Key Points
USP7 is overexpressed and regulates HRR in CLL cells.
USP7 inhibition is selectively cytotoxic to CLL cells independently of ATM and p53 and synergizes with chemotherapy.
Abstract
The role of deubiquitylase ubiquitin-specific protease 7 (USP7) in the regulation of the p53-dependent DNA damage response (DDR) pathway is well established. Whereas previous studies have mostly focused on the mechanisms underlying how USP7 directly controls p53 stability, we recently showed that USP7 modulates the stability of the DNA damage responsive E3 ubiquitin ligase RAD18. This suggests that targeting USP7 may have therapeutic potential even in tumors with defective p53 or ibrutinib resistance. To test this hypothesis, we studied the effect of USP7 inhibition in chronic lymphocytic leukemia (CLL) where the ataxia telangiectasia mutated (ATM)–p53 pathway is inactivated with relatively high frequency, leading to treatment resistance and poor clinical outcome. We demonstrate that USP7 is upregulated in CLL cells, and its loss or inhibition disrupts homologous recombination repair (HRR). Consequently, USP7 inhibition induces significant tumor-cell killing independently of ATM and p53 through the accumulation of genotoxic levels of DNA damage. Moreover, USP7 inhibition sensitized p53-defective, chemotherapy-resistant CLL cells to clinically achievable doses of HRR-inducing chemotherapeutic agents in vitro and in vivo in a murine xenograft model. Together, these results identify USP7 as a promising therapeutic target for the treatment of hematological malignancies with DDR defects, where ATM/p53-dependent apoptosis is compromised.
Introduction
The clinical heterogeneity of chronic lymphocytic leukemia (CLL) is underscored by genomic aberrations, particularly those involving the chromosomal regions 17p and 11q. These are the sites of the DNA damage response (DDR) genes ATM and TP53, respectively, deletions or mutations of which can adversely affect prognosis and treatment response.1-4 Ataxia telangiectasia mutated (ATM) and p53 play essential roles in DDR by coordinating DNA repair with apoptosis. Defects in DDR genes facilitate cellular transformation and/or tumor progression by allowing mutations and chromosomal alterations to persist and accumulate, thus increasing genomic instability.5 DDR defects also result in resistance to conventional chemotherapeutic agents. This has prompted the development of DDR-independent therapies, including monoclonal antibodies as well as Bcl-2 and B-cell receptor (BCR) signaling inhibitors.6-10 Although these agents act independently of the ATM/p53 pathway, in vitro studies have shown that CLL cells harboring del(17p) or TP53 mutations are less sensitive to the BCR signaling inhibitor ibrutinib.11 Moreover, BCR signaling inhibitors are not curative, and patients with del(17p) CLL continue to exhibit inferior clinical outcomes with these agents, indicating that alternative treatment strategies and therapeutic combinations are still needed.6,10,12
DDR-deficient cells rely on cooperating DNA repair pathways for survival. Therefore, an alternative therapeutic approach involves the selective targeting of DDR-deficient CLL cells by inhibiting these pathways, which results in synthetic lethality.13,14 Recently, we showed that poly ADP-ribose polymerase 1 (PARP1) inhibition selectively targets the homologous recombination repair (HRR) deficiency in ATM-deficient CLL and that ataxia telangiectasia and Rad3-related (ATR) inhibition can be used to exploit the replication checkpoint deficiency in ATM- or TP53-deficient CLL.15,16 Both approaches result in CLL cell death by mitotic catastrophe. Unlike DDR-independent therapies, such strategies specifically target the more genomically unstable and aggressive DDR-deficient tumor subpopulations.
Protein ubiquitylation is an essential posttranslational modification for regulating cellular DDR.17 It is critical for controlling the stability of many DDR proteins and in regulating their function and subcellular localization. Ubiquitylation involves the covalent attachment of ubiquitin residues onto target proteins through the sequential actions of E1, E2, and E3 enzymes, which activate, conjugate, and ligate ubiquitin, respectively. Conversely, deubiquitylases (DUBs) are enzymes that remove ubiquitin residues.17 USP7 is a DUB that plays a major role in regulating p53 function through its ability to stabilize MDM2, an E3-ubiquitin ligase that ubiquitylates and targets p53 for proteasomal degradation.18-20 Genetic studies have demonstrated that loss of USP7 allows MDM2 autoubiquitylation and subsequent degradation, which results in p53 stabilization and induction of G1 cell-cycle arrest or apoptosis.21
Consistent with this role, pharmacological targeting of USP7 has been shown to reverse MDM2-mediated downregulation of p53, leading to enhanced cytotoxicity of multiple myeloma cells.22 Alternatively, p53 can be stabilized by MDM2 inhibitors such as nutlins, but unlike in USP7 inhibition, their efficacy seems to be entirely p53 dependent and therefore of limited utility in p53-deficient malignancies.23,24 This difference may be a reflection of the increasing number of p53-independent roles identified for USP7.25-28 Indeed, we recently showed that the loss of USP7 destabilizes RAD18, an E3-ubiquitin ligase, compromising postreplication repair.29
In view of this, we reasoned that USP7 could represent an attractive therapeutic target for tumors containing ATM or p53 functional defects. Using CLL as a tumor model with a high frequency of ATM/TP53 aberrations, we demonstrate that inhibition of USP7 leads to selective loss of CLL viability independently of ATM and p53 function, mediated through the accumulation of unrepaired DNA double-strand breaks (DSBs). Furthermore, we show, for the first time, that USP7 inhibition compromises HRR and thus sensitizes tumor cells to HRR-inducing chemotherapeutic agents in vitro and in vivo.
Materials and methods
Patient samples and cell lines
Peripheral blood mononuclear cells (PBMCs) were isolated from CLL samples collected from patients across Birmingham, United Kingdom, and healthy volunteers. This study was approved by the South Birmingham Ethics Committee and performed according to institutional guidelines, and written consent was obtained from all participants. The 80 CLL samples all contained >90% CD19+ CD5+ cells. Analysis of IGHV, ATM, and TP53 genes and ATM/p53 function was performed as described previously (supplemental Table 1, available on the Blood Web site).30,31 All primary CLL samples and isogenic CLL cell lines (CII-, PGA-, and Mec1-expressing short hairpin RNAs (shRNAs) complementary to either green fluorescent protein [GFP] or ATM) were cultured in RPMI 1640 (Thermo Fisher Scientific, Rugby, United Kingdom) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific). HeLa and U2OS cells were grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. Primary CLL cells and healthy donor PBMCs were induced to proliferate by coculturing with mouse embryonic fibroblasts expressing CD40 ligand (CD40L), supplemented with 25 ng/mL interleukin-21 (IL-21; Thermo Fisher Scientific).32
Immunoblotting
Xenotransplantation and treatment
Animals were treated in accordance with UK Home Office guidelines, and thus, the experimental end point was at the first sign of illness exceeding license restrictions in any treatment arm. Six-week-old NOD/LtSz-SCID/IL2γtm1Wjl/SzJ mice were sublethally irradiated (1.25 Gy) before intravenous (tail vein) injection of Mec1 cells (3 × 106). Intravenous biweekly treatment with DMSO, HBX19818 (5 or 10 mg/kg), cyclophosphamide (20 mg/kg; Baxter, Newbury, United Kingdom), rituximab (Roche, West Sussex, United Kingdom), or a combination with HBX19818 commenced 4 days after Mec1 cell administration. After euthanization, splenic single-cell suspensions labeled with hCD45-FITC (Thermo Fisher Scientific) were quantified using an Accuri C6 flow cytometer. In some cases, CountBright beads (Thermo Fisher Scientific) were used to obtain absolute cell counts.
Cytotoxicity assays, immunocytochemistry, transfection, HRR, and comet assays
These procedures are described in supplemental data.
Statistical analysis
Statistical analysis was performed using SigmaPlot 12.5 software (Systat Software Inc., London, United Kingdom). P ≤ .05 was considered significant. Data are presented as mean ± standard error of the mean.
Results
Primary CLL cells overexpress USP7 and demonstrate differential sensitivity to USP7 inhibition in vitro and in vivo
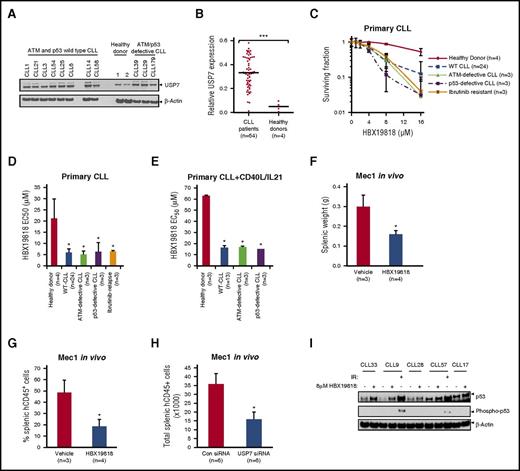
To address the feasibility of targeting USP7 in CLL, we assessed the level of USP7 protein expression in CLL cells from a patient cohort compared with PBMCs obtained from healthy donors. The panel of analyzed CLL samples (n = 64) comprised ATM/p53-proficient (n = 54) and ATM/p53-defective cases (n = 5 each). USP7 protein expression was significantly higher (P < .0005) in CLL samples (n = 64) irrespective of ATM or p53 defects in comparison with PBMCs from healthy donors (n = 4; Figure 1A-B).
USP7 is overexpressed in CLL and can be targeted with HBX19818 without activating p53 phosphorylation. Immunoblotting (A) and densitometry (B) quantification of primary CLL and healthy donor PBMCs demonstrate overexpression of USP7 in CLL samples. USP7 expression was normalized to β-actin. (C) HBX19818 dose responses of quiescent primary CLL cells and healthy donor PBMCs and the resultant 50% effective concentration (EC50) for quiescent (D) and proliferating (E) healthy donor and primary CLL cells after 72-hour treatment. HBX19818 treatment of a chemotherapy-resistant Mec1 CLL xenograft model led to reduction of splenic weight (F) and splenic tumor engraftment (G). (H) Xenotransplantation of confirmed USP7 knockdown Mec1 cells by siRNA transfection also led to reduced splenic engraftment. (I) In a panel of 5 wild-type (WT) CLLs, 6-hour HBX19818 treatment stabilized p53 but did not induce its phosphorylation. As a positive control for p53 phosphorylation, CLL9 and CLL57 were treated with 5-Gy ionising radiation (IR). Data were compared using a 2-tailed Student t test, and statistical significance denoted by: *P ≤ .05, ***P ≤ .001. siRNA, small interfering RNA.
USP7 is overexpressed in CLL and can be targeted with HBX19818 without activating p53 phosphorylation. Immunoblotting (A) and densitometry (B) quantification of primary CLL and healthy donor PBMCs demonstrate overexpression of USP7 in CLL samples. USP7 expression was normalized to β-actin. (C) HBX19818 dose responses of quiescent primary CLL cells and healthy donor PBMCs and the resultant 50% effective concentration (EC50) for quiescent (D) and proliferating (E) healthy donor and primary CLL cells after 72-hour treatment. HBX19818 treatment of a chemotherapy-resistant Mec1 CLL xenograft model led to reduction of splenic weight (F) and splenic tumor engraftment (G). (H) Xenotransplantation of confirmed USP7 knockdown Mec1 cells by siRNA transfection also led to reduced splenic engraftment. (I) In a panel of 5 wild-type (WT) CLLs, 6-hour HBX19818 treatment stabilized p53 but did not induce its phosphorylation. As a positive control for p53 phosphorylation, CLL9 and CLL57 were treated with 5-Gy ionising radiation (IR). Data were compared using a 2-tailed Student t test, and statistical significance denoted by: *P ≤ .05, ***P ≤ .001. siRNA, small interfering RNA.
Next, we used an irreversible USP7 inhibitor, HBX19818, to investigate the effect of USP7 inhibition on CLL.33 In CLL cells pretreated with HBX19818, we observed a dose-dependent reduction in the binding of HA-labeled ubiquitin to USP7, but not to USP3, a closely related DUB (supplemental Figure 1A). Furthermore, in accordance with the role of USP7 in the regulation of protein ubiquitylation, a time-dependent accumulation of polyubiquitylated proteins was observed after treatment of Mec1 CLL cells with HBX19818 (supplemental Figure 1B). Together these results demonstrate that HBX19818 specifically interacts with USP7 and functions as an inhibitor of protein deubiquitylation.
In vitro, quiescent CLL cells from patient samples (n = 33, including 3 ATM-defective, 3 p53-defective, and 3 ibrutinib-resistant patient cases) were significantly more sensitive to HBX19818 than healthy donor PBMCs (n = 4), with EC50 of 6 µM vs 21 µM, respectively (Figure 1C-D). Likewise, primary CLL samples (n = 19) induced to proliferate by coculturing with CD40L/IL-21 were significantly more sensitive to HBX19818 than healthy donor B cells induced to proliferate by the same method (Figure 1E), and the HBX19818 dose (≤10 μM) used for all subsequent experiments was cytotoxic to CLL but not healthy donor lymphocytes. This indicated that inhibition of USP7 selectively targets quiescent and proliferating CLL cells irrespective of their subtype. Notably, the HBX19818 EC50 of ATM- or p53-defective samples (n = 6), whether quiescent or proliferating, did not differ significantly from ATM/p53-proficient samples, suggesting that the cytotoxic effect of HBX19818 occurs through an ATM/p53-independent mechanism (Figure 1C-E). This finding was corroborated by data on the CII (p53 wild-type) and Mec1 (p53 defective) CLL cell lines stably expressing GFP shRNA or ATM shRNA,15 which shows that the ATM and p53 functional status of the cell lines did not influence sensitivity to HBX19818 (supplemental Figure 1C).
To examine the antitumor activity of HBX19818 in vivo, a Mec1 CLL murine xenograft model was employed. Therapeutic doses of HBX19818 were well tolerated in mice. Moreover, a 2-week course of biweekly treatment led to tumor load reduction, as evidenced by significant reduction of splenic weight accompanied by decreased splenic engraftment (Figure 1F-G). In addition, siRNA-mediated downregulation of USP7 (supplemental Figure 1D) before xenotransplantation led to a similar splenic Mec1 tumor load reduction (Figure 1H). Taken together, the in vitro and in vivo data suggest that USP7 is a promising target in CLL, and its inhibition with HBX19818 has substantial ATM/p53-independent activity.
Finally, protein-level observations provided additional support for a p53-independent mechanism underlying the activity of HBX19818 in CLL. As expected from previous reports, treatment of ATM/p53 wild-type CLL cells with HBX19818 induced the upregulation of p53 (Figure 1I). However, unlike IR, inhibition of USP7 did not induce phosphorylation of p53, suggesting that the accumulated p53 was not activated.
USP7 inhibition compromises HRR and results in accumulation of DNA damage
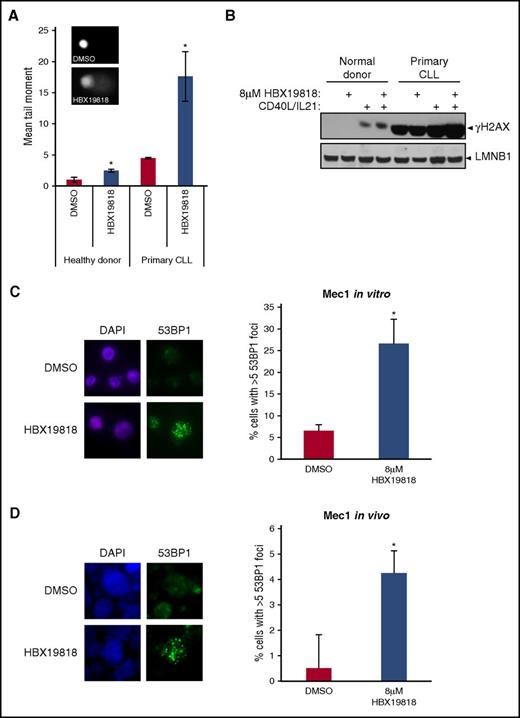
To explore the mechanisms underlying the p53-independent effects of USP7 inhibition, we first addressed whether USP7 inhibition leads to differential accumulation of unrepaired DNA damage in primary CLLs compared with healthy donor lymphocytes. We performed comet assays, which use the increased electrophoretic mobility of fragmented DNA as a measure of DNA damage,34 and we also assessed levels of γH2AX, a marker of DNA damage. Inhibition of USP7 led to increased tail moment and elevated γH2AX levels in lymphocytes derived from patients with CLL or healthy donors (Figure 2A). However, both basal and USP7 inhibition–induced DNA damage were differentially higher in CLL cells (Figure 2A-B). In agreement with this observation, siRNA-mediated depletion of USP7 in HeLa cells also increased comet tail moments (supplemental Figure 2).
Inhibition of USP7 with HBX19818 induces accumulation of DNA damage. (A) DNA damage was significantly elevated both in primary CLL cells (n = 3) and healthy donor lymphocytes (n = 3) treated with 8-µM HBX19818 for 6 hours, as quantified from Comet assay–derived tail moments. HBX-induced DNA damage was increased 3.92-fold in CLL cells and 2.42-fold in healthy donor cells, relative to DMSO. Representative images of DMSO- and HBX19818-treated cells are depicted. (B) Levels of the DNA damage marker γH2AX increased after HBX19818 treatment of quiescent and proliferating primary CLL cells and proliferating healthy donor cells. Lamin B1 (LMNB1) was the loading control. Immunofluorescence labeling showed significant induction of 53BP1 foci, a DSB marker, in Mec1 cells after 6-hour treatment with HBX19818 in vitro (C) and in vivo (D) in the chemotherapy-resistant murine xenograft model. Representative images are shown (DAPI, blue; 53BP1, green) and the mean data from 3 independent experiments are presented. Original magnification ×60 (A,C-D). Data were compared using a 2-tailed Student t test, and statistical significance denoted by: *P ≤ .05.
Inhibition of USP7 with HBX19818 induces accumulation of DNA damage. (A) DNA damage was significantly elevated both in primary CLL cells (n = 3) and healthy donor lymphocytes (n = 3) treated with 8-µM HBX19818 for 6 hours, as quantified from Comet assay–derived tail moments. HBX-induced DNA damage was increased 3.92-fold in CLL cells and 2.42-fold in healthy donor cells, relative to DMSO. Representative images of DMSO- and HBX19818-treated cells are depicted. (B) Levels of the DNA damage marker γH2AX increased after HBX19818 treatment of quiescent and proliferating primary CLL cells and proliferating healthy donor cells. Lamin B1 (LMNB1) was the loading control. Immunofluorescence labeling showed significant induction of 53BP1 foci, a DSB marker, in Mec1 cells after 6-hour treatment with HBX19818 in vitro (C) and in vivo (D) in the chemotherapy-resistant murine xenograft model. Representative images are shown (DAPI, blue; 53BP1, green) and the mean data from 3 independent experiments are presented. Original magnification ×60 (A,C-D). Data were compared using a 2-tailed Student t test, and statistical significance denoted by: *P ≤ .05.
We proceeded to investigate the nature of the DNA damage induced by USP7 inhibition. A marker of DNA DSBs, 53BP1 foci, accumulated in Mec1 cells after HBX19818 treatment both in vitro and in vivo (Figure 2C-D). Moreover, in keeping with our recent report demonstrating that USP7 activity is required to stabilize RAD18,29 RAD18 protein levels were reduced in both CLL and healthy donor cells after HBX19818 treatment (Figure 3A). The impact of USP7 inhibition on RAD18 stability was independent of ATM/p53, as evidenced by comparable RAD18 downregulation exhibited by p53 wild-type and defective isogenic CLL cell lines stably expressing either GFP (control) or ATM shRNA, when treated with HBX19818. RAD18 was also destabilized in HeLa cells after siRNA-induced USP7 depletion (Figure 3B). The specificity of this effect was demonstrated by the coexpression of siRNA-resistant FLAG-USP7, which replaced the depleted endogenous USP7 and rescued RAD18 stability (Figure 3B).
USP7-disrupted cells show reduced activation of HRR. (A) Normal healthy donor PBMCs and 3 isogenic CLL cell lines (2 p53 wild type [wt] and 1 p53 defective [def]) expressing the indicated shRNAs show diminished expression and thus destabilization of RAD18 protein after treatment with HBX19818. β-actin was the loading control (Con). (B) HeLa cells transfected with USP7 or Con siRNAs and empty (Vec) or siRNA-resistant Flag-USP7 (R-USP7)–expressing plasmid vectors demonstrate rescue of RAD18 stability in a complementation assay. The reduction of RAD18 levels in the presence of USP7 siRNA alone is prevented by the expression of R-USP7, which replaced the endogenous USP7 depleted by USP7 siRNA. (C,D) Representative immunofluorescence labeling of DNA damage markers γH2AX and RAD51 (green) with DAPI-counterstained nuclei (blue); (E-H) quantification of RAD51 foci formation from 24-hour time-course studies of 5-Gy IR-induced damage (n = 3). In MEC1 cells (C,G), normal healthy donor cells (E), and primary CLL cells (F), HBX19818 induced inhibition of USP7 1 hour before irradiation abrogated the formation of IR-induced RAD51 foci. (H) IR-induced RAD51 foci formation was reduced in HeLa cells after USP7 depletion by USP7 siRNA. (I) Analysis of HRR using the DR-GFP reporter assay in U2OS cells showed that HRR is compromised by USP7 depletion. CtIP depletion was the positive Con for HRR-defective cells. Original magnification ×60 (C-D). Data were compared using a 2-tailed Student t test, and statistical significance denoted by: *P ≤ .05, **P ≤ .01. Ub, ubiquitylated.
USP7-disrupted cells show reduced activation of HRR. (A) Normal healthy donor PBMCs and 3 isogenic CLL cell lines (2 p53 wild type [wt] and 1 p53 defective [def]) expressing the indicated shRNAs show diminished expression and thus destabilization of RAD18 protein after treatment with HBX19818. β-actin was the loading control (Con). (B) HeLa cells transfected with USP7 or Con siRNAs and empty (Vec) or siRNA-resistant Flag-USP7 (R-USP7)–expressing plasmid vectors demonstrate rescue of RAD18 stability in a complementation assay. The reduction of RAD18 levels in the presence of USP7 siRNA alone is prevented by the expression of R-USP7, which replaced the endogenous USP7 depleted by USP7 siRNA. (C,D) Representative immunofluorescence labeling of DNA damage markers γH2AX and RAD51 (green) with DAPI-counterstained nuclei (blue); (E-H) quantification of RAD51 foci formation from 24-hour time-course studies of 5-Gy IR-induced damage (n = 3). In MEC1 cells (C,G), normal healthy donor cells (E), and primary CLL cells (F), HBX19818 induced inhibition of USP7 1 hour before irradiation abrogated the formation of IR-induced RAD51 foci. (H) IR-induced RAD51 foci formation was reduced in HeLa cells after USP7 depletion by USP7 siRNA. (I) Analysis of HRR using the DR-GFP reporter assay in U2OS cells showed that HRR is compromised by USP7 depletion. CtIP depletion was the positive Con for HRR-defective cells. Original magnification ×60 (C-D). Data were compared using a 2-tailed Student t test, and statistical significance denoted by: *P ≤ .05, **P ≤ .01. Ub, ubiquitylated.
RAD18 stimulates the repair of DSBs by facilitating the relocalization of proteins required for HRR, including FANCD2 and RAD51, to sites of DNA damage. Therefore, we examined the effect of USP7 depletion or inhibition on HRR by quantifying the formation of IR-induced RAD51/FANCD2 foci. Consistent with its importance in promoting HRR, inhibition of USP7 significantly reduced the formation of IR-induced RAD51 foci in proliferating healthy donor, primary CLL, and Mec1 cells (Figure 3C,E-G). This difference was observed as early as 1 hour post-irradiation and is therefore unlikely to have been caused by cell-cycle alterations (supplemental Figure 3A). Defective localization of Rad51 was also seen in IR-treated HeLa cells depleted of USP7 (Figure 3D,H) and in USP7−/− HCT116 cells (supplemental Figure 3B). Furthermore, re-expression of USP7 rescued RAD51 foci in USP7−/− HCT116 cells (supplemental Figure 3C). In addition to IR, we examined the effect of USP7 knockdown on HRR induced by mitomycin C (MMC), an alkylating agent that causes DNA cross-links necessitating functional HRR for repair. Activation of HRR stimulates the ubiquitylation of FANCD2. In accordance with its role in HRR, knockdown of USP7 decreased FANCD2 ubiquitylation, as evidenced by the reduced ratio of ubiquitylated to nonubiquitylated FANCD2 protein, as well as the formation of FANCD2 and RAD51 foci in HeLa cells treated with MMC for 24 hours (supplemental Figure 3D-G).
Lastly, to assess the functional consequences of reduced RAD51 recruitment to sites of DNA damage after abrogation of USP7, we used the plasmid-based DR-GFP HRR reporter system to directly measure HRR. In this assay, restriction enzyme (I-Sce1)–directed cutting/DSB formation in a defective GFP gene induces HRR using homologous sequences from another differentially defective GFP gene, resulting in restoration of GFP function and the appearance of fluorescently labeled cells.35 Depletion of USP7 significantly reduced the number of GFP-positive cells, indicating decreased HRR-dependent DSB repair (Figure 3I; supplemental Figure 3H).
USP7 inhibition induces caspase-independent cell death through overactivation of PARP1
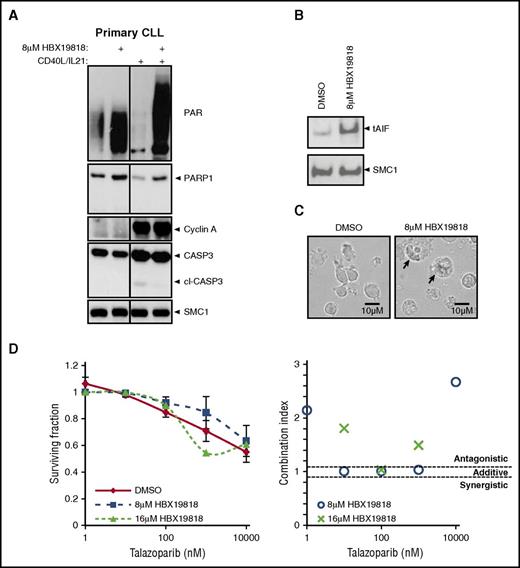
We further investigated the process through which tumor-cell death occurs when USP7 is inhibited. Consistent with a p53-independent mechanism of cellular death, inhibition of USP7 did not induce caspase-3 or PARP1 cleavage in either quiescent or proliferating primary CLL cells. Rather, after exposure of CLL cells to HBX19818, both PARP1 expression and activity increased as measured by the level of protein poly ADP-ribosylation (Figure 4A).
USP7 inhibitor HBX19818 increased PARP1, protein PARylation, and nuclear apoptosis-inducing factor. (A) HBX19818 treatment (6 hours) elevates PARP1 and PARylated protein levels in the absence of caspase-3 cleavage (cl-CASP3) in both quiescent (without CD40L/IL-21) and proliferating (with CD40L/IL-21) primary CLL cells. CD40L/IL-21–stimulated proliferation was confirmed by the presence of cyclin A expression. SMC1 was the loading control. (B) Analysis of nuclear extracts from proliferating primary CLL cells demonstrated that HBX19818-induced translocation of apoptosis-inducing factor (tAIF) accumulation in the nucleus. (C) HBX19818 treatment for 72 hours induced CLL cells to swell and disperse their contents (arrows), morphological changes that are consistent with necrotic cell death. (D) In ATM/p53-defective primary CLL induced to proliferate (n = 6) the PARP inhibitor talazoparib did not potentiate the cytotoxic activity (left) of HBX19818 and acted in an antagonistic manner, as indicated by combination indices (right).
USP7 inhibitor HBX19818 increased PARP1, protein PARylation, and nuclear apoptosis-inducing factor. (A) HBX19818 treatment (6 hours) elevates PARP1 and PARylated protein levels in the absence of caspase-3 cleavage (cl-CASP3) in both quiescent (without CD40L/IL-21) and proliferating (with CD40L/IL-21) primary CLL cells. CD40L/IL-21–stimulated proliferation was confirmed by the presence of cyclin A expression. SMC1 was the loading control. (B) Analysis of nuclear extracts from proliferating primary CLL cells demonstrated that HBX19818-induced translocation of apoptosis-inducing factor (tAIF) accumulation in the nucleus. (C) HBX19818 treatment for 72 hours induced CLL cells to swell and disperse their contents (arrows), morphological changes that are consistent with necrotic cell death. (D) In ATM/p53-defective primary CLL induced to proliferate (n = 6) the PARP inhibitor talazoparib did not potentiate the cytotoxic activity (left) of HBX19818 and acted in an antagonistic manner, as indicated by combination indices (right).
PARP1 functions as a DNA damage sensor that catalyzes postranslational modification of DNA-bound proteins. This is mediated through the formation of PAR chains that serve as docking platforms for DNA repair factors.36 Overactivation of PARP1 promotes cell death in 2 ways.36 Firstly, PAR can migrate across the cytosol from the nucleus to stimulate a reciprocal translocation of apoptosis-inducing factor from the mitochondria to the nucleus to stimulate large-scale DNA fragmentation.37 Secondly, nicotinamide adenine dinucleotide+ is the substrate required for the generation of ADP-ribose by PARP1. Consequently, overactivation of PARP1 decreases nicotinamide adenine dinucleotide+ levels, resulting in depletion of ATP and necrotic cell death. We observed evidence for both of these cytotoxic mechanisms with HBX19818 treatment. Immunoblotting of isolated nuclear fractions showed that HBX19818 was capable of inducing translocation of apoptosis-inducing factor (Figure 4B). In addition, morphological features of dying CLL cells were consistent with necrosis (Figure 4C). This form of cell death may cause inflammation. However, there was no evidence of systemic inflammation in HBX19818-treated animals (supplemental Figure 3I). Finally, the antagonistic action of PARP inhibition with HBX19818 provided further evidence for the role of PARP1 in this process (Figure 4D). Thus, the aberrant accumulation of DSBs upon USP7 inhibition leads to protein hyper–poly ADP-ribosylation and ATM/p53-independent cell death.
USP7 inhibition sensitizes ATM/p53-defective CLL cells to HRR-inducing chemotherapy
Given the role of USP7 in HRR, we hypothesized that targeting USP7 would enhance the efficacy of HRR-inducing chemotherapeutic agents such as cyclophosphamide and MMC. Therefore, we assessed the effect of combining HBX19818 with physiologically relevant concentrations of these agents.38,39
In Mec1 cells, the addition of HBX19818 led to potentiation of γH2AX induced either by MMC or the metabolically active form of cyclophosphamide, 4HC (Figure 5A). Accordingly, the combination of HBX19818 with either 4HC or MMC exacerbated the cytotoxicity of either agent in Mec1 cells and p53-defective (Figure 5B-C,E-F) primary CLL cells (supplemental Figure 4A-B). Calculation of the combination index showed that in vitro cotreatment with HBX19818 and chemotherapeutics was synergistic in p53-deficient cells (Figure 5D,G). We observed no synergy between 4HC or MMC and HBX19818 in p53 wild-type CLL (supplemental Figure 4A-B), consistent with a high efficacy of DNA damaging agents in cells with an intact p53 apoptotic pathway.
Inhibition of USP7 sensitized CLL cells to DNA cross-linking agents. (A) HBX19818 potentiated the induction of γH2AX in Mec1 cells treated with either MMC or 4HC for 72 hours. β-actin was the loading control. HBX19818 (8 µM) significantly (P < .05) increased the sensitivity of Mec1 cells (n = 3) (B,E) and proliferating p53-defective primary CLL cells (n = 3) (C,F) to either 4HC (B-C) or MMC (E-F). Inserts indicate the activity of HBX19818 alone. The accompanying combination index plots indicate synergism of HBX19818 with 4HC (D) and MMC (G) in both Mec1 and p53-defective primary CLL cells. (H) Tumor load was reduced by all treatment regimens, both single agents (5 and 10 mg/kg HBX19818, 20 mg/kg cyclophosphamide [CycloP], and 0.5 mg/kg rituximab [RTX]) and combinations with HBX1918 vs control (DMSO) treatment. Tumor load reduction induced by CycloP (20 mg/kg; n = 4) alone was significantly enhanced when coadministered with the higher dose of HBX19818 (10 mg/kg; n = 4). This demonstrated that HBX19818 increased the in vivo efficacy of CycloP in the Mec1 murine xenograft model. Data were compared using 2-way analysis of variance (B-C,E-F,) or 2-tailed Student t test (H), and statistical significance denoted by: *P ≤ .05, ***P ≤ .001.
Inhibition of USP7 sensitized CLL cells to DNA cross-linking agents. (A) HBX19818 potentiated the induction of γH2AX in Mec1 cells treated with either MMC or 4HC for 72 hours. β-actin was the loading control. HBX19818 (8 µM) significantly (P < .05) increased the sensitivity of Mec1 cells (n = 3) (B,E) and proliferating p53-defective primary CLL cells (n = 3) (C,F) to either 4HC (B-C) or MMC (E-F). Inserts indicate the activity of HBX19818 alone. The accompanying combination index plots indicate synergism of HBX19818 with 4HC (D) and MMC (G) in both Mec1 and p53-defective primary CLL cells. (H) Tumor load was reduced by all treatment regimens, both single agents (5 and 10 mg/kg HBX19818, 20 mg/kg cyclophosphamide [CycloP], and 0.5 mg/kg rituximab [RTX]) and combinations with HBX1918 vs control (DMSO) treatment. Tumor load reduction induced by CycloP (20 mg/kg; n = 4) alone was significantly enhanced when coadministered with the higher dose of HBX19818 (10 mg/kg; n = 4). This demonstrated that HBX19818 increased the in vivo efficacy of CycloP in the Mec1 murine xenograft model. Data were compared using 2-way analysis of variance (B-C,E-F,) or 2-tailed Student t test (H), and statistical significance denoted by: *P ≤ .05, ***P ≤ .001.
Subsequently, we used the Mec1 xenograft model to examine the in vivo efficacy of combining 2 doses of the USP7 inhibitor with cyclophosphamide or a non-DNA damaging agent, the CD20 antibody rituximab. Although we observed that HBX19818, cyclophosphamide, or rituximab as monotherapy exhibited antitumor activity, only the combination of cyclophosphamide with the higher dose of HBX19818 had a more potent effect than cyclophosphamide alone or the combination of HBX19818 with rituximab (Figure 5H). On the basis of this, it would seem that USP7 inhibitors are more effective when used in combination with a DNA damaging agent, thus supporting the in vitro findings.
The effect of the HBX19818-chemotherapy combination was investigated further in HeLa cells. Similar to the effect of USP7 inhibition on CLL cells, HeLa cells showed reduced viability when exposed to HBX19818 (supplemental Figure 4C) or when USP7 was depleted (supplemental Figure 4D). Pharmacological inhibition (8 µM HBX19818; supplemental Figure 4E-F) or siRNA downregulation of USP7 (supplemental Figure 4G-H) significantly increased the sensitivity of HeLa cells to MMC and 4HC. USP7 depletion also increased the sensitivity of HeLa cells to MMC, as assessed by colony formation assay (supplemental Figure 4I). Furthermore, the sensitivity of HeLa cells to MMC was increased by an alternative USP7 inhibitor, HBX41108 (supplemental Figure S4J). Taken together, these results substantiate the potentiating effect of USP7 inhibition on HRR-inducing chemotherapy.
Discussion
The treatment of hematological malignancies with DDR defects remains an unmet clinical need. In CLL, the addition of monoclonal antibodies to chemotherapeutic agents represents a therapeutic advance, with deep, sustained responses achievable in a substantial proportion of patients that are often translated into long-term treatment-free survival.40,41 Nonetheless, monoclonal antibodies have failed to overcome chemotherapy resistance in DDR-defective CLL, particularly in patients with del(17p) and/or TP53 mutation.42,43 Although BCR signaling inhibitors and Bcl-2 inhibitors have proven efficacy against DDR-defective CLL, deep remissions are infrequent with BCR signaling inhibitors, and prolonged treatment with these agents is required.7-10 The presence of residual disease provides a reservoir for eventual disease relapse, especially in ATM/p53-defective CLL with high levels of genomic instability.44-46 Indeed, emerging evidence suggests that therapies eliciting deep remissions may provide enhanced benefit for del(17p) or del(11q) CLL.47 Moreover, only a proportion of patients with CLL relapsing from BCR signaling inhibitors respond to alternative kinase inhibitors, highlighting the need for novel therapeutic approaches.48,49
In this study, we demonstrate that USP7 represents a new therapeutic target for CLL. Treatment with USP7 inhibitor, either alone or in combination with chemotherapy, led to the potent killing of CLL cells in vitro and in vivo independently of ATM and p53 function. Even when primary CLL cells were cocultured with CD40L/IL-21, they were still highly responsive to USP7 inhibition, suggesting that such an approach could overcome prosurvival microenvironmental signals. In addition, we show for the first time that this cytotoxic effect is mediated, in part, through the suppression of HRR upon USP7 inhibition, allowing accumulation of unrepaired DSBs and the induction of p53-independent cell death.
We show that CLL tumor cells exhibit a basal level of genome instability, and as a consequence, they are likely to have a higher dependence on the activity of USP7 than healthy donor lymphocytes. We also demonstrate that when USP7 is inhibited, the relative increase in DNA damage accumulation is higher in the CLL cells. Although the underlying mechanism for the differential sensitivity of CLL cells to the USP7 inhibitor is unclear, we speculate that this most likely arises through synthetic lethality, in which the HR defect (and other DNA repair deficiencies [eg, postreplication repair]) caused by inhibiting USP7 interacts with CLL-specific abnormalities in backup DNA repair pathways, such that genetic damage is induced to intolerable levels and cell death is triggered.
Previous studies of USP7 in myeloma and solid malignancies, including prostate, hepatocellular, ovarian cancers and glioma, revealed that it is frequently overexpressed in tumors, and its overexpression is associated with a worse prognosis.22,50-53 This suggests that USP7 may function as an oncogene and that aberrant USP7 expression may contribute to tumorigenesis. Consistent with this, overexpression of USP7 in hepatocellular carcinoma cells increased tumor-cell growth in vitro and in vivo.51,52 In this study, we observed high levels of USP7 expression in primary CLL samples, suggesting that USP7 deregulation could also contribute to the pathogenesis of lymphoid malignancies.
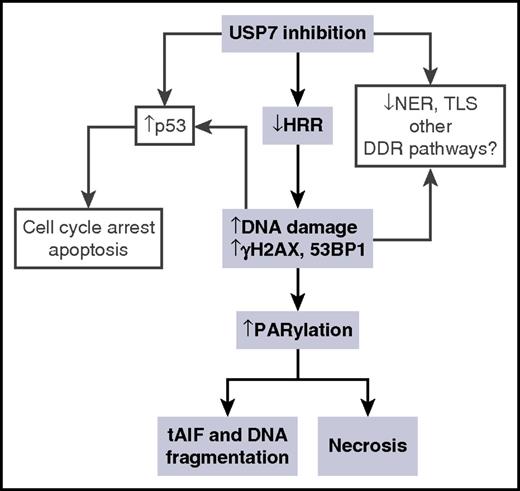
The antimyeloma activity of USP7 inhibition with the inhibitor P5091 has been shown to occur through activation of the MDM2-p53 axis.22 Here, we identified a novel mechanism whereby USP7 inhibition initiates p53-independent killing of malignant cells (Figure 6). Our data suggest that the accumulation of DNA damage in CLL cells stimulates the production of PAR through the overactivation of PARP1. This results in necrotic death, most probably because of the depletion of ATP.54 We also observed that in a proportion of CLL patient cases, PAR-stimulated nuclear translocation of AIF occurs in response to USP7 inhibition. This in turn can induce DNA fragmentation as previously reported.36,55 It has been demonstrated that the USP7 inhibitor P5019 also inhibits USP47 with a comparable EC50 to USP7.56 Although the potential off-target effects of HBX19818 have not been thoroughly tested, in this study we demonstrate that many of the cellular phenotypes resulting from USP7 inhibition with HBX19818 can be recapitulated by knocking down USP7 with siRNA. Therefore, although we cannot completely rule out the possibility that some of the antitumor activity we observed with the USP7 inhibitor HBX19818 resulted from off-target effects, we are confident that the predominant effect is mediated by inhibiting USP7.
Model for the effect of USP7 inhibition on DDR. The sequence of events inferred from the results in this study is presented in shaded boxes. Other consequences of USP7 inhibition upon DDR, inferred from other studies, are shown in clear boxes. tAIF, translocation of apoptosis-inducing factor.
Model for the effect of USP7 inhibition on DDR. The sequence of events inferred from the results in this study is presented in shaded boxes. Other consequences of USP7 inhibition upon DDR, inferred from other studies, are shown in clear boxes. tAIF, translocation of apoptosis-inducing factor.
Our recent study identified RAD18 as a novel substrate for deubiquitylation by USP7.29 RAD18 is known to protect stressed replication forks from collapse by coordinating 3 DDR pathways, namely, DNA interstrand cross-link repair, DNA damage tolerance, and HRR.57-60 In keeping with this, we showed that disruption of USP7 in epithelial, healthy donor lymphocytes or malignant lymphoid cells led to reduced RAD18 stability and failure to form DNA damage–induced RAD51 and FANCD2 foci. Therefore, we propose that replication fork instability caused by USP7 inhibition coupled with a loss of RAD18-dependent replication fork protection and repair mechanisms allows damaged forks to collapse into DSBs, the accumulation of which leads to tumor-cell death. Consistent with this, CLL cells treated with HBX19818 exhibit increased 53BP1 foci formation, which is indicative of unrepaired DSBs.61
Although we focused our analysis on the HRR pathway, there is evidence to suggest that other DNA repair pathways might be affected by USP7 inhibition. Firstly, noncycling CLL cells, in which HRR is inactive, still exhibit sensitivity to USP7 inhibition. Secondly, we observed marked synergism in noncycling CLL cells when USP7 inhibition was combined with 4HC (A.A. and E.S., unpublished data, 14 May 2014). Although DNA alkylation induced by cyclophosphamide will produce HRR-responsive DSBs in S-phase, it is most likely that this type of DNA damage is repaired through the concerted actions of the base excision, nucleotide excision, and mismatch repair pathways in noncycling cells.62 As such, the repair of cyclophosphamide-induced DNA damage requires the actions of multiple independent DNA repair pathways. Therefore, in addition to HRR, USP7 inhibition could also suppress these other pathways. These mechanisms could be explored in future studies. The activity of USP7 inhibition on quiescent cells is of clinical significance, given that the CLL population in peripheral circulation is mostly quiescent. The combined effect of USP7 inhibition on both the proliferating and quiescent populations could enhance CLL eradication.
As such, our findings have important clinical implications. Regardless of the primary type of DNA damage inflicted, many chemotherapeutic agents induce DSBs that require HRR for their resolution and cell survival.63 Consequently, the suppression of HRR by USP7 inhibition could sensitize tumor cells to HRR-inducing chemotherapeutic agents. Indeed, we showed that HBX19818 was highly synergistic when used in combination with cyclophosphamide or MMC. Thus, combination with USP7 inhibitor could potentially allow lower doses of chemotherapeutic agents to be administered to frailer patients with CLL. In the context of ATM/p53-defective CLL, the ability of USP7 inhibition to resensitize chemotherapy-resistant cells to chemotherapeutic agents supports the clinical investigation of the USP7 inhibitor–chemotherapy combination for the treatment of these tumors. Indeed, the potent in vitro cytotoxic effect of such therapeutic combinations suggests that they might produce deep and durable clinical response. Furthermore, the p53-independent tumor-cell killing by USP7 inhibition suggests that such a strategy might be useful for other hematological malignancies with ATM/p53 defects.64-68
In summary, we have provided evidence for a new p53-independent role of USP7 in regulating DNA DSB repair. In addition, we have demonstrated the preclinical efficacy of USP7 inhibition in CLL both as a single agent and in combination with chemotherapy. Collectively, our in vitro and in vivo data suggest that USP7 is a promising therapeutic target and support progression of USP7 inhibitors into clinical studies for the treatment of chemotherapy-refractory CLL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff and patients at the CLL clinics of Queen Elizabeth Hospital, Birmingham, and Heartlands Hospital, Birmingham, for CLL samples and the Biomedical Services Unit for help with animal experiments.
This work was supported grants from Bloodwise, United Kingdom (11045) (T.S.), and Worldwide Cancer Research (13-1012) (G.S.S.).
Authorship
Contribution: A.A., C.R., R.D., G.S.S., and T.S. designed the study; A.A., E.S., G.S.S., N.J.D., A.Z., C.E.O., J.M., D.D.C., S.Y., T.P., P.K., A.S., and E.Y. performed experiments; M.K., H.P., P.H., S.P., P.M., and G.P. contributed clinical samples; A.A. wrote the manuscript; and M.K., C.E.O., A.M.R.T., G.S.S., and T.S. revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tatjana Stankovic, Institute of Cancer and Genomic Sciences, University of Birmingham, Vincent Dr, Birmingham B15 2TT, United Kingdom; e-mail: t.stankovic@bham.ac.uk; Grant S. Stewart, Institute of Cancer and Genomic Sciences, University of Birmingham, Vincent Dr, Birmingham B15 2TT, United Kingdom; e-mail: g.s.stewart@bham.ac.uk; and Angelo Agathanggelou, Institute of Cancer and Genomic Sciences, University of Birmingham, Vincent Dr, Birmingham B15 2TT, United Kingdom; e-mail: a.agathanggelou@bham.ac.uk.
References
Author notes
A.A. and E.S. contributed equally to this study.

![Figure 3. USP7-disrupted cells show reduced activation of HRR. (A) Normal healthy donor PBMCs and 3 isogenic CLL cell lines (2 p53 wild type [wt] and 1 p53 defective [def]) expressing the indicated shRNAs show diminished expression and thus destabilization of RAD18 protein after treatment with HBX19818. β-actin was the loading control (Con). (B) HeLa cells transfected with USP7 or Con siRNAs and empty (Vec) or siRNA-resistant Flag-USP7 (R-USP7)–expressing plasmid vectors demonstrate rescue of RAD18 stability in a complementation assay. The reduction of RAD18 levels in the presence of USP7 siRNA alone is prevented by the expression of R-USP7, which replaced the endogenous USP7 depleted by USP7 siRNA. (C,D) Representative immunofluorescence labeling of DNA damage markers γH2AX and RAD51 (green) with DAPI-counterstained nuclei (blue); (E-H) quantification of RAD51 foci formation from 24-hour time-course studies of 5-Gy IR-induced damage (n = 3). In MEC1 cells (C,G), normal healthy donor cells (E), and primary CLL cells (F), HBX19818 induced inhibition of USP7 1 hour before irradiation abrogated the formation of IR-induced RAD51 foci. (H) IR-induced RAD51 foci formation was reduced in HeLa cells after USP7 depletion by USP7 siRNA. (I) Analysis of HRR using the DR-GFP reporter assay in U2OS cells showed that HRR is compromised by USP7 depletion. CtIP depletion was the positive Con for HRR-defective cells. Original magnification ×60 (C-D). Data were compared using a 2-tailed Student t test, and statistical significance denoted by: *P ≤ .05, **P ≤ .01. Ub, ubiquitylated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/2/10.1182_blood-2016-12-758219/4/m_blood758219f3.jpeg?Expires=1769147482&Signature=NEyugzYA0e8FyoG6bEWKc79ghDze7~z-SSlDQm4GXcEfG-Jx7QqKpGmiaswGsMbkwy1PRHrAOisFQmRzy~MJRAcVN5JaN5a3FL4xWQMOqAgul~dKtWkKx9PihNn9zrO5dK0nbF1zm2t9U9vuWzQIObYN0fTrGZjwaX2TtNkN3xfmibvUHbOv9Z1yQCiES58rQM2RgZkjMXzD0U-QCJpTygGVuIdjteWUiATgu7Fqq0W2OgkStBLCIqEY2hK92wVk3gpFunQMAzQLp02563RCRwGCjDYuOy81jPdcWQBAKwMPSoxhBiE6pgPrmtgp308CYysTKcgIxe46SX~HdsQOVQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Inhibition of USP7 sensitized CLL cells to DNA cross-linking agents. (A) HBX19818 potentiated the induction of γH2AX in Mec1 cells treated with either MMC or 4HC for 72 hours. β-actin was the loading control. HBX19818 (8 µM) significantly (P < .05) increased the sensitivity of Mec1 cells (n = 3) (B,E) and proliferating p53-defective primary CLL cells (n = 3) (C,F) to either 4HC (B-C) or MMC (E-F). Inserts indicate the activity of HBX19818 alone. The accompanying combination index plots indicate synergism of HBX19818 with 4HC (D) and MMC (G) in both Mec1 and p53-defective primary CLL cells. (H) Tumor load was reduced by all treatment regimens, both single agents (5 and 10 mg/kg HBX19818, 20 mg/kg cyclophosphamide [CycloP], and 0.5 mg/kg rituximab [RTX]) and combinations with HBX1918 vs control (DMSO) treatment. Tumor load reduction induced by CycloP (20 mg/kg; n = 4) alone was significantly enhanced when coadministered with the higher dose of HBX19818 (10 mg/kg; n = 4). This demonstrated that HBX19818 increased the in vivo efficacy of CycloP in the Mec1 murine xenograft model. Data were compared using 2-way analysis of variance (B-C,E-F,) or 2-tailed Student t test (H), and statistical significance denoted by: *P ≤ .05, ***P ≤ .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/2/10.1182_blood-2016-12-758219/4/m_blood758219f5.jpeg?Expires=1769147482&Signature=H78g0ptpNvu1bSL~EdPOZ4ClZqq1PcxkBnKZqEx7FS3gUNEIJUTyFlrIN9fC24~y1YfraP1BdSDBDxPjPDzjbhIsIwEJXPrlAIa061hFdnidfdUmBL9yR~cXEsL9IOSBEJkrRlkGEkm5c7oV~yL2oYE9SJ2whptnKRtt4ZrnltWx8HN-M3OkxvNj3tb1TD1~ENhBu1ajRrIYktXdl7her~2Ry3EAsLJmBeMMRmiH0zK7pLKqMY~codqmQxMeklo718xcim-rXVV~FTDLBmsJSaW~ENiHrP9CCvPUj0E6bVp-bT10SH1hgOPt3iUX~mwrcf79m1lJotZaXat1mYkDlQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal