Key Points
T cells from HDAC11KO mice have increased effector functions and mediate more rapid and potent GVHD.
HDAC11 associates with the Eomes and Tbet gene promoter regions in resting cells and disassociates upon activation.
Abstract
Histone acetylation and the families of enzymes responsible for controlling these epigenetic marks have been implicated in regulating T-cell maturation and phenotype. Here, we demonstrate a previously undefined role of histone deacetylase 11 (HDAC11) in regulating T-cell effector functions. Using EGFP-HDAC11 transgenic reporter mice, we found that HDAC11 expression was lower in effector relative to naive and central memory T-cell populations, and activation of resting T cells resulted in its decreased expression. Experiments using HDAC11 knockout (KO) mice revealed that T cells from these mice displayed enhanced proliferation, proinflammatory cytokine production, and effector molecule expression. In addition, HDAC11KO T cells had increased expression of Eomesodermin (Eomes) and TBX21 (Tbet), transcription factors previously shown to regulate inflammatory cytokine and effector molecule production. Conversely, overexpression of HDAC11 resulted in decreased expression of these genes. Chromatin immunoprecipitation showed the presence of HDAC11 at the Eomes and Tbet gene promoters in resting T cells, where it rapidly disassociated following T-cell activation. In vivo, HDAC11KO T cells were refractory to tolerance induction. HDAC11KO T cells also mediated accelerated onset of acute graft-versus-host disease (GVHD) in a murine model, characterized by increased proliferation of T cells and expression of interferon-γ, tumor necrosis factor, and EOMES. In addition, adoptive transfer of HDAC11KO T cells resulted in significantly reduced tumor burden in a murine B-cell lymphoma model. Taken together, these data demonstrate a previously unknown role of HDAC11 as a negative epigenetic regulator of T-cell effector phenotype and function.
Introduction
Two classes of enzymes work in opposition to regulate the chromatin state through acetylation: histone acetyltransferases and histone deacetylases (HDACs). Studies have shown that the importance of HDACs extend beyond effects upon histones, encompassing other functions important in immunoregulatory pathways.1-3 HDAC inhibitors influence T-cell production of various cytokines important for regulation of immune responses, such as interleukin-2 (IL-2), interferon-γ (IFN-γ), and IL-4.4,5 Due to their immunomodulatory effects, HDAC inhibitors have shown efficacy in the treatment of hematological malignancies and in allogeneic transplant models.6,7 However, given the lack of specificity of the HDAC inhibitors used in these investigations, the roles of individual HDACs in the observed immune responses remain to be fully elucidated.
HDACs are grouped by their phylogenetic relatedness and sequence homology into 4 main classes, with HDAC11 being the sole member of class IV.8 Relatively little is known about HDAC11, which was first identified in 2002. Previously, our group demonstrated that HDAC11 regulated IL-10 gene transcriptional activity in antigen-presenting cells.9 However, the role(s) of HDAC11 in T-cell function remains uninvestigated.
Allogeneic hematopoietic cell transplantation is an effective therapy for a variety of hematologic malignances, yet this efficacy is impeded by the development of graft-versus-host disease (GVHD). T helper 1 (Th1) cytokines produced by allogeneic T cells are the driving forces for the initiation and development of GVHD. TBET is a transcriptional activator of IFN-γ.10 TBET also has cooperative and partially redundant functions with EOMES, another T-box transcription factor, to control CD8 T-cell cytotoxicity, IFN-γ production, and memory T-cell formation.11,12 Previous work from us and others demonstrated that TBET and EOMES regulate activation and differentiation and are critical for the development of GVHD.13
Using HDAC11-EGFP transgenic14 and HDAC11 knockout (KO) mice,15 we sought to determine the roles of HDAC11 in T cells. T cells lacking HDAC11 expressed higher levels of Eomes and Tbet and produced increased levels of Th1 cytokines. Chromatin immunoprecipitation (ChIP) showed that HDAC11 was present at the Eomes and Tbet gene promoters in resting T cells, but was absent following activation. In vivo, HDAC11KO T cells mediated more potent GVHD and inhibited tumor progression in a murine lymphoma model. These results point to HDAC11 as an epigenetic regulator of T-cell phenotype and function.
Materials and methods
Mice
C57BL/6 background EGFP-HDAC11 reporter mice were obtained from the Mutant Mouse Regional Resource Centers.14 C57BL/6 background HDAC11KO mice were provided by Merck Research Laboratories and generated by a targeted deletion of floxed exon 3 of the HDAC11 gene utilizing Rosa26 promoter-driven cre-recombinase expression. OTII mice were purchased from Jackson Laboratories and bred with HDAC11KO mice for >10 generations to generate the OTII/HDAC11KO mouse strain. OTII phenotype was validated by flow cytometry of T cells for T-cell receptor (TCR) Vβ5. HDAC11KO genotyping was performed by polymerase chain reaction. Primer sequences are as in supplemental Table 1, available on the Blood Web site. C57BL/6 and BALB/c wild-type (WT) mice were purchased from the National Cancer Institute or The Jackson Laboratory. All animal studies were approved by the Institutional Animal Care and Use Committee at the University of South Florida, George Washington University, and University of South Carolina.
Cells
T cells were obtained from mouse organs by physical digestion and straining through a 70-µm filter. Red blood cells were lysed with ACK buffer (Invitrogen, Carlsbad, CA). CD3+, CD4+, or CD8+ T cells were isolated through EasySep negative magnetic separation (StemCell Technologies; Vancouver, BC, Canada) per manufacturer’s instructions. The FCmuMCL1 murine mantle-cell lymphoma cell line was provided by Mitchell Smith of Cleveland Clinic.
In vivo T-cell tolerance induction model
Anti-OVA OT-II TCR+ T cells (2.5 × 106) from OTII mice or OTII/HDAC11KO mice were transferred IV into WT C57BL/6 recipients. Two days later, half the mice were injected IV with a tolerogenic dose of OVA-peptide323-339 (100 µg) or Hanks balanced salt solution vehicle. Ten days later, splenic CD4+ T cells were harvested from the different cohorts and cocultured in vitro with OVA peptide323-339 pulsed WT splenocytes. IFN-γ production was assessed by enzyme-linked immunosorbent assay (ELISA) after 48 hours.
GVHD model
Bone marrow (BM) cells were isolated from the tibia and femurs of C57BL/6 mice. T cells were isolated from spleens and lymph nodes. BALB/c mice were lethally irradiated with 900 cGy using a 137Cs irradiator. The following day, 5 × 106 WT C56BL/6 BM cells depleted of T cells or BM supplemented with indicated numbers of T cells were administered by tail vein injection. Mice were weighed thrice weekly and monitored for survival. Recipient mice spleens were harvested 5 days posttransfer to determine T-cell expansion. To evaluate IFN-γ expression, T cells were activated by phorbol myristate acetate (PMA) and ionomycin and assessed by flow cytometry. To determine serum cytokine levels, mice were bled and analyzed by ELISA. Expression of FOXP3, TBET, and EOMES was evaluated at 14 days postadoptive transfer by flow cytometry. Liver pathology was scored independently by a pathologist as previously described.16
FCmuMCL1 lymphoma model
C57BL/6 mice were injected with 2.2 × 106 FCmuMCL1 cells subcutaneously into shaven flanks on day 0. On day 4, mice were injected IV with 5 × 106 T cells isolated from pooled lymph nodes and spleen of either syngeneic WT or HDAC11KO mice. Tumors were measured and volumes calculated by the formula: volume = (length × width2)/2.
For analysis of tumor infiltrate, on day 4, 3 mice per group were adoptively transferred WT T cells, or HDAC11KO T cells. Mice were killed on day 16, and tumors were excised and assessed for CD4+ and CD8+ tumor infiltrating cells by flow cytometry. A section of the tumor was also assessed by immunofluorescent imaging using with anti-mouse CD4+ (BD; Clone RM4-5, 1:100) or anti-mouse CD8+ (BD, Clone 53-6.7, 1:100) antibodies.
ChIP
The ChIP-IT peripheral blood mononuclear cells kit (Active Motif, Carlsbad, CA) was used, per manufacturer’s instructions. Antibodies against pan-acetylated histone 3 and HDAC11 were purchased from Active Motif. Quantification of immunoprecipitated chromatin was performed by quantitative reverse transcription polymerase chain reaction (qRT-PCR). Primer sequences are listed in supplemental Table 3. Calculations for ChIP experiments were performed by comparisons against expression values obtained from immunoglobulin G (IgG) immunoprecipitated control input samples. Input cycle threshold (Ct) values (10%) were adjusted to 100%, and results were calculated by the formula: 2^(adjusted input − Ct(IP))*100.
HDAC11 overexpression
Healthy donor peripheral blood mononuclear cells were obtained by Ficoll separation followed by EasySep CD3+ negative isolation. Samples were obtained from OneBlood (Tampa, FL) in compliance with the Institutional Review Board (Liberty #13.04.0004) at the University of South Florida. HDAC11-Flag insert was constructed as previously described9 and inserted into the pLenti CMV/TO GFP-Zeo DEST vector purchased from Addgene (Cambridge, MA). Unstimulated cells (1 × 107) were electroporated with 2.5 μg of HDAC11-encoding plasmid or control empty vector using program U14 of Nucleofector II and the Amaxa Human T Cell Nucleofector Kit (Lonza, Basel, Switzerland). Media were replaced on the following morning, and cells were stimulated with αCD3/CD28 Dynabeads at 1:1 ratio for 6 hours. Trizol lysis and RNA extraction were performed for gene expression analyses.
Statistical analyses
Statistical analyses were conducted using GraphPad Prism v6.0 software (La Jolla, CA). Unless otherwise indicated, unpaired Student t tests were performed for experiments involving 2 groups and 1-way analysis of variances using Dunnett tests were used for experiments involving 2 or more comparisons. For HDAC11 expression kinetics and overexpression experiments, paired Student t tests were performed. Survival differences were determined using log-rank tests. GVHD body weight differences were determined by multiple Student t tests corrected for false discovery rates by a 2-step setup method of Benjanmini, Krieger, and Yekutieli. Significance of tumor growth curve differences was calculated using 2-way analysis of variances with Tukey tests. P values ≤.05 were considered significant.
Additional “Materials and methods” can be found in the supplemental materials.
Results
HDAC11 expression is decreased in activated and effector T cells
We have previously demonstrated that HDAC11 regulates IL-10 gene expression in myeloid cells.9,15 To gain insight into the differential expression of HDAC11 in other immune cells in vivo, we used an EGFP-HDAC11 transgenic reporter mouse in which EGFP expression is driven by the HDAC11 promoter.14 At the steady state, ∼7% of monocytes (CD3−NK1.1−CD45R−MHCII−CD115+), 5% of dendritic cells (CD3−NK1.1−CD19−CD11C+), and 5% of B cells (CD3−NK1.1−CD45+CD19+) from mouse blood samples were EGFP+ (Figure 1A). In the T-cell compartment, we observed a higher percentage of expression relative to other immune cell populations investigated, with ∼50% EGFP-positive CD3+ T cells.
HDAC11 expression is downregulated in activated cells and Teff. (A) Utilizing EGFP-HDAC11 reporter mice, EGFP expression was assessed by flow cytometry in various immune cells. (B) Expression of EGFP-HDAC11 was further assessed in CD4+ (left panel) and CD8+ (right panel) T cells obtained from the lymph nodes. Plots shown are representative of 3 mice assessed in 3 independent experiments. (C) EGFP expressing and nonexpression cells were flow sorted and analyzed by qRT-PCR for expression of HDAC11 mRNA. Error bars are from 3 technical replicates per group. (D) Expression of EGFP was evaluated by flow cytometry in CD4+ and CD8+ T-cell subsets defined by CD44 and CD62L expression. Plots shown are representative of 3 mice assessed in 3 independent experiments. (E) WT CD3+ T cells isolated from mouse lymph nodes were left unstimulated or activated by αCD3/CD28 beads for indicated times and assessed by qRT-PCR for HDAC11 mRNA expression. Combined results from 5 mice, each represented with a unique symbol, assessed over 2 independent experiments are shown. P values of baseline vs activation comparisons are as follows: 2 hours: P < .05; 6 hours: P < .001; 24 hours: P < .01; 48 hours: P < .05.
HDAC11 expression is downregulated in activated cells and Teff. (A) Utilizing EGFP-HDAC11 reporter mice, EGFP expression was assessed by flow cytometry in various immune cells. (B) Expression of EGFP-HDAC11 was further assessed in CD4+ (left panel) and CD8+ (right panel) T cells obtained from the lymph nodes. Plots shown are representative of 3 mice assessed in 3 independent experiments. (C) EGFP expressing and nonexpression cells were flow sorted and analyzed by qRT-PCR for expression of HDAC11 mRNA. Error bars are from 3 technical replicates per group. (D) Expression of EGFP was evaluated by flow cytometry in CD4+ and CD8+ T-cell subsets defined by CD44 and CD62L expression. Plots shown are representative of 3 mice assessed in 3 independent experiments. (E) WT CD3+ T cells isolated from mouse lymph nodes were left unstimulated or activated by αCD3/CD28 beads for indicated times and assessed by qRT-PCR for HDAC11 mRNA expression. Combined results from 5 mice, each represented with a unique symbol, assessed over 2 independent experiments are shown. P values of baseline vs activation comparisons are as follows: 2 hours: P < .05; 6 hours: P < .001; 24 hours: P < .01; 48 hours: P < .05.
Next, we measured the EGFP expression in CD4+ and CD8+ T cells harvested from lymph nodes. Similar to circulating CD3+ T cells, approximately half of CD4+ and CD8+ T cells expressed EGFP (Figure 1B). Similar percentages of EGFP-positive CD4+ and CD8+ T cells were also seen in the spleen of these mice (data not shown). To verify that EGFP expression in T cells was indicative of HDAC11 messenger RNA (mRNA) transcription, CD4+ and CD8+ lymphocytes were sorted into EGFP-positive and -negative populations. As shown in Figure 1C, EGFP-positive T cells expressed markedly higher levels of HDAC11 mRNA relative to EGFP-negative cells (∼ >50-fold).
We then explored the expression of HDAC11 in CD4+ and CD8+ T-cell subsets. As shown in Figure 1D, naive (CD44−CD62L+) CD4+ and CD8+ T cells displayed the highest proportion of EGFP-positive cells (52%) followed by central memory (CD44+CD62L+) T cells (48%). Decreased percentages of EGFP-positive cells (37%) were seen in effector CD4+ and CD8+ T cells (CD44−CD62L−). In effector memory T cells (CD44+CD62L−), we observed the lowest percentage of EGFP-positive cells with ∼16% and 11% in the CD4+ and CD8+ populations, respectively.
The differences in HDAC11 expression in naive versus memory and effector T cells (Teff) led us to explore potential changes in HDAC11 expression following T-cell activation. CD3+ T cells isolated from lymph nodes of WT C57BL/6 mice were activated by αCD3/CD28 bead stimulation. As seen in Figure 1E, activated T cells displayed a decrease in HDAC11 mRNA expression as early as 2 hours postactivation (2 hours: P < .05; 6 hours: P < .001; 24 hours: P < .01; 48 hours: P < .05). A small, but nonsignificant increase (P = .7731) in HDAC11 expression was observed at 48 hours relative to 24 hours. Based on these observations, we hypothesized a negative regulatory role for HDAC11 in T-cell function/activation.
T cells lacking HDAC11 have increased proliferation and proinflammatory cytokine production
To gain insight into the functional role of HDAC11 in T cells, we performed studies in an HDAC11KO mouse model. CD4+ and CD8+ T cells were isolated, and the absence of HDAC11 was confirmed by qRT-PCR (supplemental Figure 2A). Phenotypic analysis of T-cell subpopulations from the thymus, spleen, or lymph nodes showed no gross differences between HDAC11KO and WT mice in resting cells (supplemental Figure 2B). Analysis of circulating lymphocytes revealed no differences in the total number of B cells or T cells between WT or HDAC11KO mice (supplemental Figure 2C-D). However, functional analyses of T cells lacking HDAC11 revealed differences relative to T cells from WT mice. Following stimulation with αCD3/CD28 beads, HDAC11KO T cells had an increased percentage of dividing cells, as indicated by flow cytometric analysis of cell tracking dye-stained T cells, relative to WT T cells (P < .01) (Figure 2A). HDAC11KO CD8+ T cells displayed higher proliferation index, with an average of 2.0 times vs WT cells dividing only 1.6 times (P < .001) (Figure 2B). Although in the absence of stimulation no detectable cytokine production was measured in either WT or HDAC11KO T cells (data not shown), αCD3/CD28-activated CD4+ T cells from HDAC11KO mice produced higher levels of the Th1 cytokines IFN-γ and IL-2 compared with WT T cells (P < .01) and trended toward elevated levels of tumor necrosis factor (TNF) (Figure 2C). Similarly, CD8+ T cells from HDAC11KO mice produced increased levels of IL-2 and IFN-γ (P < .001) as well as higher levels of TNF relative to WT CD8+ T cells (P < .01). No significant differences were seen in the Th2 cytokines IL-4, IL-6, IL-10, nor the Th17 cytokine IL-17A between HDAC11KO CD4+ or CD8+T cells and their WT counterparts. We then measured the expression of the effector molecules granzyme B and perforin in CD8+ T cells. Activated HDAC11KO CD8+ T cells displayed higher mRNA levels of both genes compared with WT T cells (P < .01) (Figure 2D).
HDAC11KO T cells have increased effector functions. (A) CD8+ T cells from HDAC11KO mice were stained with a proliferation tracking dye and activated for 72 hours with αCD3/CD28 stimulation. Proliferation was evaluated by flow cytometry. The percentage of cells (± standard error of the mean [SEM]) undergoing 1 or more divisions was graphed. Data shown are from 3 mice per group and representative of 3 independent experiments. (B) The proliferative index of these cells (ie, total number of divisions divided by the number of cells that went into division) was also determined. (C) CD4+ and CD8+ T cells from HDAC11KO mice were activated by CD3/CD28 stimulation for 72 hours. Supernatants were assessed by cytokine bead array for indicated cytokines. Values shown are from 3 mice per group (± SEM) and representative of 3 independent experiments. (D) CD8+ T cells from WT and HDAC11KO mice were left unstimulated or activated for 6 hours by αCD3/CD28–conjugated beads. Samples were then assessed by qRT-PCR for expression of granzyme B and perforin mRNA. Values shown are from 3 mice per group (± SEM) and representative of 3 independent experiments. (E) OVA-antigen–specific CD4+ T cells from OTII or OTII/HDAC11KO mice were cultured with splenocytes harvested from WT mice and pulsed with indicated concentrations of OVA peptide. Twenty-four and 48 hours later, IFN-γ production was assessed by ELISA. Results shown are from 3 mice per group and representative of 3 independent experiments. *P < .05; **P < .01; ***P < .001; ****P < .0001.
HDAC11KO T cells have increased effector functions. (A) CD8+ T cells from HDAC11KO mice were stained with a proliferation tracking dye and activated for 72 hours with αCD3/CD28 stimulation. Proliferation was evaluated by flow cytometry. The percentage of cells (± standard error of the mean [SEM]) undergoing 1 or more divisions was graphed. Data shown are from 3 mice per group and representative of 3 independent experiments. (B) The proliferative index of these cells (ie, total number of divisions divided by the number of cells that went into division) was also determined. (C) CD4+ and CD8+ T cells from HDAC11KO mice were activated by CD3/CD28 stimulation for 72 hours. Supernatants were assessed by cytokine bead array for indicated cytokines. Values shown are from 3 mice per group (± SEM) and representative of 3 independent experiments. (D) CD8+ T cells from WT and HDAC11KO mice were left unstimulated or activated for 6 hours by αCD3/CD28–conjugated beads. Samples were then assessed by qRT-PCR for expression of granzyme B and perforin mRNA. Values shown are from 3 mice per group (± SEM) and representative of 3 independent experiments. (E) OVA-antigen–specific CD4+ T cells from OTII or OTII/HDAC11KO mice were cultured with splenocytes harvested from WT mice and pulsed with indicated concentrations of OVA peptide. Twenty-four and 48 hours later, IFN-γ production was assessed by ELISA. Results shown are from 3 mice per group and representative of 3 independent experiments. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Antigen specific T cells lacking HDAC11 are hyperresponsive
To assess antigen-specific T-cell responses, the OTII/HDAC11KO mouse strain in which OVA-antigen–specific CD4+ T cells were devoid of HDAC11 was used. CD4+ T cells isolated from these mice or from control OTII mice were cocultured with WT antigen-presenting cells in the presence, or absence, of OVA peptide. Figure 2E shows that in the absence of cognate peptide, IFN-γ was not produced by either OTII or OTII/HDAC11KO T cells. At the lowest dose of OVA-peptide (2.5 µg/mL), only OTII/HDAC11KO T cells produce IFN-γ at 24 hours (P < .0001). At increasing peptide doses and at both time points, OTII/HDAC11KO T cells produced higher levels of IFN-γ compared with WT OTII T cells (P < .01 to P < .0001).
HDAC11KO mice T cells display increased percentages of effector subsets following activation
Given the differences in EGFP-HDAC11 expression observed among naive, effector, and memory CD4+ and CD8+ T cells, we evaluated the distribution of these populations in HDAC11KO mice at steady state and following activation. Analysis of CD4+ T cells isolated from lymph nodes of HDAC11KO and WT mice revealed no significant differences among naive, effector, effector memory, or central memory populations (supplemental Figure 3A). However, following activation by αCD3/CD28 beads, a reduction in naive CD4+ T cells was observed in HDAC11KO T cells (P < .05), accompanied by an increase in both effector (P < .01) and effector memory (P < .05) CD4+ T cells (supplemental Figure 3B). In contrast, unstimulated CD8+ T cells from the lymph nodes of HDAC11KO mice displayed decreased naive (P < .05) and increased central memory T cells (P < .05) (supplemental Figure 3C). Following αCD3/CD28 bead stimulation of HDAC11KO CD8+ T cells, we observed changes like those seen in the CD4+ T-cell compartment, with a reduction in naive T cells (P < .01) accompanied by an increase in both effector and effector memory T cells (P < .001) (supplemental Figure 3D).
HDAC11KO T cells express higher levels of the transcription factors Tbet and Eomes
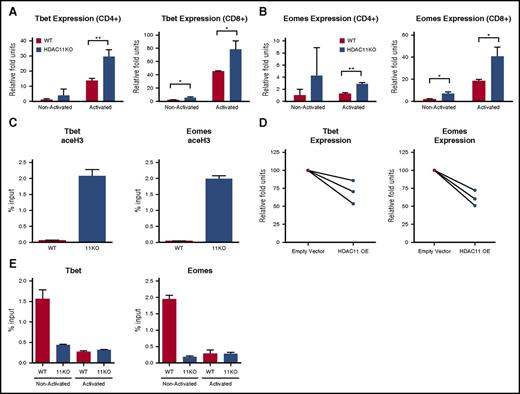
We next sought to elucidate the mechanisms contributing to the observed phenotype of HDAC11KO T cells. Given our findings that T cells devoid of HDAC11 had enhanced IFN-γ production and granzyme B expression, we investigated a potential role in transcription factors controlling these phenotypes. Previous studies have demonstrated that TBET and EOMES play a central role in the regulation of Th1 effector function12,17 and in the development of memory T cells.11,18,19 Therefore, we hypothesized that HDAC11 negatively regulate the expression of these transcription factors. To address this, we measured Tbet expression in CD4+ and CD8+ T cells from HDAC11KO mice. At baseline, higher Tbet mRNA expression was observed in HDAC11KO CD8+ T cells (P < .05) (Figure 3A). Following αCD3/CD28 activation, Tbet expression was increased in both CD4+ (P < .01) and CD8+ (P < .05) T cells lacking HDAC11 as compared with WT T cells. Similarly, enhanced Eomes expression was observed at baseline in HDAC11KO CD8+ T cells relative to WT T cells (P < .05) (Figure 3B). Following activation, Eomes mRNA levels were higher in both CD4+ (P < .01) and CD8+ (P < .05) T cells lacking HDAC11 compared with WT. Increased histone 3 acetylation levels at the Tbet and Eomes gene promoter regions were found in resting HDAC11KO CD3+ T cells, indicative of a transcriptionally permissive state (Figure 3C). To confirm that these effects were drivenby HDAC11, we overexpressed HDAC11 in human T cells (supplemental Figure 4). As shown in Figure 3D, HDAC11 overexpression resulted in decreased Tbet (P = .055) and Eomes (P < .05) mRNA expression in activated T cells as compared with empty-vector transfected cells.
HDAC11 regulates the expression of Eomes and Tbet. (A) CD4+ and CD8+ T cells isolated from WT (black bars) or HDAC11KO (white bars) mice were left unstimulated or activated for 6 hours with αCD3/CD28–conjugated beads. Expression of Tbet mRNA was assessed by qRT-PCR. (B) Expression of Eomes was likewise assessed. Results shown are for 3 mice per group and are representative of 4 independent experiments. Error bars are ± SEM. (C) CD3+ T cells isolated from WT (black bars) and HDAC11KO (white bars) mice were chromatin immunoprecipitated for pan-acetylated histone 3. Pull-down at the Eomes and Tbet promoters was assessed by qRT-PCR and calculated as percent of IgG input. Values shown are representative of 2 independent experiments of 3 pooled mice. Error bars are SEM for 3 technical replicates. (D) CD3+ T cells isolated from 3 healthy human donors in 2 separate experiments were transfected by electroporation with an empty vector or HDAC11 overexpression plasmid. Cells were allowed to rest overnight and were then activated with αCD3/CD28–conjugated beads for 6 hours, at which time cells were lysed and expression of Tbet and Eomes was determined by qRT-PCR. Empty vector expression values were normalized to 100, and donor-matched overexpressing T-cell expression was graphed as relative fold units. (E) WT and HDAC11KO T cells were left unstimulated or activated via PMA and ionomycin for 1 hour and then chromatin immunoprecipitated for HDAC11. Pull-down was assessed by qRT-PCR and calculated as percent of IgG input. Values shown are representative of 2 independent experiments. Error bars show SEM for technical replicates. *P < .05; **P < .01. OE, overexpression.
HDAC11 regulates the expression of Eomes and Tbet. (A) CD4+ and CD8+ T cells isolated from WT (black bars) or HDAC11KO (white bars) mice were left unstimulated or activated for 6 hours with αCD3/CD28–conjugated beads. Expression of Tbet mRNA was assessed by qRT-PCR. (B) Expression of Eomes was likewise assessed. Results shown are for 3 mice per group and are representative of 4 independent experiments. Error bars are ± SEM. (C) CD3+ T cells isolated from WT (black bars) and HDAC11KO (white bars) mice were chromatin immunoprecipitated for pan-acetylated histone 3. Pull-down at the Eomes and Tbet promoters was assessed by qRT-PCR and calculated as percent of IgG input. Values shown are representative of 2 independent experiments of 3 pooled mice. Error bars are SEM for 3 technical replicates. (D) CD3+ T cells isolated from 3 healthy human donors in 2 separate experiments were transfected by electroporation with an empty vector or HDAC11 overexpression plasmid. Cells were allowed to rest overnight and were then activated with αCD3/CD28–conjugated beads for 6 hours, at which time cells were lysed and expression of Tbet and Eomes was determined by qRT-PCR. Empty vector expression values were normalized to 100, and donor-matched overexpressing T-cell expression was graphed as relative fold units. (E) WT and HDAC11KO T cells were left unstimulated or activated via PMA and ionomycin for 1 hour and then chromatin immunoprecipitated for HDAC11. Pull-down was assessed by qRT-PCR and calculated as percent of IgG input. Values shown are representative of 2 independent experiments. Error bars show SEM for technical replicates. *P < .05; **P < .01. OE, overexpression.
HDAC11 interacts with the Tbet and Eomes gene promoters and disassociates upon T-cell activation
The demonstration that HDAC11 may influence the acetylation status and transcriptional activity of the Tbet and Eomes gene promoters led us to explore whether HDAC11 is recruited to these sites to regulate histone acetylation. To address this, we chromatin immunoprecipitated HDAC11 in T cells before and after activation. In nonactivated WT T cells, HDAC11 was present at the Tbet and Eomes gene promoter regions. At 1-hour postactivation with αCD3/CD28 beads, we observed negligible levels of HDAC11 at the promoter regions of these genes (Figure 3E).
HDAC11KO T cells are resistant to tolerance induction in vivo
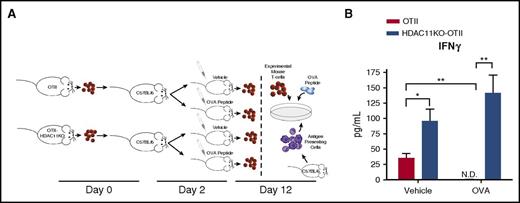
Previous studies have demonstrated that increased expression of Tbet and Eomes is correlated with decreased tolerance, so we sought to identify whether HDAC11KO T cells mediated similar effects.13,20,21 To determine the in vivo susceptibility of antigen-specific T cells lacking HDAC11 to high-dose peptide-induced tolerance, naive CD4+ T cells isolated from OTII or OTII/HDAC11KO mice were adoptively transferred into C57BL/6 mice. At day 2, mice were injected IV with a tolerogenic dose of OVA-peptide (100 µg) or vehicle. At day 12, antigen-specific T-cell responses were evaluated by measuring cytokine production ex vivo (Figure 4A). Both OTII and OTII/HDAC11KO OVA-specific CD4+ T cells that were transferred to vehicle-injected mice produced IFN-γ in response to ex vivo stimulation with OVA-peptide. The magnitude of this response was higher for OTII/HDAC11KO T cells (P < .05) (Figure 4B). OTII T cells exposed to high-dose OVA-peptide in vivo were rendered anergic, as determined by their lack of IFN-γ production upon ex vivo restimulation with OVA-peptide (P < .01). In contrast, tolerance induction was not observed in CD4+ T cells lacking HDAC11, as OTII/HDAC11KO CD4+ T cells were still capable of producing IFN-γ in response to ex vivo restimulation with OVA-peptide and displayed the highest IFN-γ production.
T cells lacking HDAC11 are resistant to tolerance induction. (A) A schematic representation of the tolerance induction model is shown. CD4+ T cells isolated from OTII or OTII/HDAC11KO mice were adoptively transferred into C57BL/6 WT recipients. Two days later, mice were challenged with 100 µg OVA323-339 peptide given IV or vehicle injection. Ten days following challenge, CD4+ T cells were harvested from mice and cocultured with WT splenocytes pulsed with OVA323-339 peptide. (B) Forty-eight hours later, IFN-γ production was assessed by ELISA. Results shown are from 3 mice per group and are representative of 2 independent experiments. *P < .05; **P < .01. N.D., not detected.
T cells lacking HDAC11 are resistant to tolerance induction. (A) A schematic representation of the tolerance induction model is shown. CD4+ T cells isolated from OTII or OTII/HDAC11KO mice were adoptively transferred into C57BL/6 WT recipients. Two days later, mice were challenged with 100 µg OVA323-339 peptide given IV or vehicle injection. Ten days following challenge, CD4+ T cells were harvested from mice and cocultured with WT splenocytes pulsed with OVA323-339 peptide. (B) Forty-eight hours later, IFN-γ production was assessed by ELISA. Results shown are from 3 mice per group and are representative of 2 independent experiments. *P < .05; **P < .01. N.D., not detected.
HDAC11KO T cells are less susceptible to suppression by regulatory T cells in vitro
We next evaluated susceptibility to regulatory T-cell (Treg) suppression. CD25− Teff from WT or HDAC11KO mice were labeled with the cell tracking dye carboxyfluorescein succinimidyl ester, cocultured with purified CD4+CD25+ Tregs from WT mice, and cultured with irradiated antigen-presenting cells and plate-bound αCD3 (1 μg/mL). In the absence of Tregs, HDAC11KO T cells displayed higher proliferation compared with WT T cells (supplemental Figure 5A-B, separate experiments shown). The addition of Tregs reduced proliferation of both HDAC11KO T cells and WT T cells. However, HDAC11KO Teff retained higher percentages of dividing cells relative to WT at all the Treg/Teff ratios evaluated (P < .001). After adjusting for differences in initial proliferation and evaluating suppression relative to proliferation in the absence of Tregs, HDAC11KO T cells still displayed increased levels of proliferating Teff relative to WT T cells (P < .01) (supplemental Figure 5C).
HDAC11KO T cells mediate potent and robust GVHD
Next, we determined the alloreactivity of T cells lacking HDAC11 in an in vivo model of acute GVHD. BALB/c mice underwent myeloablative irradiation and subsequently received BM cells with or without T cells from allogeneic WT or HDAC11KO C57BL/6 donors. Mice receiving allogeneic BM plus T cells from WT donors developed GVHD, as evidenced by decreased survival (Figure 5A squares) and reduction in body mass (Figure 5B), compared with mice that received only BM cells (ie, no T cells) (circles). However, mice receiving allogeneic WT BM plus T cells from HDAC11KO donors (triangles) developed GVHD more rapidly than WT (P < .05) and experienced a greater decrease in body mass than mice receiving allogeneic BM plus T cells from WT donors (q < 0.05). To determine the potency of GVHD induction by T cells lacking HDAC11, we titrated down the number of T cells in the allograft from WT or HDAC11KO donors. Unlike adoptively transferred WT T-cells, which at the doses of 0.5 × 106 (closed squares) or 0.25 × 106 (open squares) were no longer sufficient to induce GVHD, HDAC11KO T cells (closed triangles and open triangles, respectively) were still able to induce GVHD, resulting in a reduction in body mass (WT vs 11KO, 0.5 × 106 q < 0.0001, 0.25 × 106 q < 0.05) and impaired survival of recipient mice (WT vs 11KO, 0.5 × 106P < .05, 0.25 × 106P = .13) (Figure 5C-D).
HDAC11KO T cells mediate more potent GVHD. BALB/c mice were irradiated with 900 cGy and transplanted with 5 × 106 WT C57BL/6 mouse BM cells depleted of T cells with/without the addition of allogeneic WT or HDAC11KO T cells. (A) Survival of mice injected with BM alone (circles), BM plus 1 × 106 WT T cells (squares), or BM plus 1 × 106 HDAC11KO T cells (triangles) is displayed. (B) Body weight relative to initiation of the experiment was also monitored. Treatment groups contained 10 mice each. Results shown are from 2 independent experiments. (C) In a similar experiment, BALB/c mice were adoptively transferred with BM plus 0.5 × 106 WT T cells (closed squares), 0.5 × 106 HDAC11KO T cells (closed triangles), 0.25 × 106 WT T cells (open squares), or 0.25 × 106 HDAC11KO T cells (open triangles), and survival was monitored. (D) Body weight relative to initiation of experiment was also evaluated. (E) Serum IFN-γ and (F) TNF levels were assessed by ELISA 12 days after adoptive transfer with BM± T cells. Treatment groups contained 8 mice each. Results shown are from 2 independent experiments. (G) Splenic CD4+ and CD8+ T-cell numbers were assessed at day 5 after BM ± T-cell transplant. (H) The number of IFN-γ expressing CD4+ and CD8+ T cells was also assessed by intracellular flow cytometry after stimulation with PMA and ionomycin. (I) Fourteen days after adoptive transfer, mice were killed, and liver pathology was evaluated. (J) The number of CD8+ T cells and (K) expression levels of EOMES by CD8+ T cells in the liver were evaluated. (L) EOMES expression in splenic CD8+ T cells was likewise evaluated. Error bars graphed are SEM. *P < .05; **P < .01. BMT, bone marrow transplant; MFI, mean fluorescence intensity.
HDAC11KO T cells mediate more potent GVHD. BALB/c mice were irradiated with 900 cGy and transplanted with 5 × 106 WT C57BL/6 mouse BM cells depleted of T cells with/without the addition of allogeneic WT or HDAC11KO T cells. (A) Survival of mice injected with BM alone (circles), BM plus 1 × 106 WT T cells (squares), or BM plus 1 × 106 HDAC11KO T cells (triangles) is displayed. (B) Body weight relative to initiation of the experiment was also monitored. Treatment groups contained 10 mice each. Results shown are from 2 independent experiments. (C) In a similar experiment, BALB/c mice were adoptively transferred with BM plus 0.5 × 106 WT T cells (closed squares), 0.5 × 106 HDAC11KO T cells (closed triangles), 0.25 × 106 WT T cells (open squares), or 0.25 × 106 HDAC11KO T cells (open triangles), and survival was monitored. (D) Body weight relative to initiation of experiment was also evaluated. (E) Serum IFN-γ and (F) TNF levels were assessed by ELISA 12 days after adoptive transfer with BM± T cells. Treatment groups contained 8 mice each. Results shown are from 2 independent experiments. (G) Splenic CD4+ and CD8+ T-cell numbers were assessed at day 5 after BM ± T-cell transplant. (H) The number of IFN-γ expressing CD4+ and CD8+ T cells was also assessed by intracellular flow cytometry after stimulation with PMA and ionomycin. (I) Fourteen days after adoptive transfer, mice were killed, and liver pathology was evaluated. (J) The number of CD8+ T cells and (K) expression levels of EOMES by CD8+ T cells in the liver were evaluated. (L) EOMES expression in splenic CD8+ T cells was likewise evaluated. Error bars graphed are SEM. *P < .05; **P < .01. BMT, bone marrow transplant; MFI, mean fluorescence intensity.
To characterize the increased alloreactivity displayed by HDAC11KO T cells, we analyzed T-cell phenotypes in recipients and found that donor T cells were overwhelmingly effector memory cells regardless of HDAC11 expression (data not shown). We further evaluated the production of inflammatory cytokines of donor T cells in recipient mice. As shown in Figure 5E-F, serum levels of IFN-γ and TNF were higher in recipients of HDAC11KO T cells relative to recipients of WT T cells (P < .05 and P < .01, respectively). Increased serum inflammatory cytokine levels were accompanied by increased numbers of splenic CD4+ and CD8+ T cells in mice adoptively transferred with HDAC11KO T cells (P < .05) (Figure 5G) and an increased number of IFN-γ+ T cells reisolated from recipient mice adoptively transferred with HDAC11KO T cells (P < .05) (Figure 5H). However, no changes in the relative percentages of IFN-γ+ T cells were observed (data not shown). To determine whether differences in the presence of Tregs contributed to the enhanced GVHD mediated by HDAC11KO T cells, we evaluated splenic Tregs and found no differences in the percentages of FOXP3+ CD4+ T cells relative to WT T-cell recipients (supplemental Figure 5D). HDAC11KO T cells caused more severe liver damage in recipient mice than WT cells (Figure 5I), which was accompanied by an increase in T-cell infiltrate (P < .05) (Figure 5J) and CD8+ EOMES expression (P < .05) (Figure 5K). Similarly, increased EOMES expression was found in splenic CD8+ T cells of HDAC11KO-recipient mice (P < .05) (Figure 5L). However, no differences in TBET expression were observed (data not shown).
HDAC11KO T cells delay tumor progression in vivo
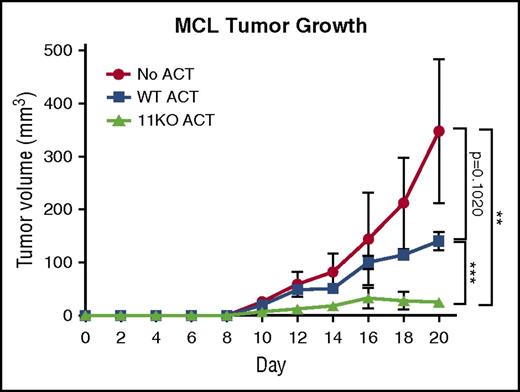
Given the observed enhanced alloreactivity, we evaluated the efficacy of HDAC11KO T cells in a syngeneic lymphoma tumor model. FCmuMCL1 tumor-bearing mice received adoptive transfers of either WT (squares) or HDAC11KO T cells (triangles) or no adoptive transfer (circles). Relative to WT recipients, HDAC11-recipient mice had a significant delay (P < .001) in tumor progression (Figure 6).
HDAC11KO T cells have enhanced antitumor efficacy in vivo. C57BL/6 mice were subcutaneously injected with 2.2 × 106 FCmuMCL1 cells on day 0. On day 4, 4 mice per group received adoptive transfer of WT T cells (squares), HDAC11KO T cells (triangles), or no adoptive transfer (circles). Group tumor volumes with standard deviations are graphed. Results shown are representative of 2 independent experiments. **P < .01; ***P < .001. ACT, adoptive cell therapy; MCL, mantle-cell lymphoma.
HDAC11KO T cells have enhanced antitumor efficacy in vivo. C57BL/6 mice were subcutaneously injected with 2.2 × 106 FCmuMCL1 cells on day 0. On day 4, 4 mice per group received adoptive transfer of WT T cells (squares), HDAC11KO T cells (triangles), or no adoptive transfer (circles). Group tumor volumes with standard deviations are graphed. Results shown are representative of 2 independent experiments. **P < .01; ***P < .001. ACT, adoptive cell therapy; MCL, mantle-cell lymphoma.
In a similar experiment, FCmuMCL1 tumors were harvested and assessed for CD4+ and CD8+ tumor-infiltrating cells by immunofluorescent staining. Although all 3 WT T-cell–recipient mice developed tumor, only 1 HDAC11KO T-cell recipient developed tumor. A representative immunofluorescence image of tumor infiltrate is shown in supplemental Figure 6A, showing increased infiltration of both CD4+ and CD8+ T cells in the HDAC11KO recipient. Quantification of WT and HDAC11KO CD8+ TIL is shown in supplemental Figure 6B.
Discussion
The results of this study demonstrate a previously unknown role of HDAC11 as a negative regulator of the effector responses of T cells. Data presented show that HDAC11 expression was downregulated in effector and effector memory T cells, and that HDAC11KO T cells displayed increased effector functions, suggesting that HDAC11 is a gatekeeper that is downregulated when T cells acquire effector function. This hypothesis was further supported by the observation that HDAC11 expression is rapidly reduced following activation of T cells. Alternatively, HDAC11 could function as a negative regulator of proliferation, as previous reports have shown that HDAC11 expression is inversely correlated with Ki67 expression.22 However, this would fail to completely explain the observed phenotypes of HDAC11KO T cells. Instead, our results support a model in which HDAC11 regulates the acquisition of T-cell activation and effector functions through maintenance of histone deacetylation of the Eomes and Tbet gene promoters, as illustrated in supplemental Figure 7.
Eomes and Tbet are paralogs, sharing 74% sequence homology in the T-box regions,12,17 have overlapping functions as regulators of IFN-γ and T-cell polarization10,12 and known roles in GVHD.13,20 In addition to the redundant regulation of IFN-γ, EOMES also modulates granzyme B and perforin expression in CD8+ T cells.12 Tbet and Eomes genes were previously shown to be regulated by histone acetylation23,24 ; however, the regulatory molecules involved were not identified.
CD4+ and CD8+ HDAC11KO T cells displayed higher basal and postactivation expression of Tbet and Eomes. However, although expression of Eomes was elevated in CD4+ HDAC11KO T cells, no notable changes in expression of Eomes were seen after activation, in line with reports showing that Eomes expression in CD4+ is negligible12 and suggests additional CD4+ T-cell–specific mechanisms acting to regulate expression of this transcription factor. Given the increased expression of Tbet and Eomes observed in HDAC11KO CD8+ T cells in the basal resting state, and the role of these 2 transcription factors in directing memory formation of T cells,11,19,25 it is plausible that their increased basal expression in T cells lacking HDAC11 might facilitate transition of these cells from naive to memory phenotype in vivo, accounting for the accumulation of CD8+ central memory T cells found in HDAC11KO mice. This increased basal expression of Tbet and Eomes appears to result in a “primed phenotype” that may explain the increased accumulation of both effector and effector-memory T cells following activation of HDAC11KO T cells. Although increased expression of EOMES in splenic and liver infiltrating CD8+ T cells was observed in our GVHD model, no differences in TBET expression were found. The lack of difference may have resulted from differences in T-cell subset homing. To support this explanation, we found splenic and liver T-cell infiltrates to be >95% effector memory (data not shown), a population with preferential homing to nonlymphoid organs26 and expressing lower levels of TBET.27
In addition to having increased proinflammatory phenotypes, T cells lacking HDAC11 were less susceptible to tolerance induction and experienced less suppression of proliferation by Tregs. Studies have suggested critical roles of Tbet and Eomes in several immunosuppressive T-cell populations,28,29 including Tregs,27,30 and have highlighted epigenetic regulation as a critical component of T-cell immunosuppression, particularly in Tregs,31 with several HDACs having demonstrated roles in development and function.32-34 Although beyond the focus of this article, identification of the role of HDAC11 and its regulation of Tbet and Eomes in T-cell immunosuppression is needed. The role of HDAC11 in Treg development and function is being addressed in a separate study.
Previous work from our group identified HDAC11 as a negative regulator of IL-10 production in antigen-presenting cells.9 However, we have recently demonstrated that even in the absence of HDAC11, HDAC6 is necessary for expression of IL-10.35 Based on those results and the data presented here, it is hypothesized that HDAC11’s ability to regulate proinflammatory and humoral immune responses is a product of its binding partners or other regulatory machinery. In the data presented herein, absence of HDAC11 alone was not sufficient to induce an inflammatory T-cell response, as activation was still required. Future identification of HDAC11 complex partners or other switches in the regulation of immune response is critical.
Studies addressing HDAC inhibition in allograft models have shown disparate effects dependent on inhibitor. For example, treatment with the HDAC inhibitor vorinostat led to increased survival in mice7,36 and increased Treg numbers and function in patients.37 Conversely, the HDAC inhibitor trichostatin A was shown to increase the expression of proinflammatory cytokines and increased histone acetylation of the granzyme B and Eomes genes in Teff,38 and treatment with the HDAC inhibitor panobinostat resulted in accelerated GVHD characterized by increased Th1 cytokines.39 Our studies have demonstrated that HDAC11KO T cells have similar increases in Th1 cytokines expression and accelerated GVHD, suggesting that previous results may be in part due to inhibition of HDAC11. Trichostatin A and panobinostat have potency against HDAC11, whereas vorinostat has none. These disparate data demonstrate that nonspecific HDAC inhibition can result in dramatically different T-cell responses and disease outcome.
Collectively, these data demonstrate that abrogation of HDAC11 augments the proinflammatory phenotype and function of T cells. Although this study was conducted mainly in murine models, human HDAC11 shares 91% sequence homology with the mus musculus gene,40 warranting evaluation as an immunotherapeutic target. These findings represent a step forward in the understanding of the mechanisms of epigenetic regulation of T-cell activation and highlight HDAC11 as a novel immunotherapeutic target.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors extend their appreciation to the Flow Cytometry Core, Tissue Core, and animal facilities at Moffitt Cancer Center for providing technical assistance.
This work was supported by National Institutes of Health, National Cancer Institute, grants RO1CA179062 (E.M.S.), RO1CA134807 (E.M.S.), P50CA168536 (E.M.S.), R01CA143821 (X.-Z.Y.), and R01CA169116 (X.-Z.Y.).
Authorship
Contribution: D.M.W. designed, conducted, and analyzed experiments, and wrote the manuscript; K.V.W. and D.W. designed, conducted, and analyzed experiments; A.L.S. designed, conducted, and analyzed experiments and assisted in manuscript preparation; F.C., Y.W., Z.W., J.C., J.P., Y.Y., Y.Z., X.W., X.Z. conducted and analyzed experiments; J.P.-I., W.W.H., E.S., and A.V. provided scientific input and direction; J.W. provided scientific input and direction and assisted in manuscript preparation; X.-Z.Y. provided scientific input and direction, provided mouse models, and oversaw aspects of the project; and E.M.S. oversaw the project, provided scientific input and direction, and assisted in manuscript preparation.
Conflict-of-interest disclosure: E.M.S., K.V.W., and D.M.W. are named on a pending patent for targeting HDAC11. The remaining authors declare no competing financial interests.
Correspondence: Eduardo M. Sotomayor, George Washington Cancer Center, 2150 Pennsylvania Ave, NW, Suite 1-401, Washington, DC 20037; e-mail: esotomayor@mfa.gwu.edu; and Xue-Zhong Yu, Medical University of South Carolina, 86 Jonathan Lucas St, Charleston, SC 29425-5090; e-mail: yux@musc.edu.
References
Author notes
D.M.W. and K.V.W. contributed equally to this study.
X.-Z.Y. and E.M.S. are joint senior authors.


![Figure 2. HDAC11KO T cells have increased effector functions. (A) CD8+ T cells from HDAC11KO mice were stained with a proliferation tracking dye and activated for 72 hours with αCD3/CD28 stimulation. Proliferation was evaluated by flow cytometry. The percentage of cells (± standard error of the mean [SEM]) undergoing 1 or more divisions was graphed. Data shown are from 3 mice per group and representative of 3 independent experiments. (B) The proliferative index of these cells (ie, total number of divisions divided by the number of cells that went into division) was also determined. (C) CD4+ and CD8+ T cells from HDAC11KO mice were activated by CD3/CD28 stimulation for 72 hours. Supernatants were assessed by cytokine bead array for indicated cytokines. Values shown are from 3 mice per group (± SEM) and representative of 3 independent experiments. (D) CD8+ T cells from WT and HDAC11KO mice were left unstimulated or activated for 6 hours by αCD3/CD28–conjugated beads. Samples were then assessed by qRT-PCR for expression of granzyme B and perforin mRNA. Values shown are from 3 mice per group (± SEM) and representative of 3 independent experiments. (E) OVA-antigen–specific CD4+ T cells from OTII or OTII/HDAC11KO mice were cultured with splenocytes harvested from WT mice and pulsed with indicated concentrations of OVA peptide. Twenty-four and 48 hours later, IFN-γ production was assessed by ELISA. Results shown are from 3 mice per group and representative of 3 independent experiments. *P < .05; **P < .01; ***P < .001; ****P < .0001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/2/10.1182_blood-2016-08-731505/4/m_blood731505f2.jpeg?Expires=1765896294&Signature=AhIdF8oZ9s9A4bXjeAoCCvavrtzBG1J9Vahq4bXXsxClssBVz-iiiLElLkXSC9DwPnerDkaUNsivULOWq0~tqOtEEW-DJGFX9GANvqQmcOgx9tG9l1yd2AdwtPxJDM90EwiQPv19JOpWz4ydo4ahBlU2ELHzB1wQMsAbWEJ3yJWGt4kWv~Unze~Jppz4hEUDjb~ybeVA0H06F3UX-mIoZnbFeyudhgVWKXnqyE29lBzQRC1ikDYSFXDkBjHhAEikznRlAW98iyHjlHAZDFvffWh92mGmZ4-olW-D3fi3UqqQr-SJCgfMufCAF6a2w5gXYWy4qQuIlcGjQo8sQVj2Sw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)