. BLIMP1 functions by suppressing the expression of GC master genes, including its direct targets PAX5 and BCL6.69 In addition, a variety of genetic and epigenetic mechanisms abrogate BLIMP1 function in ABC-DLBCL cases lacking alterations of its gene, including direct transcriptional repression by constitutively active BCL6 alleles.129 Confirming the tumor suppressor role of BLIMP1, conditional GC-specific Prdm1 deletion in the mouse leads to the development of DLBCL with ABC-like phenotype and synergizes with the constitutive activation of the canonical NF-κB pathway to accelerate lymphomagenesis.130,131
DLBCL deriving from the transformation of FL and CLL
Genomic analysis of sequential pre- and posttransformation biopsies from CLL and FL patients revealed that the transformation of CLL to DLBCL (known as Richter syndrome) derives from the dominant CLL clone through a linear pattern of progression that involves the maintenance of the original CLL-associated lesions and the acquisition of new genetic alterations affecting NOTCH1 (mutated in 35% of cases), CDKN2A/B (homozygously deleted in 30% of cases), TP53 (genetically inactivated in as many as 60% of cases), and MYC (amplified or translocated in ∼30% of cases).132 Conversely, FL and tFL arise through divergent evolution from a common mutated precursor clone that has acquired independent genetic aberrations to become an FL or a DLBCL.44 Whereas the common precursor clone is dominated by the presence of the BCL2 translocation and by inactivation of chromatin modifiers (KTMD2 and CREBBP), the DLBCL clone features lesions typical of high-grade malignancies such as CDKN2A/B loss, TP53 loss, MYC translocations/amplifications, ASHM, and HLA class-I loss. Genomic analysis of these tumors also showed that, although morphologically undistinguishable from de novo DLBCL, Richter syndrome and post-FL DLBCL are each characterized by a partially different mutational landscape and gene expression profile.44,132 This distinction has critical implications for the development of rationally designed combinatorial therapies.
Future directions
The revolutionary improvement in the understanding of the genetic basis of DLBCL achieved during the last decade is poised to have relevant implications for patient diagnosis, prognostication, and therapy. Nonetheless, only a few of the lesions identified have been adopted as routine diagnostic, prognostic, or predictive markers in the clinical settings, partly because of their heterogeneity (which requires larger studies providing robust correlates), and also due to limitations in accessing fresh tumor material from tissue biopsies. Significant advances in this respect may come from the use of circulating tumor DNA, which proved to be a reliable surrogate for simultaneously tracking multiple somatic mutations in DLBCL, with high specificity and sensitivity.133,134 This approach outperformed conventional IG sequencing and radiographic imaging for the detection of minimal residual disease and was independently predictive of clinical outcome.134 Moreover, circulating tumor DNA genotyping allows the identification of distinct patterns of clonal evolution, which makes it a valuable tool in the setting of transformation or relapse, by informing individualized therapy. The discovery of genes and pathways that are recurrently disrupted in DLBCL reveals vulnerabilities in the lymphoma cells that are often uniquely associated with distinct lymphoma subtypes and could thus be exploited for the design of more effective, targeted therapeutic approaches. Indeed, the information gained from these studies is already being translated into the development and clinical testing (or repositioning) of novel drugs or drug combinations directed against specifically dysregulated programs in DLBCL. This is the case for small-molecule inhibitors of EZH2 and BCL2, as well as inhibitors of BCR and NF-κB activity.2,4 Among these drugs, the BTK inhibitor ibrutinib is rapidly emerging as a novel paradigm for the treatment of ABC-DLBCL.2 It is expected that additional therapeutic targets will be identified as more mutational events become functionally dissected. Although these strategies are expected to impact the standard of care for this malignancy, the multitude of the dysregulated circuits, the overall complexity of the disease (which may also involve non-genetic mechanisms), and the interplay with the microenvironment underscore the need of precise patient stratification in order to identify those that are most likely to respond.
Acknowledgments
The authors thank all the members of the Dalla-Favera and Pasqualucci laboratories for their contribution to the generation of data reported in this manuscript and apologize to those colleagues whose work could not be described or cited due to space limitations.
This work was supported in part by National Institutes of Health, National Cancer Institute grants R01CA172492 (L.P.) and R35CA210105 and P50CA192937 (R.D.-F.).
Authorship
Contribution: L.P. and R.D.-F. conceived and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laura Pasqualucci, Institute for Cancer Genetics, Columbia University, 1130 St. Nicholas Ave, Room 507B, New York, NY 10032; e-mail: lp171@columbia.edu; Riccardo Dalla-Favera, Institute for Cancer Genetics, Columbia University, 1130 St. Nicholas Ave, Room 508B, New York, NY 10032; e-mail: rd10@columbia.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal