Key Points
Somatic mutations of HLA-B*40:02 are very frequently detected in granulocyte of patients with acquired aplastic anemia.
Antigen presentation via HLA-B4002 may play a critical role in the pathophysiology of acquired aplastic anemia.
Abstract
To identify HLA alleles closely involved in the autoantigen presentation in acquired aplastic anemia (AA), we studied the HLA allelic loss frequencies of 312 AA patients, including 43 patients with loss of heterozygosity of 6p chromosome (6pLOH). An analysis of the HLA alleles contained in the lost haplotype revealed HLA-B*40:02 to be the most frequently lost allele. When we examined 28 AA (12 6pLOH[+] and 16 6pLOH[−]) patients with HLA-B*40:02 for the presence of leukocytes lacking HLA-B4002 (B4002−) using a new monoclonal antibody specific to this allele, B4002− granulocytes were detected not only in all 6pLOH(+) patients but also in 9 (56%) of the 16 6pLOH(−) patients. Furthermore, 10 (83%) of the 12 6pLOH(+) patients possessed 1.0% to 78% B4002− granulocytes that retained the HLA-A allele on the same haplotype (B4002−A+), suggesting the frequent coexistence of granulocytes that underwent mutations restricted to HLA-B*40:02 with 6pLOH(+) (B4002−A−) granulocytes. Deep sequencing of the HLA-B*40:02 of sorted B4002−A+ granulocytes revealed various somatic mutations, such as frameshift, nonsense, and splice site mutations, in all 15 patients studied. Surprisingly, missense mutations in the α-3 domain of HLA-B*40:02 that are not involved in the antigen presentation were detected exclusively in the B4002+ granulocytes of 3 patients possessing B4002− granulocytes. The markedly high prevalence of leukocytes lacking HLA-B4002 as a result of either 6pLOH or structural gene mutations, or both, suggests that antigen presentation by hematopoietic stem/progenitor cells to cytotoxic T cells via the HLA-B allele plays a critical role in the pathogenesis of AA.
Introduction
Acquired aplastic anemia (AA) is thought to be a T-cell-mediated autoimmune disease based on a good response to immunosuppressive therapy, such as antithymocyte globulin (ATG) or cyclosporine (CsA).1 A large body of in vitro evidence suggests the essential role of T cells in the development of AA, which includes the presence of T cells with particular T-cell receptor clonotypes,2-7 cytotoxic T-cell clones capable of killing hematopoietic stem/progenitor cells (HSPCs),2,4 and leukocytes that lack HLA class I antigens because of copy-number neutral loss of heterozygosity in the short arm of chromosome 6 (6pLOH).8-12 Among these suggestive findings, the presence of 6pLOH(+) leukocytes is considered to be the most compelling evidence that cytotoxic T cells (CTLs) are involved in the development of bone marrow failure, because it represents the escape of HSPCs with 6pLOH from the attack of CTLs that are specific to autoantigens presented by the lacked HLA class I alleles. However, the incidence of 6pLOH in AA patients shown by several studies was at most 13%, and it is therefore unclear to what extent the HSPC-specific CTLs contribute to the development of AA and which HLA proteins are the most critically involved in the autoantigen presentation.
We previously determined the missing frequencies of individual HLA alleles by analyzing the alleles contained in the missing haplotype of 6pLOH(+) AA patients and found that 4 alleles (HLA-A*02:01, A*02:06, A*31:01, and B*40:02) were significantly more likely to be lost than were the other HLA alleles.9 However, it was difficult to determine which allele in the missing haplotype was actually responsible for the antigen presentation because the lost fragment of chromosome 6p usually contained 2 or more HLA alleles, and allele-specific monoclonal antibodies (mAbs) useful for detecting leukocytes lacking single alleles were not commercially available, except for mAbs specific to some HLA-A alleles.
Among the 4 HLA class I alleles, an important role of HLA-B*40:02 in autoantigen presentation in AA has been suggested by other studies. A congress abstract13 reported a markedly high prevalence of HLA-B61, a corresponding serotype to HLA-B*40:02, in Japanese pediatric AA patients. Inaguma et al14 established a CTL clone capable of suppressing HSPCs in a HLA-B*40:02-restricted manner from an AA patient who possessed HLA-B*40:02-lacking leukocytes because of 6pLOH. Osumi et al15 reported a case of AA whose leukocytes had a nonsense mutation in HLA-B*40:02, suggesting the presence of leukocytes lacking only HLA-B4002. Given these findings, a flow cytometry (FCM) analysis using HLA-B4002-specific mAbs may reveal leukocytes that lack HLA-B4002 due to mechanisms other than 6pLOH in AA patients carrying HLA-B*40:02. Many researchers have tried to generate a mAb specific for HLA-B61 by immunizing transgenic mice, but all attempts have failed for some reason. If anti-HLA-B61 mAbs were generated, it would greatly facilitate the understanding of the mechanisms underlying the lack of HLA-B4002 from leukocytes as well as of the immune mechanisms of AA.
We recently succeeded in generating mAbs specific to HLA-B61, taking advantage of complementary DNA (cDNA) derived from B lymphocytes from an AA patient possessing anti-HLA-B61 antibody as a result of multiple transfusions.16 The mAbs successfully identified HLA-B4002-missing leukocytes not only in 6pLOH(+) patients but also in 6pLOH(−) patients in the majority of AA patients possessing HLA-B*40:02, and deep sequencing of HLA-B4002-missing granulocyte-derived DNA revealed various mutations in the structural gene region of HLA-B*40:02, strongly suggesting a critical role of HLA-B4002 in the autoantigen presentation of AA.
Patients and methods
Subjects
A total of 312 Japanese patients with AA (age, 15 to 90 years old, median 65 years old; 132 males and 180 females; 115 severe AA and 197 nonsevere AA) were enrolled in an observational study to determine the prevalence of HLA allele-lacking leukocytes (HLA-LLs) between 2011 and 2014 (supplemental Figure 1, available on the Blood Web site). The patients’ blood samples were subjected to HLA typing and droplet digital polymerase chain reaction (ddPCR) for detecting 6pLOH. Severe AA was diagnosed when at least 2 of the following criteria were met: the neutrophil count was <0.5 × 109/L, the platelet count was <20 × 109/L, and the reticulocyte count was <20 × 109/L.17 Very severe AA was defined as a neutrophil count <0.2 × 109/L in addition to the criteria for severe AA.18 The response criteria were as previously described.19 All patients were genotyped for HLA-A, HLA-B, HLA-C, and HLA-DRB1 alleles using the polymerase chain reaction (PCR) sequence-specific oligonucleotide method.20 All patients provided their informed consent to the HLA-typing and genetic analyses. The study protocols were approved by the ethical committee of Kanazawa University Institute of Medical, Pharmaceutical, and Health Sciences.
Preparation of anti-HLA-B61 antibody
We developed a new method for generating human mAbs specific to HLA alleles by taking advantage of B cells from patients who had anti-HLA antibodies following repeated transfusions.16 Briefly, mononuclear cells isolated from 20 mL of peripheral blood from an anti-HLA-B61 antibody-positive patient were stimulated to differentiate into antibody-producing cells with a cytokine cocktail containing R848, CpG2006, anti-CD40, recombinant human interleukin (hIL)-2, hIL-4, hIL-17, hIL-21, and human B-cell activating factor in RPMI1640 containing 10% fetal calf serum for 6 days. CD138+ antibody-producing cells were then isolated with anti-CD138 antibody-conjugated microbeads using AutoMACS Pro Separator (Miltenyi Biotec, Bergisch Gladbach, Germany). Anti-HLA-B61 immunoglobin G (IgG) antibody-producing cells were isolated using a microwell array chip coated with purified HLA-B61 proteins and subjected to RNA extraction from single cells, as previously described.21,22 The antibody cDNA fragments for heavy and light chain variable domain fragments were amplified using the single-cell 5′-RACE method and inserted into expression vectors. Thereafter, HEK293 cells were transfected with the heavy and light chain vectors and cultured to obtain a supernatant containing complete mAb molecules. The specificity of the mAbs was tested by enzyme-linked immunosorbent assay and FCM. To prevent nonspecific binding of human IgG to human Fc receptors, we replaced the Fc portion of the anti-HLA-B61 mAb with the constant domain of mouse IgG1.
Detection of HLA-LLs and cell sorting
HLA-LLs were detected using BD FACSCanto II (BD Biosciences, San Jose, CA) and analyzed with the FlowJo (version 10.1) software program (Tree Star, Ashland, OR). Leukocytes lacking HLA-B4002 were referred to as B4002− cells, including both 6pLOH(+) leukocytes that lack both HLA-B4002 and 1 HLA-A allele on the same haplotype (B4002−A−) and leukocytes lacking only HLA-B4002 (B4002−A+). B4002−A+ granulocytes and HLA-B4002-positive (B4002+) granulocytes or B4002+ T cells were sorted using BD FACSAria Fusion (BD Biosciences). The mAbs used for this study are provided in supplemental Table 1.
ddPCR to detect 6pLOH involving HLA genes
The presence of 6pLOH(+) leukocytes and their percentages of total leukocytes were determined with ddPCR using a QX200 AutoDG Droplet Digital PCR System (Bio-Rad, Hercules, CA) by comparing the copy number of each HLA allele in individuals heterozygous for the HLA allele. The 6pLOH that involves HLA genes gives rise to a copy number imbalance between the 2 different alleles. The reaction mixtures of ddPCR included 2 TaqMan probes labeled with different fluorochromes (6-FAM or VIC) complementary to the allele-specific sequences, 2 primers complementary to consensus sequences surrounding the allele-specific sequence, and 40 ng of genomic DNA in 1× ddPCR Supermix for Probes (no deoxyuridine triphosphate; Bio-Rad). We designed 4 primer and probe mixture that can detect 6pLOH involving HLA-A (A31/33 mixture), HLA-B (B1 and B2 mixture), and HLA-C (C3 mixture; supplemental Tables 2 and 3). The detailed protocols for ddPCR are shown in the supplemental Methods.
When the lacking alleles contained in the haplotype of 6pLOH(+) patients could not be completely determined by ddPCR or FCM, they were estimated with a haplotype database of the Japanese population including 18 604 individuals from 5824 families.23 An allele was judged to be missing as a result of 6pLOH if the presence of the allele in the lost haplotype was estimated with >95% accuracy.
Detection of HLA-B gene mutations
To identify the somatic mutations of HLA-B*40:02 in B4002−A+ granulocytes, we performed target sequencing of HLA-B in sorted B4002−A+ granulocytes using a next-generation sequencer (NGS, MiSeq; Illumina, San Diego, CA). B4002+ granulocytes, B4002+ T cells, or both from identical individuals were used as a control. HLA genes were enriched from genomic DNA using sequence capture (SeqCap EZ System; Roche Sequencing, Pleasanton, CA), a hybridization-based gene enrichment method. Potential mutations responsible for missing HLA-B4002 were identified when variant reads were found only in B4002− granulocytes. All of the mutations were validated using deep sequencing of HLA-B locus-specific long-range PCR, as previously described.24 HLA-B alleles carrying those mutations were determined using the nearest allele-specific single-nucleotide polymorphisms. Reference sequences of HLA-B*40:02 were obtained from the Immuno Polymorphism Database International ImMunoGeneTics Project HLA database.25
Statistical analyses
The clinical parameters of the patients were compared using Fisher’s exact test for categorical variables and the Mann-Whitney U test for continuous variables with the EZR software package, a graphical user interface for R (The R Foundation for Statistical Computing, version 2.13.0).26
Results
Allelic frequency of HLA genes in this study’s subjects
When the allelic frequency of HLA genes in 312 AA patients genotyped in this study was compared with those in the Japanese general population,23 HLA-B*40:02 was the fifth most frequent allele observed in AA patients; the top 5 alleles were HLA-DRB1*15:02 (22% vs 11%, odds ratio [OR] = 2.3, P = 3.0 × 10−15), B*52:01 (19% vs 11%, OR = 1.9, P = 6.0 × 10−9), DRB1*15:01 (14% vs 7.7%, OR = 2.0, P = 4.5 × 10−8), C*12:02 (19% vs 11%, OR = 1.9, P = 2.8 × 10−8), and B*40:02 (13% vs 7.9%, OR = 1.8, P = 9.3 × 10−6).
Validation of the 6pLOH(+) cell measurement using ddPCR
The ddPCR generated 18 745 ± 1553 (mean ± 2 standard deviations) droplets per 1 reaction mixture, which allowed us to detect at least 3% to 4% 6pLOH(+) leukocytes with a specificity of 99%. The specificity of the ddPCR in detecting 6pLOH(+) leukocytes was verified by showing negativity for 50 healthy volunteers or AA patients who were known to be negative for HLA-A-allele lacking leukocytes using FCM. To determine the reliability of estimating the 6pLOH(+) cell percentage by ddPCR, we mixed leukocytes from an individual carrying HLA-A2 and Cw3 with leukocytes from an individual not carrying A2 or Cw3 at various ratios, and the mixed-leukocyte populations were examined by FCM and ddPCR. The percentages of 6pLOH(+) cells estimated by ddPCR were almost identical to those determined by FCM, suggesting sufficient reliability of ddPCR in measuring the percentage of 6pLOH(+) cells in the total leukocyte populations (supplemental Figure 2).
Detection of 6pLOH and the frequency of HLA alleles involved in the lost haplotype
Using any of the 4 different ddPCR mixtures, the presence of 6pLOH was evaluable in 224 (72%) of 312 patients with AA, because they were available for ddPCR in at least 1 of the 3 HLA class I loci. The 6pLOH(+) leukocytes that accounted for 3.9% to 84% (median, 10%) of the total leukocytes were detected in 43 (19%) of the 224 patients analyzed by the ddPCR. The 6pLOH(+) patients had significantly more severe disease (51%) than did the 6pLOH(−) patients (33%, P = .03), whereas age (P = .27), sex (P = .39), and prior immunosuppressive therapy (IST, P = .44) did not differ markedly between the 2 groups (Table 1). When the prevalence of 6pLOH was compared between patients carrying and not carrying each HLA allele, including alleles in HLA-A, HLA-B, HLA-C, and HLA-DRB1 loci, consistent with a previous report,9 HLA-B*40:02 was most strongly involved in 6pLOH (34% [25 out of 74], OR = 3.7, P = .0002), followed by HLA-DRB1*08:02 (40% [8 of 20], OR = 3.2, P = .03). Of note, the HLA-B*40:02 allele was included in the lost haplotype in all 25 6pLOH(+) patients possessing this allele. Seven of the 8 patients with HLA-DRB1*08:02 alleles possessed HLA-B*40:02 in the same lost haplotype, and the remaining patient had HLA-DRB1*08:02 in the retained haplotype, suggesting that the higher frequency of HLA-DRB1*08:02 in 6pLOH(+) patients is due to the linkage disequilibrium between HLA-B*40:02 and DRB1*08:02.
Comparison between 6pLOH(+) and 6pLOH(−) patients
| . | 6pLOH(+) . | 6pLOH(−) . | P . |
|---|---|---|---|
| Number of patients | 43 | 181 | |
| Age, median years (range) | 69 (18-82) | 64 (15-90) | .27 |
| Sex | |||
| Male | 21 | 73 | .39 |
| Female | 22 | 108 | |
| Severity | |||
| Severe | 22 | 60 | .03 |
| Nonsevere | 21 | 121 | |
| Prior IST | |||
| ATG/CsA | 5 | 12 | .44 |
| CsA | 6 | 22 | |
| None | 32 | 147 | |
| HLA-B*40:02 | |||
| (+) | 25 | 49 | .0002 |
| (−) | 18 | 132 | |
| HLA-DRB1*08:02 | |||
| (+) | 8 | 12 | .03 |
| (−) | 35 | 169 | |
| HLA-A*02:01 | |||
| (+) | 11 | 43 | .84 |
| (−) | 32 | 138 | |
| HLA-A*02:06 | |||
| (+) | 7 | 45 | .32 |
| (−) | 36 | 136 | |
| HLA-A*31:01 | |||
| (+) | 8 | 43 | .55 |
| (−) | 35 | 138 |
| . | 6pLOH(+) . | 6pLOH(−) . | P . |
|---|---|---|---|
| Number of patients | 43 | 181 | |
| Age, median years (range) | 69 (18-82) | 64 (15-90) | .27 |
| Sex | |||
| Male | 21 | 73 | .39 |
| Female | 22 | 108 | |
| Severity | |||
| Severe | 22 | 60 | .03 |
| Nonsevere | 21 | 121 | |
| Prior IST | |||
| ATG/CsA | 5 | 12 | .44 |
| CsA | 6 | 22 | |
| None | 32 | 147 | |
| HLA-B*40:02 | |||
| (+) | 25 | 49 | .0002 |
| (−) | 18 | 132 | |
| HLA-DRB1*08:02 | |||
| (+) | 8 | 12 | .03 |
| (−) | 35 | 169 | |
| HLA-A*02:01 | |||
| (+) | 11 | 43 | .84 |
| (−) | 32 | 138 | |
| HLA-A*02:06 | |||
| (+) | 7 | 45 | .32 |
| (−) | 36 | 136 | |
| HLA-A*31:01 | |||
| (+) | 8 | 43 | .55 |
| (−) | 35 | 138 |
Among 150 HLA-B*40:02-negative patients, 6pLOH was detected in 18 (12%) patients. HLA alleles that were more likely to be possessed by 6pLOH(+) patients were HLA-DRB1*15:01 (23% [9 of 40], OR = 3.3, P = .02), C*01:02 (24% [8 of 34], OR = 3.3, P = .03), and B*54:01 (29% [5 of 17], OR = 3.8, P = .03). HLA-B*54:01 was missing in all 5 6pLOH(+) patients carrying this allele, whereas 7 of 9 HLA-DRB1*15:01 and 2 of 8 C*01:02 in the 6pLOH(+) patients were in the retained haplotype. Three HLA alleles (HLA-A*02:01, A*02:06, and A*31:01) that had been suggested to be frequently lost due to 6pLOH by our previous study9 were not significantly associated with 6pLOH in this study cohort; each allele was found at similar frequencies between 6pLOH(+) and 6pLOH(−) patients (HLA-A*02:01, 20% [11 of 54], OR = 1.1, P = .84; A*02:06, 13% [7 of 52], OR = 0.59, P = .32; and A*31:01, 16% [8 of 51], OR = 0.73, P = .55). The trend was consistent in HLA-B*40:02-negative patients (HLA-A*02:01, 11% [4 of 35], OR = 0.93, P = 1.0; A*02:06, 14% [5 of 37], OR = 1.2, P = .77; and A*31:01, 7.5% [3 of 38], OR = 0.55, P = .56).
B4002− granulocytes in patients with HLA-B*40:02
Peripheral blood samples were available for FCM using the anti-HLA-B61 antibody in 28 patients with AA carrying HLA-B*40:02, including 12 6pLOH(+) and 16 6pLOH(−) patients. B4002− granulocytes that accounted for 24% to 99% of the total granulocytes were detected in all of the 12 6pLOH(+) patients. Unexpectedly, 1.0% to 99% B4002− granulocytes were also detected in 9 (56%) of the 16 6pLOH(−) patients. None of the 12 individuals carrying HLA-B*40:02 (6 healthy individuals and 6 patients with hematological malignancies) were positive for B4002− granulocytes. The 21 patients possessing B4002− granulocytes had more severe disease (67%, 14 of 21), whereas only 1 of the 7 patients (14%, P = .03) not possessing B4002− granulocytes had severe disease.
Because 10 of the 12 6pLOH(+) patients were heterozygous for the HLA-A allele, the HLA-A allelic expression by their B4002− granulocytes was able to be evaluated. As is shown in Figure 1 and Table 2, 8.0% to 99% of granulocytes of all of the 10 6pLOH(+) patients lacked both HLA-B4002 and the HLA-A allele (B4002−A−) on the same haplotype containing HLA-B*40:02 and were therefore thought to represent 6pLOH(+) granulocytes. In contrast, 8 of the 10 patients also had 1.0% to 78% granulocytes that lacked HLA-B4002 but retained the HLA-A allele (B4002−A+), indicating that these B4002− cells lacked HLA-B4002 via mechanisms other than 6pLOH.
Detection of HLA-B4002-lacking granulocytes using an anti-HLA-B61 mAb in AA patients with HLA-B*40:02. (A) Representative dot plots of 3 patients with HLA-A2, A24, and B4002 are shown. (i) Case 22, which had only wild-type, B4002+A24+A2+ cells. (ii) Case 12, which showed B4002− cells that lacked HLA-B4002 but retained both HLA-A alleles (A24 and A2), designated as B4002−A24+A2+, in addition to 6pLOH(+) cells lacking both HLA-B4002 and A24 on the same haplotype. (iii) Case 7, which showed B4002−A24+A2+ cells but did not show either B4002−A24− or B4002−A2− (6pLOH[+]) cells. (B) Four different patterns of HLA-B4002-lacking cell status and their proportions in 28 AA patients with HLA-B*40:02. FITC, fluorescein isothiocyanate; PE, phycoerythrin.
Detection of HLA-B4002-lacking granulocytes using an anti-HLA-B61 mAb in AA patients with HLA-B*40:02. (A) Representative dot plots of 3 patients with HLA-A2, A24, and B4002 are shown. (i) Case 22, which had only wild-type, B4002+A24+A2+ cells. (ii) Case 12, which showed B4002− cells that lacked HLA-B4002 but retained both HLA-A alleles (A24 and A2), designated as B4002−A24+A2+, in addition to 6pLOH(+) cells lacking both HLA-B4002 and A24 on the same haplotype. (iii) Case 7, which showed B4002−A24+A2+ cells but did not show either B4002−A24− or B4002−A2− (6pLOH[+]) cells. (B) Four different patterns of HLA-B4002-lacking cell status and their proportions in 28 AA patients with HLA-B*40:02. FITC, fluorescein isothiocyanate; PE, phycoerythrin.
Clinical characteristics of 28 patients tested using anti-HLA-B61 antibody
| Case . | Age, years . | Sex . | Severity . | Before/after IST . | Years after IST . | IST . | Response to IST . | % of B4002−granulocytes . | Phenotype of B4002−granulocytes . | |
|---|---|---|---|---|---|---|---|---|---|---|
| 6pLOH (%) . | B4002−A+ (%) . | |||||||||
| 1 | 68 | M | Nonsevere | After | 17 | CsA | CR | 24 | 0 | 6pLOH |
| 2 | 59 | M | Severe | After | 6.2 | ATG/CsA | CR | 99 | 0 | 6pLOH |
| 3 | 41 | M | Severe | After | 17 | ATG/CsA | CR | 0 | 60 | B4002−A+ |
| 4 | 72 | M | Very severe | After | 2.3 | ATG/CsA | PR | 0 | 2 | B4002−A+ |
| 5 | 30 | M | Severe | After | 1.9 | ATG/CsA | PR | 0 | 8 | B4002−A+ |
| 6 | 87 | F | Severe | After | 23 | ATG/CsA | CR | 0 | 10 | B4002−A+ |
| 7 | 15 | M | Severe | After | 3.7 | ATG/CsA | PR | 0 | 53 | B4002−A+ |
| 8 | 57 | F | Severe | After | 2.5 | ATG/CsA | PR | 0 | 1 | B4002−A+ |
| 9 | 54 | F | Nonsevere | After | 14 | CsA | PR | 0 | 99 | B4002−A+ |
| 10 | 56 | F | Nonsevere | After | 0.4 | CsA | NR | 0 | 70 | B4002−A+ |
| 11 | 57 | M | Severe | After | 1.9 | CsA | PR | 0 | 3 | B4002−A+ |
| 12 | 74 | F | Nonsevere | After | 25 | CsA | CR | 13 | 15 | B4002−A+ and 6pLOH |
| 13 | 69 | F | Nonsevere | After | 6.4 | CsA | PR | 97 | 3 | B4002−A+ and 6pLOH |
| 14 | 49 | F | Severe | After | 0.6 | ATG/CsA | PR | 43 | 1 | B4002−A+ and 6pLOH |
| 15 | 79 | F | Nonsevere | After | 1.5 | CsA | CR | 17 | 17 | B4002−A+ and 6pLOH |
| 16 | 21 | F | Very severe | Before | NA | ATG/CsA | CR | 8 | 75 | B4002−A+ and 6pLOH |
| 17 | 38 | M | Nonsevere | After | 2.2 | CsA | PR | 36 | 58 | B4002−A+ and 6pLOH |
| 18 | 81 | F | Severe | Before | NA | No IST | NA | 67 | 33 | B4002−A+ and 6pLOH |
| 19 | 28 | M | Very severe | After | 14 | ATG/CsA | CR | 39 | 40 | B4002−A+ and 6pLOH |
| 20 | 32 | M | Very severe | Before | NA | ATG/CsA | PR | 21 | 52 | B4002−A+ and 6pLOH |
| 21 | 81 | M | Severe | Before | NA | CsA | NR | 19 | 25 | B4002−A+ and 6pLOH |
| 22 | 66 | F | Nonsevere | After | 8.1 | ATG/CsA | CR | 0 | 0 | None |
| 23 | 68 | M | Nonsevere | After | 1.7 | CsA | CR | 0 | 0 | None |
| 24 | 25 | M | Nonsevere | After | 0.6 | ATG/CsA | NR | 0 | 0 | None |
| 25 | 39 | M | Severe | After | 2.8 | ATG/CsA | CR | 0 | 0 | None |
| 26 | 71 | F | Nonsevere | After | 1.4 | ATG/CsA | PR | 0 | 0 | None |
| 27 | 24 | F | Nonsevere | After | 2.2 | CsA | NR | 0 | 0 | None |
| 28 | 56 | M | Nonsevere | Before | NA | No IST | NA | 0 | 0 | None |
| Case . | Age, years . | Sex . | Severity . | Before/after IST . | Years after IST . | IST . | Response to IST . | % of B4002−granulocytes . | Phenotype of B4002−granulocytes . | |
|---|---|---|---|---|---|---|---|---|---|---|
| 6pLOH (%) . | B4002−A+ (%) . | |||||||||
| 1 | 68 | M | Nonsevere | After | 17 | CsA | CR | 24 | 0 | 6pLOH |
| 2 | 59 | M | Severe | After | 6.2 | ATG/CsA | CR | 99 | 0 | 6pLOH |
| 3 | 41 | M | Severe | After | 17 | ATG/CsA | CR | 0 | 60 | B4002−A+ |
| 4 | 72 | M | Very severe | After | 2.3 | ATG/CsA | PR | 0 | 2 | B4002−A+ |
| 5 | 30 | M | Severe | After | 1.9 | ATG/CsA | PR | 0 | 8 | B4002−A+ |
| 6 | 87 | F | Severe | After | 23 | ATG/CsA | CR | 0 | 10 | B4002−A+ |
| 7 | 15 | M | Severe | After | 3.7 | ATG/CsA | PR | 0 | 53 | B4002−A+ |
| 8 | 57 | F | Severe | After | 2.5 | ATG/CsA | PR | 0 | 1 | B4002−A+ |
| 9 | 54 | F | Nonsevere | After | 14 | CsA | PR | 0 | 99 | B4002−A+ |
| 10 | 56 | F | Nonsevere | After | 0.4 | CsA | NR | 0 | 70 | B4002−A+ |
| 11 | 57 | M | Severe | After | 1.9 | CsA | PR | 0 | 3 | B4002−A+ |
| 12 | 74 | F | Nonsevere | After | 25 | CsA | CR | 13 | 15 | B4002−A+ and 6pLOH |
| 13 | 69 | F | Nonsevere | After | 6.4 | CsA | PR | 97 | 3 | B4002−A+ and 6pLOH |
| 14 | 49 | F | Severe | After | 0.6 | ATG/CsA | PR | 43 | 1 | B4002−A+ and 6pLOH |
| 15 | 79 | F | Nonsevere | After | 1.5 | CsA | CR | 17 | 17 | B4002−A+ and 6pLOH |
| 16 | 21 | F | Very severe | Before | NA | ATG/CsA | CR | 8 | 75 | B4002−A+ and 6pLOH |
| 17 | 38 | M | Nonsevere | After | 2.2 | CsA | PR | 36 | 58 | B4002−A+ and 6pLOH |
| 18 | 81 | F | Severe | Before | NA | No IST | NA | 67 | 33 | B4002−A+ and 6pLOH |
| 19 | 28 | M | Very severe | After | 14 | ATG/CsA | CR | 39 | 40 | B4002−A+ and 6pLOH |
| 20 | 32 | M | Very severe | Before | NA | ATG/CsA | PR | 21 | 52 | B4002−A+ and 6pLOH |
| 21 | 81 | M | Severe | Before | NA | CsA | NR | 19 | 25 | B4002−A+ and 6pLOH |
| 22 | 66 | F | Nonsevere | After | 8.1 | ATG/CsA | CR | 0 | 0 | None |
| 23 | 68 | M | Nonsevere | After | 1.7 | CsA | CR | 0 | 0 | None |
| 24 | 25 | M | Nonsevere | After | 0.6 | ATG/CsA | NR | 0 | 0 | None |
| 25 | 39 | M | Severe | After | 2.8 | ATG/CsA | CR | 0 | 0 | None |
| 26 | 71 | F | Nonsevere | After | 1.4 | ATG/CsA | PR | 0 | 0 | None |
| 27 | 24 | F | Nonsevere | After | 2.2 | CsA | NR | 0 | 0 | None |
| 28 | 56 | M | Nonsevere | Before | NA | No IST | NA | 0 | 0 | None |
B4002− granulocytes are granulocytes that lost HLA-B4002 expression as a result of 6pLOH or HLA-B*40:02 mutations. B4002−A+ granulocytes are B4002− granulocytes that retain the HLA-A allele on the same haplotype.
CR, complete response; F, female; M, male; NA, not applicable; NR, no response; PR, partial response.
Two 6pLOH(+) patients (cases 16 and 18) were homozygous for the HLA-A allele, and the percentage of 6pLOH(+) granulocytes was estimated by using ddPCR. Only 9% and 66% of the sorted B4002− granulocytes were 6pLOH(+), suggesting that these patients also had B4002−A+ granulocytes that accounted for 91% and 34% of B4002− granulocytes. None of the 20 (10 6pLOH[+] and 10 6pLOH[−]) patients with HLA-B*40:02 had granulocytes that lacked an HLA-A allele but retained HLA-B4002. The prevalence of missing HLA-B4002 from granulocytes in the 28 patients was 75% (21 of 28, 9 with B4002−A+ cells alone, 10 with both B4002−A+ and B4002−A− [6pLOH] cells, and 2 with B4002−A− cells alone; Figure 1).
Mutations of HLA-B alleles in B4002−A+ granulocytes
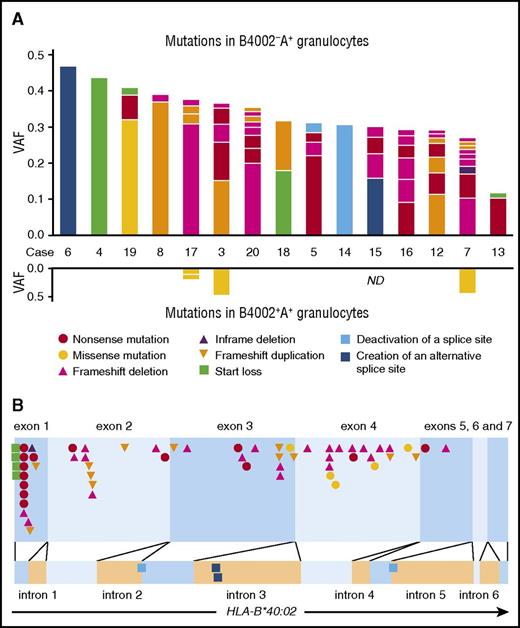
B4002−A+ granulocytes were available for mutation analyses of HLA-B alleles in 15 of the 19 patients who possessed the aberrant granulocytes. The mean coverage of the HLA-B gene was 426× for the capture method and 32 077× for the PCR method. Somatic mutations were judged to be present when the frequency exceeded 0.01. In total, 59 different somatic mutations of HLA-B were identified in the sorted B4002−A+ granulocytes, all of which were present in HLA-B*40:02 and not in any of the other HLA-B alleles. The median variant allele frequency (VAF) was 3.7% (range, 1.0% to 47%), and the number of mutations in each patient ranged from 1 to 9 (median, 4; Figure 2; supplemental Table 4).
Somatic mutations of HLA-B*40:02 in 15 patients with AA. (A) The kinds and VAFs of somatic mutation in individual patients identified in B4002−A+ (upper graph) and B4002+A+ (lower graph) granulocytes are shown. The kinds of mutations are designated in different colors. (B) The positions of all somatic mutations in the HLA-B*40:02 region are shown. The color and shape of each symbol correspond to those of panel A.
Somatic mutations of HLA-B*40:02 in 15 patients with AA. (A) The kinds and VAFs of somatic mutation in individual patients identified in B4002−A+ (upper graph) and B4002+A+ (lower graph) granulocytes are shown. The kinds of mutations are designated in different colors. (B) The positions of all somatic mutations in the HLA-B*40:02 region are shown. The color and shape of each symbol correspond to those of panel A.
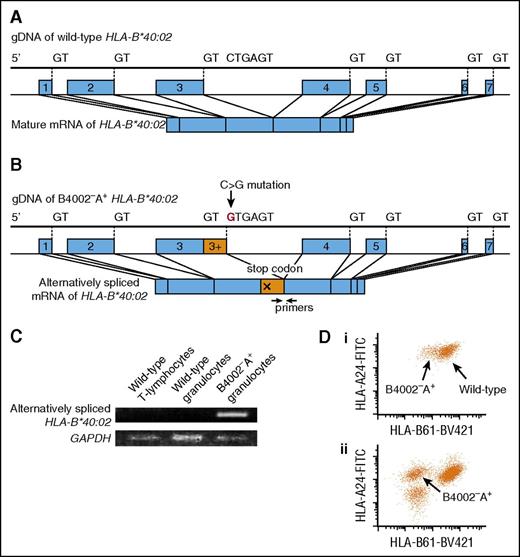
Fifty-five mutations were exonic, and 4 were intronic. The exonic mutations were frameshift duplications (n = 12), frameshift deletions (n = 23), an in-frame deletion (n = 1), nonsense mutations (n = 14), a missense mutation (n = 1), and start losses (n = 4). All exonic mutations were expected to result in a lack of HLA-B4002 expression, except for 1 missense mutation in case 19, which could potentially lead to a structural change in the epitope recognized by anti-HLA-B61 mAb. All 4 intronic mutations were considered to be splice site mutations; 2 mutations deactivated 5′ or 3′ splice sites, whereas the other 2 were single-base substitutions within intron 3, creating alternative 5′ splicing sites with a strong consensus sequence, GTGAGT: TTCCTGAGT> TTCGTGAGT (c.619+133C>G) in case 6 and GGCATGAGT> GGCGTGAGT (c.619+123A>G) in case 15. The presence of alternative splice sites potentially inhibits normal splicing, leading to a decreased expression of HLA-B*40:02. The presence of alternatively spliced messenger RNA (mRNA) solely in B4002−A+ granulocytes was confirmed in case 6, whose cDNA of both sorted B4002−A+ and B4002+ granulocytes was available using a PCR specific to alternatively spliced transcripts (Figure 3).
A mutation creating an alternative splice site in case 6. (A) Normal splicing of HLA-B*40:02. (B) The C>G mutation in intron 3 created an alternative 5′ splicing site with a strong consensus sequence, GTGAGT, which can change the splicing of HLA-B*40:02, leading to a stop codon formation. (C) mRNA from 3 different leukocyte subsets was reverse-transcribed and amplified with alternatively spliced mRNA-specific primers. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was amplified as an internal control. (D) The expression level of HLA-B4002 by B4002−A+ granulocytes of case 6 (i) was higher than that by B4002−A+ granulocytes of case 12 (ii), which lost HLA-B4002 because of nonsense and frameshift mutations. gDNA, genomic DNA.
A mutation creating an alternative splice site in case 6. (A) Normal splicing of HLA-B*40:02. (B) The C>G mutation in intron 3 created an alternative 5′ splicing site with a strong consensus sequence, GTGAGT, which can change the splicing of HLA-B*40:02, leading to a stop codon formation. (C) mRNA from 3 different leukocyte subsets was reverse-transcribed and amplified with alternatively spliced mRNA-specific primers. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was amplified as an internal control. (D) The expression level of HLA-B4002 by B4002−A+ granulocytes of case 6 (i) was higher than that by B4002−A+ granulocytes of case 12 (ii), which lost HLA-B4002 because of nonsense and frameshift mutations. gDNA, genomic DNA.
Missense mutations in the HLA-B*40:02 allele of B4002+ granulocytes
B4002+ granulocytes were subjected to deep sequencing of HLA-B as a negative control in 14 of the 15 patients possessing B4002−A+ granulocytes. Surprisingly, 4 different missense mutations of HLA-B*40:02, which are not involved in the antigen presentation by this class I allele, were detected in B4002+ granulocytes from 3 (cases 3, 7, and 17) of the 14 patients (Figure 2). The same mutations were not detected in the corresponding B4002−A+ granulocytes of all 3 patients, B4002+ T cells of 2 patients (cases 7 and 17), or the reference sequence of HLA-B*40:02. The VAFs of the 4 missense mutations were 9.5% (case 17), 9.6% (case 17), 43% (case 7), and 47% (case 3), respectively. The 4 missense mutations in B4002+ cells and 1 missense mutation in B4002−A+ cells occurred close together at an α-3 domain region in the protein 3-dimensional structure (supplemental Figure 3), suggesting the inability of the mutant HSPCs to interact with CTLs.
The presence of mutations in the α-3 domain region of HLA-B*40:02 in B4002+ granulocytes of 3 patients possessing B4002−A+ granulocytes prompted us to study 5 AA patients with HLA-B*40:02 who were negative for B4002−A+ granulocytes. The B4002+ granulocytes from 4 patients (cases 23, 24, 25, and 26) without B4002− granulocytes and 1 patient (case 1) with 6pLOH(+) granulocytes were subjected to deep sequencing, with T cells as a negative control. No mutations were found in the 5 patients’ samples.
Response to IST in patients carrying HLA-B*40:02
Of the 28 patients carrying HLA-B*40:02, 15 patients were treated with ATG plus CsA therapy before (n = 13) or after (n = 2) sampling for the study (Table 2). All 11 (100%) patients possessing B4002− granulocytes showed a good response (complete response [CR] in 5 and partial response [PR] in 6), while 3 of the 4 (75%) patients not possessing B4002− granulocytes responded (CR in 2 and PR in 1). Eleven patients were treated with CsA monotherapy before (n = 10) and after (n = 1) sampling; 7 of the 9 patients possessing B4002− granulocytes showed a response (CR in 3 and PR in 4); 1 of 2 patients not possessing B4002− granulocytes showed a response (CR). Two patients (one with B4002− cells and the other without B4002− cells) were not treated with IST.
Chronological analysis of B4002− granulocytes
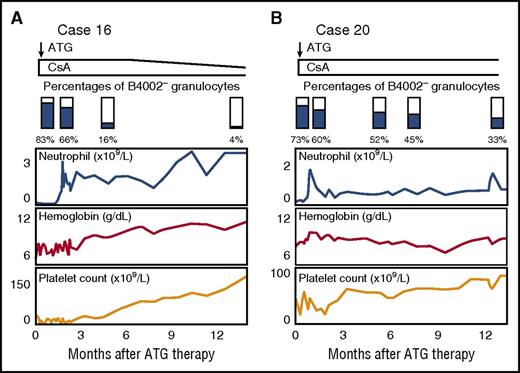
The percentages of B4002− granulocytes in 2 patients (cases 16 and 20) were determined several times after ATG/CsA therapy. Both patients responded to ATG therapy, and their hematological recovery was associated with a decrease in the B4002− granulocyte percentage (83% to 4.3% and 73% to 33% after 13 and 12 months of the therapy, respectively; Figure 4). In contrast, the B4002− granulocyte percentages of 4 patients (cases 2, 3, 6, and 19) whose first samples were tested after the recovery of hematopoiesis following 6.2 to 23 years of ATG/CsA therapy did not change markedly when their second samples were examined 6 to 10 months (median, 9 months) after the first examination: 99% to 98%; 60% to 59%; 10% to 10%; and 79% to 85%, respectively.
Chronological analysis of B4002−granulocytes. The percentages of B4002− granulocytes in case 16 (A) and case 20 (B) were determined several times after ATG/CsA therapy. Both patients responded to the therapy, and their hematological recovery was associated with a decrease in the B4002− granulocyte percentage.
Chronological analysis of B4002−granulocytes. The percentages of B4002− granulocytes in case 16 (A) and case 20 (B) were determined several times after ATG/CsA therapy. Both patients responded to the therapy, and their hematological recovery was associated with a decrease in the B4002− granulocyte percentage.
Discussion
The current study in a large number of AA patients using a novel ddPCR, similar to our previous studies, detected 6pLOH in 19% of the patients and confirmed a significantly higher frequency of lacking HLA-B*40:02 than of lacking other HLA class I alleles, including HLA- A*02:01, A*02:06, and A*31:01. Furthermore, a new mAb specific to HLA-B61 revealed the presence of B4002− granulocytes in 75% of patients carrying HLA-B*40:02. These findings suggest that HLA-B*40:02 is critically involved in the autoantigen presentation to T cells in patients with AA. The prevalence of HLA-LLs may have been underestimated because of a lack of mAbs specific to many other HLA alleles.
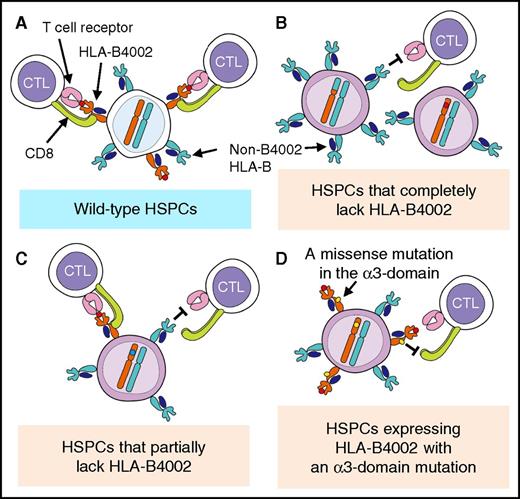
6pLOH has been considered to be the sole or a major method of lacking class I HLAs in HSPCs targeted by CTLs of AA9,10,12 and in acute myeloid leukemia cells relapsed after HLA-mismatched hematopoietic stem cell transplantation,27,28 although some reports have shown a possible lack of HLA alleles due to structural gene mutations.15,29,30 Our deep sequencing in the present study of HLA class I genes derived from sorted B4002−A+ granulocytes revealed various mutations solely in the HLA-B*40:02 gene. The HLA-B*40:02 mutations are apparently somatic because B4002+A+ leukocytes were present in all patients possessing B4002− granulocytes, and neither the B4002+A+ granulocytes nor B4002+ T lymphocytes from each individual showed the same mutations. The high number of mutations per patient with B4002− granulocytes (median, 4) suggests that HSPC clones with various mutations arose in the bone marrow of the patients prior to the AA development and might have been selected to contribute to hematopoiesis under strong pressure by CTLs specific to autoantigens presented by HLA-B4002 at the onset of AA (Figure 5).
Deduced mechanisms responsible for the escape of HSPCs from cytotoxic T cells specific for autoantigens presented by HLA-B4002. Normal HSPCs are killed by CTLs that recognize autoantigens presented by HLA-B4002 (A), but HSPCs that have undergone various mutations of HLA-B4002 escape this CTL attack in different ways, such as via the complete loss of HLA-B4002 expression due to 6pLOH, nonsense and frameshift mutations, or deactivation of splice sites (B), partial loss of HLA-B4002 protein due to alternative splice site formation (C), and failure of CD8+ T-cell binding to the α-3 domain of HLA-B4002 (D).
Deduced mechanisms responsible for the escape of HSPCs from cytotoxic T cells specific for autoantigens presented by HLA-B4002. Normal HSPCs are killed by CTLs that recognize autoantigens presented by HLA-B4002 (A), but HSPCs that have undergone various mutations of HLA-B4002 escape this CTL attack in different ways, such as via the complete loss of HLA-B4002 expression due to 6pLOH, nonsense and frameshift mutations, or deactivation of splice sites (B), partial loss of HLA-B4002 protein due to alternative splice site formation (C), and failure of CD8+ T-cell binding to the α-3 domain of HLA-B4002 (D).
Recent studies using NGS have revealed clonal hematopoiesis with various somatic mutations in the granulocytes of AA patients, but they failed to show somatic mutations in HLA genes except for 6pLOH, which was detected in 13% of patients.10,31 This is mainly due to insufficient depth of the previous analyses, which did not focus on HLA gene regions. The somatic mutations in HLA genes do not imply a propensity toward myelodysplastic syndrome but may instead represent the selection by immune pressure, such as PIGA mutations. The decrease in the B4002− granulocyte percentage associated with successful ATG therapy in 2 patients and the higher prevalence of B4002− granulocytes in severe AA than in nonsevere AA patients support this hypothesis.
One unexpected and important finding of this study is that B4002+ granulocytes from 3 patients with B4002− granulocytes also had missense mutations in the limited α-3 domain region of HLA-B*40:02, which is not involved in the antigen presentation by this class I allele. CTLs require binding of their CD8 molecules to the α-3 domain of HLA class I to exert their function.32 The single amino acid change in the α-3 domain observed in the 3 patients may have made the mutant HSPCs resistant to the attack by CTLs.33,34 These findings also suggest that detecting HLA protein expression with specific mAbs may not be enough to screen HSPCs or leukemic cells that evade CTL attack; deep sequencing of HLA genes including the α-3 domain coding region is necessary.
The allelic frequency of HLA-B*40:02 in healthy individuals is reported to be higher in Asian populations, with rates of 7.9% in Japanese, 8.7% in South Korean, 2.0% in Chinese, 2.4% in Thai, and 7.7% in American Filipino populations, in comparison with rates of 1.6% in German, 0.3% in Northern Irish, 1.8% in Italian, 0.9% in Swiss, and 1.5% in U.S. Caucasian populations.35 The higher frequency of this class I allele may partly explain the higher incidence of AA in Asian countries than in Western countries. Although the incidence of AA in the United States is much lower than in Japan, a study of 11 American AA patients with somatic HLA loss by Babushok et al36 also revealed that the most frequently affected allele was HLA-B*40:02, suggesting that an autoantigen presented by this HLA-B is commonly involved in the development of AA in Japan and the United States.
The markedly high prevalence of allele-lacking granulocytes in AA patients carrying HLA-B*40:02 suggests that antigens presented by HLA-B4002 may serve as autoantigens eliciting T-cell responses to HSPCs. We previously isolated a T-cell clone from a 6pLOH(+) patient that was capable of inhibiting HSPCs in a HLA-B*40:02-restricted manner but failed to identify target antigens on HSPCs because of the inability to propagate the T-cell clone.14 Current technology has enabled the preparation of T-cell receptor vectors and their transfectants by taking advantage of predominantly expanding T-cell clones in patients’ bone marrow in response to unknown antigens.37 Studying T cells from AA patients possessing B4002− granulocytes is therefore warranted to identify autoantigens of AA.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and donors and their physicians, including T. Endo and K. Kahata of Hokkaido University; M. Hirao and M. Kida of NTT Medical Center of Tokyo; H. Hiramatsu of Kyoto University; H. Taniguchi of Sasebo City General Hospital; M. Yamazaki of Keiju Medical Center; C. Sugimori of Ishikawa Prefectural Central Hospital; and M. Tanabe, K. Ohata, K. Ishiyama, Y. Kondo, and H. Yamazaki of Kanazawa University for contributing to this study and Advanced Preventive Medical Sciences Research Center, Kanazawa University, for the use of their facilities.
Y.Z. is a doctoral candidate at Kanazawa University, and this work is submitted in partial fulfillment of the requirements for the PhD.
This work was supported in part by Japan Society for Promotion of Science KAKENHI (Grants-in-Aid for Scientific Research) (Grants JP16H05335 and JP16H06502) and by the Platform Project for Supporting Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Japan Agency for Medical Research and Development.
Authorship
Contribution: Y.Z., N.N., T.I., H.M., and T.K. collected clinical data and blood samples; K.K. performed HLA genotyping; Y.Z. performed ddPCR; Y.Z. and T.I. performed cell sorting; K.H. and A.T. performed deep sequencing of HLA-B; Y.Z., H.T., T.O., H.K., and A.M. generated an anti-HLA-B61 mAb; Y.Z., H.T., and S.N. designed the research and wrote the manuscript; and all authors critically reviewed the manuscript and checked the final version of it.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shinji Nakao, 13-1 Takaramachi, Kanazawa, Ishikawa 920-8640 Japan; e-mail: snakao8205@staff.kanazawa-u.ac.jp.
References
Author notes
Y.Z., H.T., and K.H. contributed equally to this study.

![Figure 1. Detection of HLA-B4002-lacking granulocytes using an anti-HLA-B61 mAb in AA patients with HLA-B*40:02. (A) Representative dot plots of 3 patients with HLA-A2, A24, and B4002 are shown. (i) Case 22, which had only wild-type, B4002+A24+A2+ cells. (ii) Case 12, which showed B4002− cells that lacked HLA-B4002 but retained both HLA-A alleles (A24 and A2), designated as B4002−A24+A2+, in addition to 6pLOH(+) cells lacking both HLA-B4002 and A24 on the same haplotype. (iii) Case 7, which showed B4002−A24+A2+ cells but did not show either B4002−A24− or B4002−A2− (6pLOH[+]) cells. (B) Four different patterns of HLA-B4002-lacking cell status and their proportions in 28 AA patients with HLA-B*40:02. FITC, fluorescein isothiocyanate; PE, phycoerythrin.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/21/10.1182_blood-2016-11-752378/4/m_blood752378f1.jpeg?Expires=1769132028&Signature=dcKqJL4IAvvWq8jBHiXao7fBbYA49BcwKTMS-a56OzSrzKVoOZ93LT7SujG2LwJnjySjpMgZKOJcjfI6wlz-II18e-y0cjIv3WszcsGCTosNfk4gu9QNWIp2tAFMazJbZ-9M3LRLFkLqXMn68lqpptDwx2DejWjV~JiS-uJLqFH9lk9HoitnVOl2u0Jb7I8j5xy1nheRuPmoonWcz4PDnj2DjKIP63Vy693~RngK5x1CT~UDrJIkBOs476TvJSIubmk2N7ntRsHNWORo52-Dah76~vxbu6e5g4~elFcokLbdRFX2Fdrl4gXHLtoWq9cTJBv~1wWSRkh8g~N2t6SgDA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)