In this issue of Blood, Choi et al demonstrated an oncogenic role of RUNX1 in T-cell acute lymphoblastic leukemia (T-ALL),1 which is in marked contrast to its tumor-suppressive role identified earlier in T-ALL, acute myeloid leukemia (AML), and myelodysplastic syndrome (MDS).
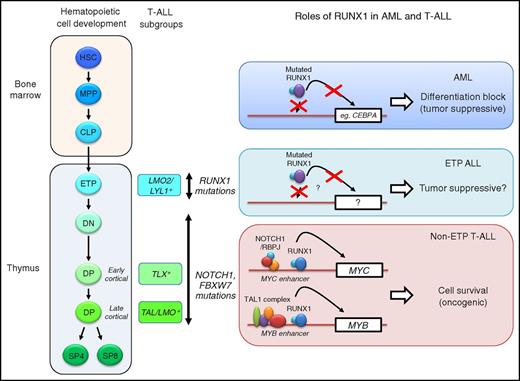
Hematopoietic cell development, T-ALL subgroups and the roles of RUNX1. Different subgroups of T-ALL (TAL/LMO+, TLX+, and LMO2/LYL+) are associated with specific stages of T-cell differentiation. RUNX1 mutations are enriched in ETP ALL. Mutations in the NOTCH1 signaling pathways (NOTCH1 and FBXW7) are enriched in non-ETP T-ALL. In AML cells, mutated RUNX1 fails to bind to DNA or activate transcription.2 This leads to differentiation block via repression of downstream target genes such as CEBPA (top). In non-ETP T-ALL, RUNX1 binds to the MYC and MYB enhancers with the NOTCH1/RBPJ complex and the TAL1 complex, respectively, and activates their expression, thereby promoting T-ALL cell survival (bottom).1 In this context, RUNX1 serves as an oncogenic factor. In ETP ALL, mutated RUNX1 possibly acts as a tumor suppressor, although detailed mechanisms have not yet been elucidated (middle). CLP, common lymphoid progenitor; DN, double negative (CD4−CD8−); DP, double positive (CD4+CD8+); ETP, early T-cell precursor; HSC, hematopoietic stem cell; MPP, multipotent progenitor; SP, single positive (CD4+ or CD8+).
Hematopoietic cell development, T-ALL subgroups and the roles of RUNX1. Different subgroups of T-ALL (TAL/LMO+, TLX+, and LMO2/LYL+) are associated with specific stages of T-cell differentiation. RUNX1 mutations are enriched in ETP ALL. Mutations in the NOTCH1 signaling pathways (NOTCH1 and FBXW7) are enriched in non-ETP T-ALL. In AML cells, mutated RUNX1 fails to bind to DNA or activate transcription.2 This leads to differentiation block via repression of downstream target genes such as CEBPA (top). In non-ETP T-ALL, RUNX1 binds to the MYC and MYB enhancers with the NOTCH1/RBPJ complex and the TAL1 complex, respectively, and activates their expression, thereby promoting T-ALL cell survival (bottom).1 In this context, RUNX1 serves as an oncogenic factor. In ETP ALL, mutated RUNX1 possibly acts as a tumor suppressor, although detailed mechanisms have not yet been elucidated (middle). CLP, common lymphoid progenitor; DN, double negative (CD4−CD8−); DP, double positive (CD4+CD8+); ETP, early T-cell precursor; HSC, hematopoietic stem cell; MPP, multipotent progenitor; SP, single positive (CD4+ or CD8+).
RUNX1 is one of the best-characterized hematopoietic transcription factors.2 This factor is essential for the differentiation of hematopoietic stem cells into different cell lineages as well as for the maintenance of adult hematopoiesis. Since its discovery as a translocation partner gene identified in AML, RUNX1 has also been recognized as a tumor suppressor in the context of AML and MDS.2,3 Point mutations in the runt domain or chromosomal translocations that lack the transactivation domain of RUNX1 (eg, RUNX1-RUNX1T1) are frequently observed in AML and MDS. Clinical and basic studies indicated that these loss-of-function mutations of RUNX1 are associated with differentiation block and contribute to malignant transformation.
Mutations of RUNX1 have also been reported in T-ALL in several cohorts.4-8 All of these mutations are heterozygous and clustered at the RUNT domain and the transactivation domain. Most are missense or frameshift mutations, suggesting that they are loss-of-function mutations. Studies in Germany revealed that RUNX1 mutations are associated with poor prognosis.5,6 RUNX1 target genes are downregulated in T-ALL cells that express the T-ALL oncogene TLX1 or TLX3.4 Overexpression of RUNX1 in TLX-positive T-ALL cell lines inhibits cell growth. Hence, these studies implicated RUNX1 as a tumor suppressor in T-ALL. In contrast, several studies suggested potential oncogenic roles of RUNX1 in T-cell malignancies. Prior studies using retroviral mutagenesis screening identified Runx genes, including Runx1, as one of common insertion sites in the Myc-induced murine T-cell lymphoma model, suggesting that RUNX proteins act as collaborating oncogenic factors.9 In human T-ALL cells, the RUNX1 protein co-occupies regulatory elements with the oncogenic transcription factor TAL1/SCL at key downstream target genes,10 similar to that observed in normal hematopoietic stem and progenitor cells, thus serving as a collaborator of TAL1.
Interestingly, Choi et al demonstrated that RUNX1 is required for T-ALL cell survival by activating several key oncogenes with TAL1 and NOTCH1.1 The authors tested their hypothesis by generating a T-ALL mouse model in which the Runx1 gene was conditionally deleted. They first isolated leukemic cells from the Tal1/Lmo2 double-transgenic mice and transplanted them into recipient mice, followed by tamoxifen treatment to delete Runx1. Strikingly, a loss of Runx1 abolished T-ALL development, and Runx1-deficient leukemia cells underwent apoptosis. Consistently, inhibition of the RUNX1 protein by short hairpin RNA knockdown or treatment with small molecules in human T-ALL cell lines induced apoptosis. The authors found that RUNX1 co-occupies the MYB and MYC regulatory elements with TAL1 and NOTCH1, respectively, in both mouse and human T-ALL cells, and transcriptionally activates their expressions. The majority of human T-ALL cell lines and primary leukemia cells are sensitive to the pharmacological inhibition of RUNX1, thus implicating RUNX1 as a therapeutic target. It is noteworthy that the RUNX1 gene was not mutated in most of the tested samples. Thus, these results indicated that the wild-type RUNX1 protein is functionally required for leukemia development and the survival of T-ALL cells as a collaborator of 2 driver T-ALL oncogenes, TAL1 and NOTCH1.
This study clearly demonstrated the oncogenic roles of RUNX1 in T-ALL. However, this study also challenged previous studies on RUNX1 in T-ALL. Is RUNX1 generally oncogenic in T-ALL? What are the consequences of RUNX1 mutations in T-ALL? One factor that possibly affects these effects may be the differentiation status of T cells where leukemia arises. T-ALL can be classified into several subgroups based on the expression of oncogenic transcription factors such as TAL (TAL1, TAL2), TLX (TLX1, TLX3), HOXA, NKX2-1, and LMO2/LYL1.8,11 These subgroups are closely associated with specific stages of T-cell development. For example, TAL1-positive T-ALL cases typically show the immunophenotype of the late cortical stage of T cells, whereas LMO2/LYL1-positive T-ALL cases show the early T-cell precursor (ETP) phenotype (see figure). Notably, earlier studies indicated that most of the RUNX1-mutated T-ALL cases show an ETP phenotype.4-7 Consistently, recent genome-wide sequencing studies indicated that RUNX1 mutations are enriched in LMO2/LYL1-positive T-ALL cases and are less prevalent in TAL1-positive cases.8 In the LMO2/LYL1-positive T-ALL subgroup, genetic mutations that are commonly found in AML, such as mutations of FLT3 and NRAS, are also enriched, while mutations in the NOTCH1 signaling pathway are less frequently found.8 Thus, the mutational profile in ETP ALL is different from that of non-ETP T-ALL. Although further investigations using animal models are necessary, RUNX1 possibly serves as a tumor suppressor in the context of ETP ALL, similar to that reported in AML, while it serves as an oncogenic collaborator in “typical” non-ETP T-ALL cases, which are driven by TAL1 and NOTCH1. In support of this, Choi et al noted that one of the T-ALL cell lines (LOUCY) representing ETP ALL is relatively more resistant to RUNX inhibitors than non-ETP T-ALL cell lines.1
Another important consideration is the functional redundancy between the RUNX1 and RUNX3 proteins. Choi et al showed that RUNX3 is expressed together with RUNX1 in some cell lines and half of primary T-ALL cases.1 RUNX3 can also bind at the MYC and MYB enhancers and positively regulates those expressions in one T-ALL cell line (KOPT-K1). Knockdown of either RUNX1 or RUNX3 induced apoptosis in this cell line, indicating that those T-ALL cells are dependent on both RUNX1 and RUNX3 for their survival. In other words, pharmacological inhibition of RUNX proteins by small molecules that inhibit both RUNX1 and RUNX3 would be an ideal approach to treat T-ALL cells, as the authors demonstrate.1 It is also of interest to elucidate the roles of RUNX3 in ETP ALL with RUNX1 mutations. Taken together, the study by Choi et al elucidated novel aspects of RUNX proteins in the pathogenesis of T-ALL.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal