Key Points
RUNX1 maintains Myb and Myc enhancer activity and is required for leukemogenesis in vivo.
RUNX1 inhibition impairs the growth of primary T-ALL patient cells without an effect on normal human hematopoietic cells.
Abstract
The gene encoding the RUNX1 transcription factor is mutated in a subset of T-cell acute lymphoblastic leukemia (T-ALL) patients, and RUNX1 mutations are associated with a poor prognosis. These mutations cluster in the DNA-binding Runt domain and are thought to represent loss-of-function mutations, indicating that RUNX1 suppresses T-cell transformation. RUNX1 has been proposed to have tumor suppressor roles in T-cell leukemia homeobox 1/3–transformed human T-ALL cell lines and NOTCH1 T-ALL mouse models. Yet, retroviral insertional mutagenesis screens identify RUNX genes as collaborating oncogenes in MYC-driven leukemia mouse models. To elucidate RUNX1 function(s) in leukemogenesis, we generated Tal1/Lmo2/Rosa26-CreERT2Runx1f/f mice and examined leukemia progression in the presence of vehicle or tamoxifen. We found that Runx1 deletion inhibits mouse leukemic growth in vivo and that RUNX silencing in human T-ALL cells triggers apoptosis. We demonstrate that a small molecule inhibitor, designed to interfere with CBFβ binding to RUNX proteins, impairs the growth of human T-ALL cell lines and primary patient samples. We demonstrate that a RUNX1 deficiency alters the expression of a crucial subset of TAL1- and NOTCH1-regulated genes, including the MYB and MYC oncogenes, respectively. These studies provide genetic and pharmacologic evidence that RUNX1 has oncogenic roles and reveal RUNX1 as a novel therapeutic target in T-ALL.
Introduction
Core binding transcription factors are heterodimeric complexes composed of a common CBFβ subunit bound to 1 of 3 DNA-binding CBFα subunits, RUNX1, RUNX2, or RUNX3. RUNX1 is predominantly expressed in the hematopoietic lineage and has central roles in embryonic and adult hematopoiesis.1-4 All 3 RUNX proteins share a highly conserved DNA-binding runt domain and the C-terminal VWRPY motif. The association of RUNX proteins with CBFβ increases binding to RUNX consensus sites. In adult mice, Runx1 deletion impairs lymphoid and megakaryocytic maturation and induces myeloid cell expansion and hematopoietic stem cell exhaustion.2-4 Germ line RUNX1 mutations are associated with familial platelet disorder and an increased risk of myelodysplastic syndrome and acute myeloid leukemia (AML).5,6 RUNX1 and CBFβ are the most frequent targets of chromosomal translocation in AML, resulting in the generation of novel AML1-ETO and CBFβ-MYH11 fusion proteins.7,8 These fusion proteins are thought to interfere with RUNX function, block myeloid differentiation, and thereby contribute to myeloid leukemia.9,10
Heterozygous frameshift and missense mutations in RUNX1 are found in a subset of T-cell acute lymphoblastic leukemia (T-ALL) patients and are associated with poor overall survival.11-13 Most of the mutations (L29S, H58N, H78Y, S115fs, and G138fs) cluster in the runt domain of RUNX114-18 and are thought to affect DNA binding and result in loss-of-function mutations. Published data in human T-ALL cell lines and mouse models also suggest that RUNX1 functions to suppress T-cell leukemia.11,19,20 For example, N-ethyl-N-nitrosourea treatment of chimeric Runx1-deficient mice results in the development of T-ALL.20 In human T-ALL cell lines, the T-cell leukemia homeobox 1 (TLX1) and TLX3 oncogenes have been shown to directly repress the RUNX1 transcriptional network in T-ALL, and retroviral RUNX1 expression in these leukemic cells impairs growth.11 Similarly, oncogenic NOTCH1 has been shown to indirectly repress RUNX1 expression in leukemia-initiating or stem cell populations.19
In contrast, RUNX1 has also been shown to support the expression of the TAL1 oncogene, part of an autoregulatory feedback loop involving GATA3 and MYB.21 The basic helix-loop-helix transcription factor TAL1 and its LIM-domain–only partners LMO1 or LMO2 are frequently misexpressed in pediatric and adult T-ALL patients. We have modeled human T-ALL by directing the expression of Tal1 and Lmo2 in developing mouse thymocytes and have demonstrated that these mice develop fully penetrant T-ALL.22 Mutations in the heterodimerization domain and the PEST regulatory region of NOTCH1 often co-occur with TAL1 activation in T-ALL patients.23 Similarly, the mouse Tal1/Lmo2 leukemias acquire spontaneous mutations in Notch1.24 We have shown that the NOTCH1-MYC pathway is required for mouse T-ALL growth in vivo and for leukemia-initiating cell activity.24,25 Hence, the TAL1/MYB and NOTCH1/MYC pathways are critical nodes in T-cell transformation.
To elucidate whether RUNX1 potentiates or suppresses T-cell leukemogenesis, we generated Tal1/Lmo2/Rosa26-CreERT2Runx1f/f mice and reveal a crucial, prosurvival role for RUNX1 in T-ALL. We demonstrate that Runx1 deletion in mouse T-ALL cells interferes with Myb and Myc enhancer activity, resulting in significant delays in leukemogenesis in vivo. Similarly, we demonstrate that RUNX1/3 knockdown in human T-ALL cell lines or treatment with a recently developed CBFβ/RUNX allosteric inhibitor mimics the effects of Runx1 deletion in mouse T-ALL cells and induces apoptosis. These data provide genetic and pharmacologic evidence that RUNX1 has critical survival roles in T-ALL and support the idea that RUNX1 inhibition may have therapeutic benefit for T-ALL patients.
Methods
Mice
A cohort of Tal1/Lmo2/Rosa26-CreERT2Runx1f/f mice was generated by mating Tal1/Lmo2 mice with Rosa26-CreERT2Runx1f/f mice. Tal1/Lmo2/Rosa26-CreERT2Runx1f/f leukemic cells were transplanted into F1 (FVB/N × C57BL/6J) recipient mice, and corn oil (MilliporeSigma, C-8267) or Tamoxifen (1 mg, MilliporeSigma, T-5648) was intraperitoneally injected for 3 days 1 week after transplantation. Mouse Tal1/Lmo2 T-ALL cells were infected with retroviruses expressing short hairpin RNAs (shRNAs) to c-Myb or Renilla luciferase and effects on disease latency and penetrance determined as described.25 All animal procedures used in this study were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee.
Primary mouse and patient T-ALL cells and cell lines
Mouse Tal1/Lmo2/Rosa26-CreERT2Runx1f/f T-ALL cells were treated with ethanol or 5 or 10 nM of 4-hydroxytamoxifen (4-OHT; MilliporeSigma) for 24 hours, washed with phosphate-buffered saline and cultured for 1 or 2 days prior to further analyses. Primary human T-ALL samples were obtained from children with T-ALL enrolled in clinical trials at the Dana-Farber Cancer Institute, collaborating institutions, or the University of Massachusetts Memorial Hospital. Samples were collated with informed consent and with approval of the institutional review board. Leukemic blasts were isolated from peripheral blood or bone marrow as described.26
RUNX silencing
The lentiviral pLKO.1-puro vectors carrying shRNA targeting RUNX1 and RUNX3 were generously provided by Marjorie Brand (Ottawa Hospital Research Institute). Viruses were generated and human T-ALL cell lines infected as previously described.27 The level of knockdown was determined by using quantitative real-time polymerase chain reaction (qRT-PCR) and immunoblotting 4 days after infection.
Genomic DNA and RNA analyses
Total RNA was extracted by using Trizol, and complementary DNA was synthesized by using Superscript First-Strand Synthesis System (Invitrogen). qRT-PCR were performed on the AB7300 Detection System (Applied Biosystems) by using POWER SYBR Green Master Mix (Applied Biosystems) and gene-specific primers. Gene expression was determined by using the ΔΔ cycle threshold method normalized to GAPDH for human or β-Actin for mouse transcripts, unless otherwise specified. By using isolated genomic DNA, Runx1 deletion was determined by polymerase chain reaction (PCR) as described previously.2
Immunoblotting and coimmunoprecipitation
To examine protein expression in human T-ALL cells, cells were lysed in modified radioimmunoprecipitation assay buffer, transferred to a membrane, and probed with antibodies to RUNX1 (ab23980, Abcam), RUNX3 (MAB3765, R&D Systems), TAL1 (sc-12984, Santa Cruz Biotechnology), MYB (05-175, EMD Millipore), NOTCH1 (Val1744, Cell Signaling Technology), MYC (N262, Santa Cruz Biotechnology), or extracellular signal-regulated kinase 1/2 (9102, Cell Signaling Technology). Blots were imaged by using ImageLab Software (Bio-Rad). To determine the effect of the AI-10-104 inhibitor on CBFβ/RUNX heterodimers, the human T-ALL cell lines were treated with AI-10-104 or dimethyl sulfoxide (DMSO) and lysed in modified radioimmunoprecipitation assay buffer. RUNX1 or RUNX3 was immunoprecipitated by using anti-RUNX1 (39000, Active Motif) or anti-RUNX3 (9647, Cell Signaling Technology) antibody and protein A agarose beads (Roche Applied Science) according to the manufacturer’s instructions. The membrane was probed with anti-CBFβ28 , anti-RUNX1, or anti-RUNX3 antibodies, and proteins were detected by using Clean-Blot IP Detection Reagents (Thermo Fisher Scientific).
Chromatin immunoprecipitation-quantitative PCR
Chromatin immunoprecipitation was performed as previously described.29 Mouse T-ALL cells treated with ethanol or 10 nM 4-OHT were lysed, and nuclei were fragmented to a size of 150–300 bp by using Bioruptor (Diagenode). Fragmented chromatin was incubated overnight at 4°C with normal immunoglobulin G (sc-2027, Santa Cruz Biotechnology) or anti-TAL1 (C-21, Santa Cruz Biotechnology), anti-RUNX1 (ab23980, Abcam), anti-RUNX3 (9647, Cell Signaling Technology), anti-NOTCH1 (C-20, Santa Cruz Biotechnology), anti-Histone 3 (ab1791, Abcam), or anti-H2K27ac (ab4729, Abcam). Chromatin-antibody complexes were pulled down by incubating with magnetic beads (Dynal) for 4 hours. The enrichment of DNA fragments was tested by using quantitative PCR with primers specific for the site of interest.
Cell viability and death assays
Human T-ALL cell lines or T-ALL patient samples were cultured for 3 days in the presence of DMSO or various concentrations of AI-10-104 or AI-4-88. Metabolic activity was assayed by MTS (CellTiter 96 AQueous One Solution Cell Proliferation Assay, Promega) or CellTiter-Glo (CellTiter-Glo Luminescent Cell Viability Assay, Promega) and measured by using a Beckman Coulter DTX880 plate reader. Absorbance values were normalized to the DMSO control. Human T-ALL cell lines transduced with lentiviruses or treated with AI-10-104 or AI-4-88 were stained with Annexin V- fluorescein isothiocyanate (FITC) and 7-aminoactinomycin D (7AAD) to detect apoptotic cells or with anti-CD4 antibody and analyzed by flow cytometry.
Results
RUNX activity is required for the growth and survival of T-ALL cells
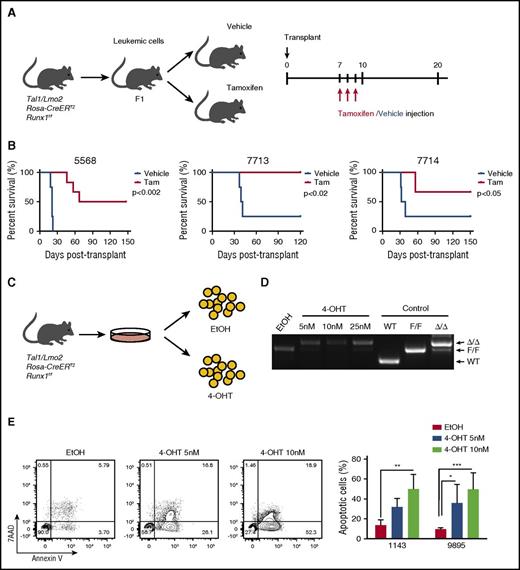
In the human T-ALL cell lines and patient samples, TAL1 induces a core transcriptional regulatory circuit that involves RUNX1 and GATA3 that culminates in MYB expression.21 These data suggest that RUNX1 is required to support TAL1-mediated leukemogenesis. To address this, we generated Tal1/Lmo2/Rosa26(R26)-CreERT2Runx1f/f mice and transplanted mouse leukemic cells into secondary recipients that were treated with vehicle or tamoxifen (Figure 1A). These studies revealed that RUNX1 is essential for T-ALL maintenance in vivo and for leukemic survival in vitro (Figure 1). We demonstrate that Runx1 deletion interferes with or prevents leukemic growth in vivo (Figure 1B). Some tamoxifen-treated mice transplanted with Tal1/Lmo2/R26-Cre-ERT2 Runx1f/f leukemias (designated 5568 and 7714) developed disease; however, these leukemic cells retained a floxed Runx1 allele that likely escaped Cre-mediated deletion in vivo (supplemental Figure 1A, available on the Blood Web site). To rule out any potential effects of tamoxifen- or Cre-mediated toxicity on leukemogenesis, we generated Tal1/Lmo2/R26-CreERT2 mice and treated them with vehicle or tamoxifen, but observed no significant effects on disease progression or leukemic cell survival (supplemental Figure 1B-C). Consistent with the in vivo data, Runx1 deletion induced by 4-OHT treatment in vitro (Figure 1C-D) resulted in apoptosis of mouse T-ALL cells (Figure 1E).
RUNX1 is required for the maintenance of leukemic growth in vivo, and Runx1 deletion in vitro results in apoptosis of leukemic cells. (A) Experimental strategy used to determine the effects of Runx1 deletion on leukemia progression in vivo. Three independent mouse T-ALLs from Tal1/Lmo2/Rosa26-CreERT2Runx1f/f mice were transplanted into mice and treated 1 week later with vehicle or tamoxifen for 3 days. (B) Kaplan-Meier survival curves are shown for 3 mouse T-ALLs. The difference in overall survival between the vehicle- and tamoxifen (Tam)-treated groups was assessed by the log-rank test (n = 4 for the vehicle group, n = 6 for Tam group in all 3 experiments). (C) Experimental strategy used to determine the effects of Runx1 deletion on mouse T-ALL survival in vitro. (D) Genomic DNA was isolated from mouse T-ALL cells 48 hours after EtOH or 4-OHT treatment to examine Runx1 deletion by genomic PCR. (E) Mouse T-ALL cell lines 1143 and 9895 were treated with vehicle or 4-OHT for 72 hours, stained with Annexin V-FITC and 7-AAD and analyzed by flow cytometry. The quantifications of Annexin-V–positive cells from 4 independent experiments are shown as means ± standard deviations (SD) (right). *P < .05; **P < .005; ***P < .0005; two-way analysis of variance (ANOVA) multiple comparisons test.
RUNX1 is required for the maintenance of leukemic growth in vivo, and Runx1 deletion in vitro results in apoptosis of leukemic cells. (A) Experimental strategy used to determine the effects of Runx1 deletion on leukemia progression in vivo. Three independent mouse T-ALLs from Tal1/Lmo2/Rosa26-CreERT2Runx1f/f mice were transplanted into mice and treated 1 week later with vehicle or tamoxifen for 3 days. (B) Kaplan-Meier survival curves are shown for 3 mouse T-ALLs. The difference in overall survival between the vehicle- and tamoxifen (Tam)-treated groups was assessed by the log-rank test (n = 4 for the vehicle group, n = 6 for Tam group in all 3 experiments). (C) Experimental strategy used to determine the effects of Runx1 deletion on mouse T-ALL survival in vitro. (D) Genomic DNA was isolated from mouse T-ALL cells 48 hours after EtOH or 4-OHT treatment to examine Runx1 deletion by genomic PCR. (E) Mouse T-ALL cell lines 1143 and 9895 were treated with vehicle or 4-OHT for 72 hours, stained with Annexin V-FITC and 7-AAD and analyzed by flow cytometry. The quantifications of Annexin-V–positive cells from 4 independent experiments are shown as means ± standard deviations (SD) (right). *P < .05; **P < .005; ***P < .0005; two-way analysis of variance (ANOVA) multiple comparisons test.
During mouse thymocyte development, RUNX1 is expressed in immature double-negative and double-positive (DP) thymocytes, whereas RUNX3 expression occurs later in more mature CD8+ single-positive thymocytes.30 Consistently, we found RUNX1 expressed predominantly in mouse DP leukemic cells, with no RUNX3 protein expression detected (supplemental Figure 2A), thereby explaining the RUNX1 dependency observed in mouse T-ALL.
To determine whether human T-ALL cells were similarly RUNX1-dependent, we examined CBFβ, RUNX1, and RUNX3 expression in human T-ALL cell lines and primary patient samples (Figure 2A; supplemental Figure 2B). All the human T-ALL cell lines examined expressed CBFβ, and most expressed RUNX1 with low to undetectable levels of RUNX3 (Figure 2A). However, RUNX1 and RUNX3 were coexpressed in KOPTK1 and LOUCY cell lines and in 5 of 8 primary pediatric T-ALL samples examined (Figure 2A; supplemental Figure 2B). We reduced RUNX1 expression in human T-ALL cell lines (Jurkat, KOPTK1, PF382, and RPMI8402) by expressing 2 independent RUNX1-specific shRNAs and, as reported previously,21 observed significant decreases in cell viability and increases in apoptotic cells (Figure 2B-C; supplemental Figure 2C-D). CBFβ knockdown also induced apoptosis (Figure 2D), revealing prosurvival roles for the CBFβ/RUNX1 heterodimer in T-ALL.
RUNX1 is ubiquitously expressed in human T-ALL cells, and RUNX1 or CBFβ knockdown results in apoptosis. (A) Protein was isolated from human T-ALL cell lines and RUNX1, RUNX3, CBFβ, TAL1, MYB, NOTCH1, and MYC protein levels were determined by immunoblotting. Extracellular signal–regulated kinase 1/2 (ERK1/2) was used as a loading control. (B) The human T-ALL cell line Jurkat was infected with lentiviruses expressing a control shRNA or 2 shRNAs specific for RUNX1. RUNX1 mRNA and protein levels were examined by qRT-PCR and immunoblotting. (C) RUNX1 knockdown results in leukemic cell apoptosis. Control (GFP) and RUNX1 shRNA-transduced Jurkat cells were stained with Annexin V-FITC and 7AAD and analyzed by flow cytometry 6 days after infection. A representative flow profile is shown (left). The percentage of apoptotic cells was determined by Annexin V/7AAD staining and analyzed by flow cytometry. Four independent experiments were performed, and data are shown as means ± SD (right). (D) CBFβ knockdown also induces apoptosis. Control (GFP) or CBFβ shRNA-transduced Jurkat cells were stained with Annexin V-FITC and 7AAD and analyzed by flow cytometry. Four independent experiments were performed, and data are shown as means ± SD (right). (E) CBFβ protein levels in control and knockdown cells were analyzed by immunoblotting. **P < .005; ***P < .0005; ****P < .0001, one-way ANOVA multiple comparisons test.
RUNX1 is ubiquitously expressed in human T-ALL cells, and RUNX1 or CBFβ knockdown results in apoptosis. (A) Protein was isolated from human T-ALL cell lines and RUNX1, RUNX3, CBFβ, TAL1, MYB, NOTCH1, and MYC protein levels were determined by immunoblotting. Extracellular signal–regulated kinase 1/2 (ERK1/2) was used as a loading control. (B) The human T-ALL cell line Jurkat was infected with lentiviruses expressing a control shRNA or 2 shRNAs specific for RUNX1. RUNX1 mRNA and protein levels were examined by qRT-PCR and immunoblotting. (C) RUNX1 knockdown results in leukemic cell apoptosis. Control (GFP) and RUNX1 shRNA-transduced Jurkat cells were stained with Annexin V-FITC and 7AAD and analyzed by flow cytometry 6 days after infection. A representative flow profile is shown (left). The percentage of apoptotic cells was determined by Annexin V/7AAD staining and analyzed by flow cytometry. Four independent experiments were performed, and data are shown as means ± SD (right). (D) CBFβ knockdown also induces apoptosis. Control (GFP) or CBFβ shRNA-transduced Jurkat cells were stained with Annexin V-FITC and 7AAD and analyzed by flow cytometry. Four independent experiments were performed, and data are shown as means ± SD (right). (E) CBFβ protein levels in control and knockdown cells were analyzed by immunoblotting. **P < .005; ***P < .0005; ****P < .0001, one-way ANOVA multiple comparisons test.
RUNX1 supports the expression of a subset of TAL1- and NOTCH1-regulated genes
We hypothesized that Runx1 deletion, although unlikely to influence transgenic Tal1 messenger RNA levels, may suppress TAL1/LMO2-regulated genes that are important in mouse thymocyte survival, proliferation, and differentiation. RUNX1 regulates genes that are important in thymocyte development and represses CD4 expression during the DP to single-positive thymocyte transition.31 In addition to significant decreases in the RUNX1-regulated genes Cxcr4 and Bcl2, we observed increases in Cd4 and Cdkn1a mRNA expression in Runx1-deleted mouse T-ALLs (Figure 3). Similarly, RUNX1 suppression in human T-ALL cell lines resulted in a partial derepression of the CD4 coreceptor, resulting in statistically significant increases in the mean fluorescent intensity of cell surface CD4 staining in RUNX1-deficient human T-ALL cells (supplemental Figure 2E). These data suggest that, in mouse and human T-ALL cells, RUNX1 depletion may stimulate leukemic cell differentiation prior to the induction of apoptosis.
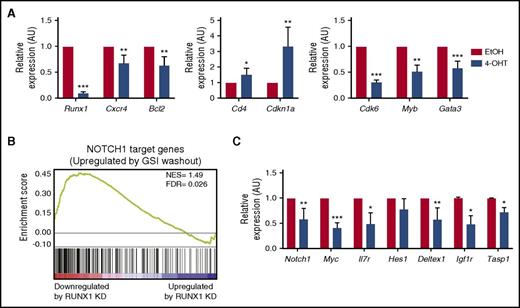
RUNX1 regulates a subset of TAL1- and NOTCH1-regulated genes. (A) mRNA was isolated from mouse T-ALL cells 48 hours after vehicle or 4-OHT treatment, and the expression of a subset of a RUNX1- and TAL1-regulated genes was determined by qRT-PCR. Three to 4 independent experiments were performed, and data are shown as means ± standard errors of the means (SEM). (B) Gene set enrichment analysis of RUNX1-regulated genes and genes changed on reactivation of NOTCH1 by γ-secretase inhibitor (GSI) washout (51). RUNX1 target genes that were significantly downregulated by RUNX1 knockdown in Jurkat cells were used as a data set (22). (C) The expression of a subset of NOTCH1-regulated genes in Runx1-deleted mouse T-ALL cells was determined by qRT-PCR. Three to 4 independent experiments were performed and data are shown as means ± SEM. *P < .05; **P < .005; ***P < .0005, Student t test.
RUNX1 regulates a subset of TAL1- and NOTCH1-regulated genes. (A) mRNA was isolated from mouse T-ALL cells 48 hours after vehicle or 4-OHT treatment, and the expression of a subset of a RUNX1- and TAL1-regulated genes was determined by qRT-PCR. Three to 4 independent experiments were performed, and data are shown as means ± standard errors of the means (SEM). (B) Gene set enrichment analysis of RUNX1-regulated genes and genes changed on reactivation of NOTCH1 by γ-secretase inhibitor (GSI) washout (51). RUNX1 target genes that were significantly downregulated by RUNX1 knockdown in Jurkat cells were used as a data set (22). (C) The expression of a subset of NOTCH1-regulated genes in Runx1-deleted mouse T-ALL cells was determined by qRT-PCR. Three to 4 independent experiments were performed and data are shown as means ± SEM. *P < .05; **P < .005; ***P < .0005, Student t test.
Significant reductions in Myb, Gata3 and Cdk6 expression were also observed in Runx1-deleted mouse T-ALL cells and in the human TAL1-positive T-ALL cell line Jurkat (Figure 3A, supplemental Figure 2F). These data reveal that the TAL1-RUNX1-GATA3 autoregulatory loop is conserved in this mouse T-ALL model driven by the TAL1 oncogene. Moreover, we demonstrate that TAL1/LMO2-mediated mouse leukemic growth requires MYB in vitro and in vivo (supplemental Figure 3).
Using a RUNX1-regulated gene set and genes induced on NOTCH1 reactivation,21,32 we performed gene set enrichment analysis and identified a subset of NOTCH1-regulated genes that were also affected by RUNX1 knockdown in human T-ALL cells (Figure 3B; normalized enrichment score = 1.49; false discovery rate = 0.026). We observed significant reductions in the expression of Notch1, Myc, Il7rα, Igf1r, and Deltex1 mRNAs in the Runx1-deleted mouse T-ALL cell line (Figure 3C). This is, to our knowledge, the first report demonstrating that RUNX1 regulates NOTCH1 expression in mouse T-ALL cells. Runx1 deletion had no effect, however, on Hes1 mRNA levels or on intracellular NOTCH1 binding to the mouse Hes1 promoter (Figure 3C; supplemental Figure 4A). Similarly, no significant decrease in human HES1 expression was observed on RUNX1 knockdown in Jurkat cells (supplemental Figure 2F), indicating that a subset of NOTCH1-regulated genes is RUNX1-dependent. RUNX1 depletion in human T-ALL cell lines consistently decreased the expression of MYC and IL7Rα. These data are consistent with published chromatin immunoprecipitation sequencing studies demonstrating that RUNX1 co-occupies a subset of NOTCH1-regulated genes and prior findings that RUNX1 and NOTCH1 regulate IL7R expression.33
Although the features that predict a RUNX1 dependency remain unclear, several of the TAL1- and NOTCH1-regulated genes supported by RUNX1 are associated with super-enhancers in human T-ALL cells,34 suggesting that enhancer-regulated genes may be uniquely sensitive to the effects of RUNX1 depletion.
RUNX1 is required for TAL1 and NOTCH1 binding to oncogene enhancers
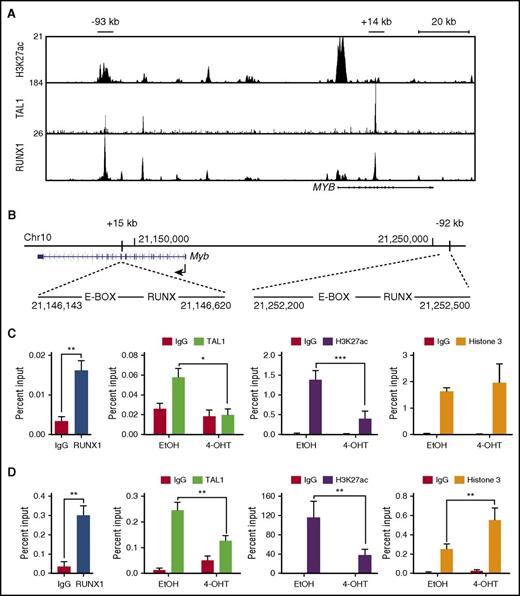
Comparisons between the mouse and human MYB genes reveal the presence of conserved locus-control–like regions located ∼−92 kb and +15 kb from the mouse Myb promoter and −93 kb and +14 kb from human MYB promoter (Figure 4B)21,35 . These sites possess several features associated with enhancer activity, including the presence of multiple transcription factors (TAL1, RUNX1, HEB, GATA3, and ETS1) as well as RNA polymerase II, mediator, BRD4, and acetylated H3K27.21,34,36 The mouse Myb (–92 kb and +15 kb) regions each harbor 1 canonical RUNX binding site, and RUNX1 binding to these conserved regions is observed in mouse T-ALL cells (Figure 4C-D). To determine if Runx1 deletion in mouse T-ALL cells affects TAL1 binding to these regions, we performed chromatin immunoprecipitation followed by qRT-PCR (ChIP-qPCR). We observed statistically significant reductions in TAL1 binding to the Myb +15-kb and −92-kb enhancer elements (Figure 4C-D) and decreases in Myb mRNA levels (Figure 3A) in the Runx1-deleted T-ALL cells. Reductions in TAL1 occupancy were accompanied by significant depletion of the active chromatin mark H3K27ac at these sites (Figure 4-D).
RUNX1 is required for TAL1 binding to the Myb enhancers and for the retention of active chromatin marks. (A) H3K27ac, TAL1, and RUNX1 enrichment at the MYB locus by chromatin immunoprecipitation sequencing is shown in genome browser tracks (genome.ucsc.edu, human hg19). (B) The mouse genomic region (mm10) around the Myb locus is shown, depicting the E-BOX (TAL1)- and RUNX-binding sites at positions +15 kb and −92 kb from the Myb TSS. (C-D) Enrichment of RUNX1, TAL1, H3K27ac, and H3 to the +15-kb (C) and −92-kb (D) Myb enhancer regions determined by ChIP-qPCR in control or Runx1-deleted mouse T-ALL cells. Data are shown as the mean of 3 or 4 independent experiments with error bars representing ± SEM. *P < .05; ** P < .005; P < .0005, two-way ANOVA multiple comparisons test.
RUNX1 is required for TAL1 binding to the Myb enhancers and for the retention of active chromatin marks. (A) H3K27ac, TAL1, and RUNX1 enrichment at the MYB locus by chromatin immunoprecipitation sequencing is shown in genome browser tracks (genome.ucsc.edu, human hg19). (B) The mouse genomic region (mm10) around the Myb locus is shown, depicting the E-BOX (TAL1)- and RUNX-binding sites at positions +15 kb and −92 kb from the Myb TSS. (C-D) Enrichment of RUNX1, TAL1, H3K27ac, and H3 to the +15-kb (C) and −92-kb (D) Myb enhancer regions determined by ChIP-qPCR in control or Runx1-deleted mouse T-ALL cells. Data are shown as the mean of 3 or 4 independent experiments with error bars representing ± SEM. *P < .05; ** P < .005; P < .0005, two-way ANOVA multiple comparisons test.
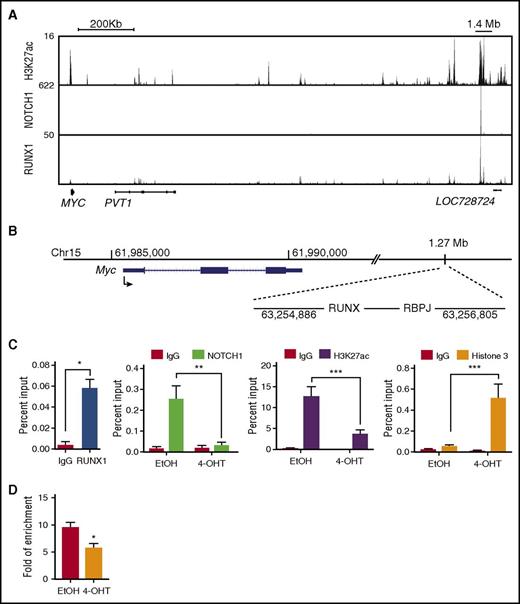
NOTCH1 contributes to T-ALL growth via its direct regulation of MYC.37-39 NOTCH1 regulation of MYC is mediated through a distal enhancer located 1.27 Mb 3′ from the transcriptional start site (TSS) of the mouse Myc gene and 1.4 Mb from the TSS of the human MYC gene.29,40 This region was designated the NOTCH1-bound MYC enhancer (N-Me) and was shown to be essential for NOTCH1-mediated MYC expression during mouse thymocyte development and for NOTCH1-mediated leukemic transformation.29 We examined intracellular NOTCH1 binding to the N-Me in the Runx1-deleted mouse TAL1/LMO2 T-ALL cells. Consistent with the observed reductions in Myc mRNA (Figure 3C), intracellular NOTCH1 binding at the N-Me and H3K27ac levels were significantly reduced in the Runx1-deficient mouse T-ALL cells (Figure 5C), whereas no differences in TAL1 or intracellular NOTCH1 binding to gene desert regions were observed (supplemental Figure 4B). We also found the histone 3 (H3) levels increased at the enhancer regions examined (Figures 4D and 5C), suggesting that Runx1 deletion results in increased H3 loading and a closed chromatin configuration. We used an assay for transposase accessible chromatin (ATAC) and observed decreased ATAC-quantitative PCR enrichment at the N-Me in Runx1-deleted leukemic cells (Figure 5D). These data suggest that a RUNX1 deficiency results in transcription factor depletion and reduced chromatin accessibility at the N-Me.
RUNX1 is required for intracellular NOTCH1 binding and for chromatin accessibility at the N-Me. (A) H3K27ac, NOTCH1, and RUNX1 enrichment at the human MYC super-enhancer is shown in genome browser tracks (genome.ucsc.edu, human hg19). (B) The mouse genomic region (mm10) encompassing Myc and its enhancer loci located 1.27 Mb from the TSS are shown. The RBPJ- and RUNX-binding sites are depicted. (C) Recruitment of RUNX1, intracellular NOTCH1, H3K27ac, and H3 to the mouse Myc enhancer was determined by ChIP-qPCR in control or Runx1-deleted mouse T-ALLs. (D) The degree of open chromatin at the N-Me enhancer region in control or Runx1-deleted mouse T-ALLs was determined by ATAC-quantitative PCR. Data are the mean of 3 or 4 independent experiments and error bars represent ± SEM. *P < .05; **P < .005; ***P < .0005, two-way ANOVA multiple comparisons test.
RUNX1 is required for intracellular NOTCH1 binding and for chromatin accessibility at the N-Me. (A) H3K27ac, NOTCH1, and RUNX1 enrichment at the human MYC super-enhancer is shown in genome browser tracks (genome.ucsc.edu, human hg19). (B) The mouse genomic region (mm10) encompassing Myc and its enhancer loci located 1.27 Mb from the TSS are shown. The RBPJ- and RUNX-binding sites are depicted. (C) Recruitment of RUNX1, intracellular NOTCH1, H3K27ac, and H3 to the mouse Myc enhancer was determined by ChIP-qPCR in control or Runx1-deleted mouse T-ALLs. (D) The degree of open chromatin at the N-Me enhancer region in control or Runx1-deleted mouse T-ALLs was determined by ATAC-quantitative PCR. Data are the mean of 3 or 4 independent experiments and error bars represent ± SEM. *P < .05; **P < .005; ***P < .0005, two-way ANOVA multiple comparisons test.
RUNX dependency extends to TAL1-negative, TLX1/3-transformed human T-ALL cells
These data demonstrating that RUNX1 supports the TAL1 and NOTCH1 transforming signatures predict that a RUNX1 dependency should extend to TAL1-negative, mutant NOTCH1-transformed T-ALL cells. We examined this in the HPB-ALL cell line, which is negative for TAL1, expresses TLX3 and activated NOTCH1 (Figure 2A), and where RUNX1 was previously proposed to have tumor suppressor functions.11 Reductions in RUNX1 expression induced apoptosis and significantly decreased MYC, IL7R, and IGF1R expression (Figure 6A,B). Reductions in MYB expression were also observed (Figure 6B), suggesting that RUNX1 may regulate MYB expression in the absence of TAL1.
RUNX1 mediates survival in a TAL1-negative, TLX3-transformed human T-ALL cell line, and RUNX3 also contributes to human T-ALL survival. (A) The human T-ALL cell line HPB-ALL was transduced with lentiviruses expressing shRNAs against GFP or RUNX1. Apoptotic cells were quantified by Annexin V/7AAD staining followed by flow cytometry. Data are shown as the mean of 3 independent experiments with error bars representing ± SEM. (B) Gene expression in control or RUNX1 knockdown cells was determined by qRT-PCR. Three independent experiments were performed, and data are shown as means with error bars representing ± SEM. (C) RUNX1 or RUNX3 knockdown in KOPTK1 cells induces cell death. The human T-ALL KOPTK1 cell line was transduced with lentiviruses expressing shRNAs against GFP, RUNX1, or RUNX3. Apoptotic cells were quantified by Annexin V/7AAD staining followed by flow cytometry. (D) RUNX1 and RUNX3 binding to N-Me, MYB +14-kb, and MYB –93-kb enhancer loci was determined by ChIP-qPCR. Data are shown as the mean of 4 independent experiments with error bars representing ± SEM (*P < .05, multiple Student t tests). (E-F) The expression of MYB and MYC in RUNX1- or RUNX3-silenced KOPTK1 cells was determined by qRT-PCR. Data are shown as the mean of 3 or 4 independent experiments with error bars representing ± SEM. *P <.05; **P < .005; ***P < .0005, one-way ANOVA multiple comparisons test.
RUNX1 mediates survival in a TAL1-negative, TLX3-transformed human T-ALL cell line, and RUNX3 also contributes to human T-ALL survival. (A) The human T-ALL cell line HPB-ALL was transduced with lentiviruses expressing shRNAs against GFP or RUNX1. Apoptotic cells were quantified by Annexin V/7AAD staining followed by flow cytometry. Data are shown as the mean of 3 independent experiments with error bars representing ± SEM. (B) Gene expression in control or RUNX1 knockdown cells was determined by qRT-PCR. Three independent experiments were performed, and data are shown as means with error bars representing ± SEM. (C) RUNX1 or RUNX3 knockdown in KOPTK1 cells induces cell death. The human T-ALL KOPTK1 cell line was transduced with lentiviruses expressing shRNAs against GFP, RUNX1, or RUNX3. Apoptotic cells were quantified by Annexin V/7AAD staining followed by flow cytometry. (D) RUNX1 and RUNX3 binding to N-Me, MYB +14-kb, and MYB –93-kb enhancer loci was determined by ChIP-qPCR. Data are shown as the mean of 4 independent experiments with error bars representing ± SEM (*P < .05, multiple Student t tests). (E-F) The expression of MYB and MYC in RUNX1- or RUNX3-silenced KOPTK1 cells was determined by qRT-PCR. Data are shown as the mean of 3 or 4 independent experiments with error bars representing ± SEM. *P <.05; **P < .005; ***P < .0005, one-way ANOVA multiple comparisons test.
In addition to RUNX1, we detected RUNX3 expression in a subset of human T-ALL cell lines and primary patient samples (Figure 2A; supplemental Figure 2B). Reductions in RUNX1 or RUNX3 expression in KOPTK1 resulted in apoptosis (Figure 6C), indicating that both RUNX1 and RUNX3 support the survival of these human T-ALL cells. Consistent with these data, we detected RUNX1 and RUNX3 binding at the N-Me and found MYC expression significantly reduced in the RUNX1- or RUNX3-suppressed KOPTK1 cells (Figure 6D-F). Although we detected RUNX1 and RUNX3 binding at the MYB −93-kb enhancer, neither protein was detected at the +14-kb enhancer (Figure 6D). Suppression of RUNX1 or RUNX3 reduced MYB expression, however, statistical significance was achieved only in the RUNX1-silenced cells (Figure 6E-F). Unlike RUNX1, RUNX3 suppression in Jurkat cells did not induce apoptosis and RUNX3 binding was not detected at the MYC or MYB enhancer elements bound by RUNX1 (supplemental Figure 5A-C). These data reveal that KOPTK1 cells rely on RUNX1 and RUNX3 to maintain MYC and MYB levels, whereas in Jurkat cells, RUNX1 supports MYC and MYB expression, and RUNX3 does not contribute. Our findings suggest that the relative levels of RUNX1 and RUNX3 may dictate their roles in MYC and MYB regulation and that T-ALL survival requires a certain threshold level of CBFβ/RUNX1 and/or RUNX3.
CBFβ/RUNX inhibition induces apoptosis of human T-ALL cells and patient samples
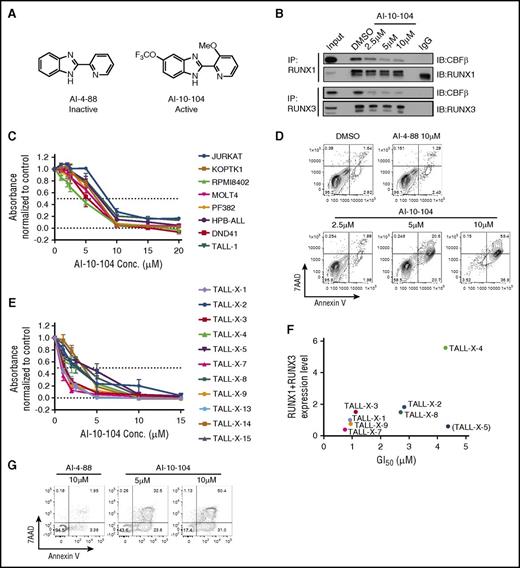
To examine RUNX proteins as therapeutic targets in T-ALL, a series of small molecule inhibitors designed to interfere with CBFβ binding to RUNX proteins were developed by the Bushweller laboratory (Figure 7A and Illendula et al41 ). The inhibitor AI-10-104, which is designed to interfere with RUNX transcriptional activity by preventing CBFβ binding to RUNX proteins, thereby leaving them in an autoinhibited state, induced a dose-dependent decrease in the CBFβ/RUNX1 and CBFβ/RUNX3 heterodimers detected in human T-ALL cells (Figure 7B), but had no detectable effects on CBFβ, RUNX1, or RUNX3 protein levels (supplemental Figure 6A). These data suggest that the AI-10-104 inhibitor reduces RUNX transcriptional activity by interfering with the formation of the CBFβ/RUNX1 and/or CBFβ/RUNX3 heterodimers in T-ALL cells. Treatment of human T-ALL cell lines with AI-10-104 induced apoptosis in a dose-dependent manner, whereas treatment with 10 μM of the inactive analog AI-4-88 had no effect on leukemic growth or viability (Figure 7C-D; supplemental Figure 6B-C). Consistent with the depletion data, the RUNX inhibitor AI-10-104 induced apoptosis in the TLX3-transformed T-ALL cell lines HPB-ALL and DND-41 (supplemental Figure 6B). Treatment of mutant NOTCH1 human T-ALL cells with the RUNX inhibitor also resulted in statistically significant reductions in MYC mRNA levels, suggesting that AI-10-104 interferes with NOTCH1/MYC enhancer activity (supplemental Figure 6D). Notably, LOUCY cells, which do not express TAL1 or mutant NOTCH1 (supplemental Table 1) were resistant to AI-10-104 treatment (supplemental Table 1: GI50 (the concentration for 50% of maximal inhibition of cell proliferation) = 11 μM).
Treatment with a RUNX-CBFβ inhibitor impairs the growth of human T-ALL cell lines and primary pediatric T-ALL samples. (A) Structures of inactive (AI-4-88) and active (AI-10-104) inhibitors are shown. (B) Protein lysates from the human T-ALL cell line KOPTK1 treated with DMSO or increasing concentrations of AI-10-104 for 6 hours were immunoprecipitated with RUNX1 or RUNX3 antibodies and immunoblotted with CBFβ, RUNX1, and RUNX3 antibodies. (C) Eight human T-ALL cell lines were treated with increasing concentrations of AI-10-104 for 3 days, and cell growth/metabolism was analyzed by MTS assay. (D) The human T-ALL cell line Jurkat was treated with vehicle, 10 μM of the inactive analog AI-4-88, or with increasing concentrations of AI-10-104 for 4 days. Cells were stained with Annexin V-FITC and 7AAD and analyzed by flow cytometry. A representative flow profile of 3 independent experiments is shown. (E) Eleven pediatric T-ALL patient samples were treated with vehicle or increasing concentrations of AI-10-104 (1-15 μM) for 3 days, and cell growth/metabolism was analyzed by CellTiterGlo assay. Absorbance values were normalized to those obtained with vehicle control. (F) The sensitivity of patient samples to AI-10-104 (GI50) correlates with RUNX1 and RUNX3 expression levels (Pearson’s r = 0.8781, P = .0093, sample TALL-X-5 excluded). (G) Patient sample TALL-X-15 was treated with 10 μM of AI-4-88 or with 5 or 10 μM of AI-10-104 for 6 days. Cells were stained with Annexin V-FITC and 7AAD and analyzed by flow cytometry.
Treatment with a RUNX-CBFβ inhibitor impairs the growth of human T-ALL cell lines and primary pediatric T-ALL samples. (A) Structures of inactive (AI-4-88) and active (AI-10-104) inhibitors are shown. (B) Protein lysates from the human T-ALL cell line KOPTK1 treated with DMSO or increasing concentrations of AI-10-104 for 6 hours were immunoprecipitated with RUNX1 or RUNX3 antibodies and immunoblotted with CBFβ, RUNX1, and RUNX3 antibodies. (C) Eight human T-ALL cell lines were treated with increasing concentrations of AI-10-104 for 3 days, and cell growth/metabolism was analyzed by MTS assay. (D) The human T-ALL cell line Jurkat was treated with vehicle, 10 μM of the inactive analog AI-4-88, or with increasing concentrations of AI-10-104 for 4 days. Cells were stained with Annexin V-FITC and 7AAD and analyzed by flow cytometry. A representative flow profile of 3 independent experiments is shown. (E) Eleven pediatric T-ALL patient samples were treated with vehicle or increasing concentrations of AI-10-104 (1-15 μM) for 3 days, and cell growth/metabolism was analyzed by CellTiterGlo assay. Absorbance values were normalized to those obtained with vehicle control. (F) The sensitivity of patient samples to AI-10-104 (GI50) correlates with RUNX1 and RUNX3 expression levels (Pearson’s r = 0.8781, P = .0093, sample TALL-X-5 excluded). (G) Patient sample TALL-X-15 was treated with 10 μM of AI-4-88 or with 5 or 10 μM of AI-10-104 for 6 days. Cells were stained with Annexin V-FITC and 7AAD and analyzed by flow cytometry.
We also examined primary pediatric T-ALL samples for their sensitivity to the CBFβ/RUNX inhibitor AI-10-104. Treatment of diagnostic and relapsed pediatric T-ALL samples with AI-10-104 in vitro inhibited growth with an average GI50 of 2.4μM (Figure 7E) and induced apoptosis (Figure 7G; Supplemental Figure 6E), whereas treatment with the inactive compound AI-4-88 had no effect on the growth or viability of primary T-ALL samples (supplemental Figure 6C). Moreover, AI-10-104 sensitivity correlated with RUNX1/3 expression levels in 7 of 8 T-ALL patient samples selected at random (Figure 7F).
RUNX1 is required for hematopoietic stem and progenitor cell development and survival,2,3 raising the possibility that RUNX inhibition in leukemic patients may result in on-target effects on normal hematopoietic stem and progenitor cells. We performed dose response studies on bone marrow samples from 3 independent, healthy donors. Treatment of normal human hematopoietic cells with AI-10-104 resulted in an average GI50 of 15.4 μM (supplemental Figure 6F), which exceeded the average GI50 observed for primary patient leukemic samples by sevenfold. Unfortunately, the pharmacokinetics of the current AI-10-104 inhibitor preclude its preclinical testing in vivo. Nonetheless, these data suggest a therapeutic window may exist for optimized derivatives of AI-10-104 in T-ALL patients.
Discussion
We provide genetic evidence that RUNX1 and 3 have crucial prosurvival roles in T-ALL and are required to support critical nodes of the TAL1- and NOTCH1-transforming signatures. We found that mouse T-ALL cells rely on RUNX1 to maintain TAL1 and intracellular NOTCH1 occupancy at the Myb and Myc enhancers, respectively. These findings define novel functions for RUNX1 in T-cell leukemogenesis in maintaining oncogene enhancer activity by elevating constituent transcription factor binding to these regions.
Our data are supported by the demonstration that a recently developed CDK7 inhibitor (THZ1) exhibited selectivity for human T-ALL cells and was shown to act via suppression of the RUNX transcriptional network.36 Although CDK7 is a component of the general transcription factor IIH complex, low-dose THZ1 treatment of human T-ALL cells affected the transcription of a subset of genes, with RUNX1 expression most profoundly affected.
We show that RUNX1 deficiency reduces transcription factor binding at the mouse Myb +15-kb and −92-kb enhancers and the (N-Me. The reductions in TAL1 binding to the mouse Myb enhancer regions in Runx1-deleted T-ALL cells are particularly noteworthy because the proximal lck promoter drives Tal1 expression and, consequently, reductions in TAL1 binding at the Myb enhancer do not reflect RUNX1 effects on endogenous Tal1 transcription. These data suggest that, in addition to regulating TAL1 expression,21 RUNX1 supports TAL1 binding to the Myb enhancer. We also found that Cdk6 expression depends on RUNX1 (Figure 3) and, consistent with our data, Palii et al.27 show that RUNX1/3 suppression in Jurkat cells reduced TAL1 binding to several genes that are important in thymocyte differentiation, including the CDK6 locus. In this study, however, RUNX1/3 knockdown had no detectable effect on TAL1 expression, suggesting that RUNX1 primarily regulates TAL1 binding. These findings are relevant to T-ALL patients because most patients activate TAL1 expression via chromosomal rearrangements that displace the TAL1 promoter and thereby subvert RUNX1-mediated effects on TAL1 transcription.
Precisely how RUNX1 deficiency interferes with TAL1 and intracellular NOTCH1 binding to these enhancer regions is unclear. RUNX1 has been shown to interact with TAL1 and intracellular NOTCH1 in T-ALL cells27,42 suggesting that RUNX1 may be a component of both transcriptional complexes. However, the E-box, RUNX, and NOTCH1/CSL/RBJκ consensus sites are dispersed throughout the conserved Myb and Myc enhancer regions examined, making it unlikely that TAL1/RUNX1 or intracellular NOTCH1/RUNX1 bind as single complexes.
RUNX1 deficiency results in decreases in the active chromatin mark H3K27ac and increases in H3 loading (Figures 4,5), raising the possibility that RUNX1 directly regulates chromatin and/or recruits histone-modifying enzymes and/or other chromatin regulators to these enhancer regions. RUNX1 has been shown to interact with histone acetyltransferase p30043 and BRG1,44 the ATPase subunit of the SWI/SNF chromatin remodeling complex. BRG1 knockdown led to marked reductions in transcription factor binding and disruption of MYC 1.7-Mb enhancer-promoter interaction in AML cells.45 Similarly, NOTCH1 inhibition interferes with N-Me interactions with the MYC promoter and suppresses MYC mRNA levels.40 Our data show that a RUNX1 deficiency evicts TAL1 and NOTCH1 from the Myb and Myc enhancers, respectively, leading us to speculate that RUNX1 depletion destabilizes enhancer-promoter interactions at these loci.
Importantly, we demonstrate that the prosurvival roles for RUNX1 revealed in our mouse TAL1/LMO2 T-ALL model translate to human T-ALL cells transformed by TAL1, TLX3, and/or NOTCH1. What remains unclear is whether T-ALL cells that do not express TAL1 or activated NOTCH1 also depend on the RUNX transcription factors for survival. We attempted to address this issue in LOUCY cells (TAL1- and NOTCH1-negative), which proved relatively resistant to AI-10-104 treatment (supplemental Table 2), suggesting that the TAL1 and/or NOTCH1 status determines RUNX dependency. Based on the prevalence of TAL1 and NOTCH1 activation in T-ALL, we expect most T-ALLs to be sensitive to RUNX inhibition. Consistent with our findings, Jenkins and Weng found mouse T-ALLs transformed by activated NOTCH1 and all human T-ALL cell lines examined (n = 15) depend on RUNX1 for their survival (Catherine Jenkins and Andrew Weng, manuscript submitted August 2017). However, there are clear TAL1 and NOTCH1 independent mechanisms to overexpress and/or deregulate MYB and MYC in T-ALL,46,47 and the role of RUNX proteins in these cases remains to be tested.
Attempts have been made to target enhancers in cancer therapy by using BET bromodomain inhibitors or histone-modifying enzymes. The obvious concern is that such treatments would have toxic side effects due to inhibition of enhancer activity in normal cells. Although the BRD4 inhibitor JQ1 has clear anti-leukemic activity via its effects on MYC,25,26,48 toxicities have been observed, and RNA interference–mediated inhibition of BRD4 in mice has deleterious effects on tissue homeostasis.49 These findings predict that targeting broad regulators of enhancer activity may interfere with normal tissue repair and regeneration and may not be tolerated long term in patients.
Our genetic and pharmacologic experiments reveal that targeting RUNX1 might be an alternative strategy to disrupt oncogenic MYB and MYC enhancers in T-ALL and elicit antileukemia activity. With the development of more potent and stable AI-10-104 analogs, the effects of RUNX1 inhibition can be tested in preclinical mouse and human T-ALL models for efficacy and ensure the safety of the therapeutic strategy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Kelliher and Castilla laboratories and Thomas Fazzio for technical advice and helpful suggestions. The authors also thank the Flow Cytometry Core and the Department of Molecular, Cell and Cancer Biology Bioinformatics Core at the University of Massachusetts for their support.
This work was supported by the National Cancer Institute, National Institutes of Health (grant RO1CA96899) (M.A.K.). This work was partially supported by a Hyundai Hope On Wheels Award and an Innovator Award from Alex’s Lemonade Stand (M.A.K.).
Authorship
Contribution: A.C. performed the experiments and analyzed the data in this manuscript, except where indicated; J.Y. and L.J.Z. performed the Gene Set Enrichment Analysis shown in Figure 3; J.T. provided the data shown in supplemental Figure 3; N.H. provided the data shown in supplemental Figure 2B; A.I. and J.H.B. provided the AI-4-88 and AI-10-104 compounds and provided advice regarding their use; J.E.R. performed the studies using patient samples shown in Figure 7E; J.A.P. performed the coimmunoprecipitation studies shown in Figure 7B; J.H.B., J.A.P., J.E.R., and L.H.C. analyzed and interpreted the data and provided helpful discussions; and M.A.K. designed and supervised the study and wrote the paper.
Conflict-of-interest disclosures: The authors declare no competing financial interests.
Correspondence: Michelle A. Kelliher, Department of Molecular, Cell and Cancer Biology, University of Massachusetts Medical School, Lazare Research Building, 364 Plantation St, Worcester, MA 01605; e-mail: michelle.kelliher@umassmed.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal