Key Points
Mass cytometry reveals a Treg immune signature for AA and for response to antithymocyte globulin.
AA Tregs in vitro are expandable, stable, and functional, with potential for future therapeutic options.
Abstract
Idiopathic aplastic anemia (AA) is an immune-mediated and serious form of bone marrow failure. Akin to other autoimmune diseases, we have previously shown that in AA regulatory T cells (Tregs) are reduced in number and function. The aim of this study was to further characterize Treg subpopulations in AA and investigate the potential correlation between specific Treg subsets and response to immunosuppressive therapy (IST) as well as their in vitro expandability for potential clinical use. Using mass cytometry and an unbiased multidimensional analytical approach, we identified 2 specific human Treg subpopulations (Treg A and Treg B) with distinct phenotypes, gene expression, expandability, and function. Treg B predominates in IST responder patients, has a memory/activated phenotype (with higher expression of CD95, CCR4, and CD45RO within FOXP3hi, CD127lo Tregs), expresses the interleukin-2 (IL-2)/STAT5 pathway and cell-cycle commitment genes. Furthermore, in vitro–expanded Tregs become functional and take on the characteristics of Treg B. Collectively, this study identifies human Treg subpopulations that can be used as predictive biomarkers for response to IST in AA and potentially other autoimmune diseases. We also show that Tregs from AA patients are IL-2–sensitive and expandable in vitro, suggesting novel therapeutic approaches such as low-dose IL-2 therapy and/or expanded autologous Tregs and meriting further exploration.
Introduction
Treatment options for idiopathic aplastic anemia (AA) include allogeneic hematopoietic stem cell transplantation or immunosuppressive therapy (IST), and recently eltrombopag for refractory severe AA.1-3 Some patients are ineligible for hematopoietic stem cell transplantation because of older age or lack of a suitable donor, and after IST, one-third of patients fail to respond and 35% relapse after responding. Up to 20% of patients transform to myelodysplastic syndrome or acute myeloid leukemia after IST.3-7 Additional novel therapeutic approaches are needed for AA patients who fail to respond to IST, and along with this, more robust diagnostic tests that predict response to IST.
We, and others, have shown a reduction in the number and function of regulatory T cells (Tregs) in AA.8-11 Tregs from AA patients also secrete pro-inflammatory cytokines.8 Correlation between number and function of Tregs and response to standard IST has not been fully investigated, and it is unclear whether the dominant Treg subpopulation in AA is more of a T conventional (Tcon) subtype or are genuinely functional Tregs.
The aims of this study were to (1) identify an immune signature for AA compared with healthy individuals, based on Treg subpopulations and Tcon; (2) identify the immune signature that predicts response to IST at time of diagnosis of AA; and (3) examine the expandability and characteristics of Treg subpopulations that may form the basis for a novel therapeutic approach to AA patients who are refractory to IST. For this, we used a novel deep-phenotyping strategy using multiparameter mass cytometry (known as cytometry by time-of-flight [CyTOF]) and automated clustering method including t-distributed stochastic neighbor embedding (t-SNE)12 to use SNE visually (viSNE) as a means of identifying cell populations13 in combination with spanning-tree progression analysis of density-normalized events (SPADE).14 This enabled us to identify 2 distinct subpopulations of human Tregs in AA and healthy age-matched donors (HDs) and to demonstrate clear differences in AA that predicted response to IST. We also showed that AA Tregs can be expanded in vitro and that they are stable and functional.
Methods
Patients and healthy donors
Thirty-nine AA patients at diagnosis and 31 HDs were recruited (Table 1). Peripheral blood mononuclear cells (PBMCs) from 16 patients (12 IST responders and 4 nonresponders) were used for initial CyTOF analysis; samples from another 15 patients (11 IST responders and 4 nonresponders) were used as validation cohort. PBMCs from an additional 8 patients were used for functional assays in addition to 4 patients from the initial cohort. Response to IST was evaluated at 6 months posttherapy. Median age was 45 years (range, 20-72 years). IST-eligible AA patients were randomly invited to participate in this study before commencement of therapy. All patients <35 years of age were screened for Fanconi anemia.
Patients’ characteristics
| Characteristic . | Value . |
|---|---|
| Number of patients | 39 |
| Median age, y (range) | 45 (20-72) |
| Sex | |
| M | 19 |
| F | 20 |
| Disease severity at diagnosis | |
| VSAA | 11 |
| SAA | 16 |
| NSAA | 12 |
| PNH clone at the time of study | |
| Yes | 28 |
| No | 11 |
| Size of PNH clone, % (range) | |
| Red cells | 0.402 (0-87.3) |
| Granulocytes | 3.86 (0-96.7) |
| Monocytes | 4.517 (0-91.8) |
| Etiology | |
| Idiopathic | 39 |
| Response to treatment | |
| CR | 6 |
| PR | 19 |
| NR | 14 |
| Characteristic . | Value . |
|---|---|
| Number of patients | 39 |
| Median age, y (range) | 45 (20-72) |
| Sex | |
| M | 19 |
| F | 20 |
| Disease severity at diagnosis | |
| VSAA | 11 |
| SAA | 16 |
| NSAA | 12 |
| PNH clone at the time of study | |
| Yes | 28 |
| No | 11 |
| Size of PNH clone, % (range) | |
| Red cells | 0.402 (0-87.3) |
| Granulocytes | 3.86 (0-96.7) |
| Monocytes | 4.517 (0-91.8) |
| Etiology | |
| Idiopathic | 39 |
| Response to treatment | |
| CR | 6 |
| PR | 19 |
| NR | 14 |
Thirty-nine AA patients were recruited in this study, and PB samples were used for mass cytometry and/or in vitro experiments. AA patients who were eligible for IST were randomly selected and invited to participate in this study. A small to moderate PNH clone was detected in 28 patients, with no cases of hemolytic PNH. PBMCs from 16 patients (12 IST responders and 4 nonresponders) were used for initial CyTOF analysis; samples from another 15 patients (11 IST responders and 4 nonresponders) were used as a validation cohort. PBMCs from additional 8 patients were used for functional assays. Among nonresponder patients (n = 14), 7 underwent transplant later and the remaining were treated with cyclosporine A and supportive care.
CR, complete response; F, female; M, male; NR, non-response; NSAA, nonsevere aplastic anemia; PNH, paroxysmal nocturnal hemoglobinuria; PR: partial response; VSAA: very severe AA.
Antibodies and cell staining
We designed a panel of antibodies based on surface markers, transcription factors, and cytokines (see supplemental Table 1, available on the Blood Web site). Each antibody was tagged with a rare metal isotope and its function verified by conventional flow cytometry before mass cytometry (supplemental Methods). The CyTOF-2 mass cytometer (Fluidigm) was used for data acquisition. Acquired data were normalized based on normalization beads (Ce 140, Eu151, Eu153, Ho165, and Lu175).15 Automated clustering was performed on a subset of 800 000 cells sampled from all individuals. The number of cells sampled from each individual was proportional to the total number of cells in that sample. Collected cells were stained with metal-conjugated antibodies with or without a 4-hour stimulation with phorbol myristate acetate (PMA) and ionomycin in the presence of brefeldin. Intracellular staining for transcription factors and cytokines was performed after fixation and permeabilization according to the manufacturer's instructions (eBioscience). Conventional flow cytometry data were analyzed by polychromatic flow cytometer (LSRFortessa, BD Biosciences).
Data processing, scale transformation, automated clustering, and distance computations
Data were initially processed and analyzed using Cytobank.16 The stand-alone analysis tool cyt was also used for performing t-SNE dimensionality reduction and merging distinct FCS files.13 We analyzed mass cytometry complex data using viSNE12 to visually identify delineated subpopulations13 in combination with SPADE14 and heat maps15 to distinguish CD4+ T-cell subpopulations, in particular Tregs15 (see supplemental Methods).
Treg expansion
Freshly isolated live Tregs were stimulated with anti-CD3/CD28 beads (1:1 ratio) (Dynabeads human T-activator CD3/CD28, Life Technologies) and high-dose (1000 IU/mL) interleukin-2 (IL-2)(Proleukin, Novartis) for 4 weeks with all-trans-retinoic acid 2 μM (Sigma Aldrich) and rapamycin 100 nM (Rapa; Alfa-Aesar). Culture medium (XV Prime, Irvine Scientific, supplemented with AB serum 10%) and beads were replenished every week. After 4 weeks of expansion, cells were rested with decreasing doses of IL-2 for 5 days.17
DNA methylation analysis by deep amplicon bisulfite sequencing
DNA isolation method, primer sequences, and cycling conditions are shown in supplemental Figure MS1 and the supplemental Methods.
T-cell receptor diversity
Amplification and sequencing of T-cell receptor B (TCR-B) complementarity determining region 3 (CDR3) was performed using immunoSEQ Platform (Adaptive Biotechnologies, Seattle, WA), as previously described (supplemental Methods). A power geometric (PG) index with Horvitz-Thompson type was used as correction for undersampling and Good-Turing coverage adjustment.18-20
Gene expression
Statistical analysis
Different statistical methods have been used that are explained in the “Results” and/or figure legends. P < .05 was considered as statistically significant in all cases. Statistics were calculated using SPSS, version 22, or R, version 3.2.2.
Study approval
King's College Hospital Local Research Ethics Committee and Institutional Review Board of the National Heart, Lung, and Blood Institute approved the clinical studies of IST and sample collection, and informed written consent was obtained from patients.
Results
Identification of an immune signature for AA compared with healthy individuals based on distinct Treg subpopulations
We analyzed 31 AA patients at diagnosis and 5 HDs. Of these 31 patients, 16 were part of an initial test cohort and a further 15 part of a validation cohort. Metal-tagged antibodies against surface and intracellular markers were used in 2 separate panels (with or without stimulation with PMA/ionomycin) to stain T cells, including known markers for Tregs (CD25, CD127, FOXP3), naïve/memory subsets, homing/trafficking receptors, and differentiation/activation markers (supplemental Table 1). CD4+ and CD8+ T cells and B cells were clustered using this “whole panel” clustering approach (34 markers in panel 1 [supplemental Table 1]) to minimize bias. FOXP3+ cells were also clustered within CD4+ T cells (Figure 1; supplemental Figure 1). After gating for CD3+, CD4+, and CD8−, and merging all samples, viSNE was performed and cells clustered, based on 13 markers that most clearly clustered Tregs (Figure 2; supplemental Figure 2). Treg subpopulations were clustered and identified by high expression of CD25 and FOXP3 and low expression of CD127 (Figure 2A; supplemental Figure 1B). Identified Tregs expressed CD27hi, CD45RAlo, CD45ROhi, CD95hi, CD7lo, CD28hi, and CCR4hi compared with the total CD4+ T-cell subpopulation (Figure 2). The frequency of total Tregs was significantly lower in AA patients compared with HDs (2.7% vs 5.7% of CD4+ T cells, P < .01), confirming our previously published findings.8 The numbers of Tregs were not significantly different between patients with severe AA/very severe AA, and nonsevere AA.
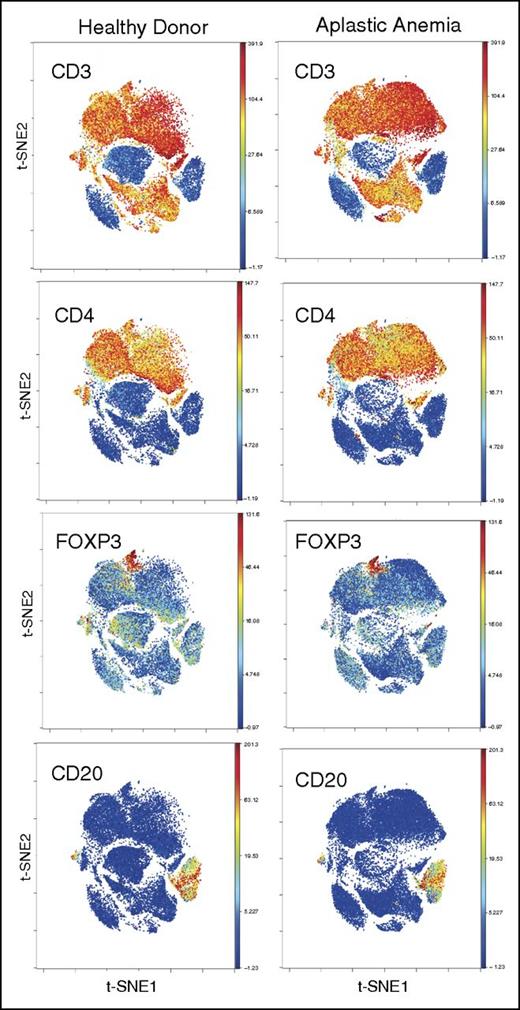
PBMC staining and clustering. PBMCs from AA and HDs were clustered using 34 surface and intracellular markers as our panel 1 (supplemental Table 1). Intact cells were gated based on Ir-191 and event length, followed by Ir-191 and Ir-193 gating. Viable cells were selected based on CD45 expression and negativity for Rh. All FCS files were first normalized using control beads and analyzed using Cytobank web-based software (see “Methods”). CD4+ and CD8+ T and B cells clustered together in both AA and HDs. Figures are representative of 16 AA and 5 HD samples.
PBMC staining and clustering. PBMCs from AA and HDs were clustered using 34 surface and intracellular markers as our panel 1 (supplemental Table 1). Intact cells were gated based on Ir-191 and event length, followed by Ir-191 and Ir-193 gating. Viable cells were selected based on CD45 expression and negativity for Rh. All FCS files were first normalized using control beads and analyzed using Cytobank web-based software (see “Methods”). CD4+ and CD8+ T and B cells clustered together in both AA and HDs. Figures are representative of 16 AA and 5 HD samples.
Identification of Treg subset by automated clustering. (A) After initial gating for CD3+, CD4+, and CD8− T cells, the gated cells were clustered using viSNE (Cytobank). Treg populations were identified based on high expression of CD25 and FOXP3 and low expression of CD127. (B) Median expression of the 7 most discriminative parameters between total CD4+ cells and Tregs as identified by the automated clustering algorithm FLOCK on a subset of 700 000 cells proportionally selected from all samples. Tregs were defined as clusters whose median expression was simultaneously higher than the 90% quantile of FOXP3 expression, higher than the 90% quantile of CD25 expression, and lower than the 50% quantile of CD127 expression across all CD3+CD8− cells. Heat map plot is based on 19 AA samples (pre- and post-IST) and 5 HD samples. (C) Median expression of the 8 most discriminative parameters between the 2 Treg subpopulations identified by the automated clustering algorithm FLOCK. Expression values were transformed using the asinh function in a cofactor of 5. Heat map plot is based on 19 AA samples (pre- and post-IST) and 5 HD samples. (D) The density plot of viSNE plots revealed 2 subpopulations within Tregs, designated as Treg A and B (arrows). The frequencies of Treg A and B were different between HD and AA patients. Patients who did not respond to IST (IST NR) had a higher number of Treg A at the time of diagnosis compared with responder patients (IST R) and HDs. The viSNE plots (right) are an overlay of Tregs’ contour plots colored by density and CD4+ T cells uncolored contour plots. (E) At the time of diagnosis and before treatment, Treg A frequency was higher in responder as well as nonresponder patients compared with HDs (38.8% ± 5% and 63.5% ± 4.5% vs 20.3% ± 6.6%, P < .05, P < .0001), whereas the frequency of Treg B was lower in both responder and nonresponder AA patients at the time of diagnosis compared with HDs (48.8% ± 6.1% and 28.9% ± 2.7% vs 72.2% ± 6.7%, P = .005, P < .0001). The nonresponder patients, however, had significantly higher Treg A and lower Treg B compared with responder patients (63.5% ± 4.5% vs 38.8% ± 5.0, P < .005 for Treg A; 28.9% ± 2.7% vs 48.8% ± 6.1%, P < .05 for Treg B). Error bars are standard error of mean. Kruskal-Wallis 1-way analysis of variance test was used for statistical analysis. ****P < .0001, ***P < .001, **P < .01, *P < .05. (F) The overlap between the Treg subpopulations identified using viSNE and manually gated Treg populations based on CD45RA and FOXP3 expression. Although subpopulations A and B mainly overlap with subpopulations I (CD45RAhi, FOXP3lo) and II (CD45RAlo, FOXP3hi), respectively, subpopulation III (CD45RAlo, FOXP3lo) was spread over population B as well as outside the Treg area. Figures are overlays of manually gated Treg populations on viSNE plots of total CD4+ T cells from an IST responder AA patient.
Identification of Treg subset by automated clustering. (A) After initial gating for CD3+, CD4+, and CD8− T cells, the gated cells were clustered using viSNE (Cytobank). Treg populations were identified based on high expression of CD25 and FOXP3 and low expression of CD127. (B) Median expression of the 7 most discriminative parameters between total CD4+ cells and Tregs as identified by the automated clustering algorithm FLOCK on a subset of 700 000 cells proportionally selected from all samples. Tregs were defined as clusters whose median expression was simultaneously higher than the 90% quantile of FOXP3 expression, higher than the 90% quantile of CD25 expression, and lower than the 50% quantile of CD127 expression across all CD3+CD8− cells. Heat map plot is based on 19 AA samples (pre- and post-IST) and 5 HD samples. (C) Median expression of the 8 most discriminative parameters between the 2 Treg subpopulations identified by the automated clustering algorithm FLOCK. Expression values were transformed using the asinh function in a cofactor of 5. Heat map plot is based on 19 AA samples (pre- and post-IST) and 5 HD samples. (D) The density plot of viSNE plots revealed 2 subpopulations within Tregs, designated as Treg A and B (arrows). The frequencies of Treg A and B were different between HD and AA patients. Patients who did not respond to IST (IST NR) had a higher number of Treg A at the time of diagnosis compared with responder patients (IST R) and HDs. The viSNE plots (right) are an overlay of Tregs’ contour plots colored by density and CD4+ T cells uncolored contour plots. (E) At the time of diagnosis and before treatment, Treg A frequency was higher in responder as well as nonresponder patients compared with HDs (38.8% ± 5% and 63.5% ± 4.5% vs 20.3% ± 6.6%, P < .05, P < .0001), whereas the frequency of Treg B was lower in both responder and nonresponder AA patients at the time of diagnosis compared with HDs (48.8% ± 6.1% and 28.9% ± 2.7% vs 72.2% ± 6.7%, P = .005, P < .0001). The nonresponder patients, however, had significantly higher Treg A and lower Treg B compared with responder patients (63.5% ± 4.5% vs 38.8% ± 5.0, P < .005 for Treg A; 28.9% ± 2.7% vs 48.8% ± 6.1%, P < .05 for Treg B). Error bars are standard error of mean. Kruskal-Wallis 1-way analysis of variance test was used for statistical analysis. ****P < .0001, ***P < .001, **P < .01, *P < .05. (F) The overlap between the Treg subpopulations identified using viSNE and manually gated Treg populations based on CD45RA and FOXP3 expression. Although subpopulations A and B mainly overlap with subpopulations I (CD45RAhi, FOXP3lo) and II (CD45RAlo, FOXP3hi), respectively, subpopulation III (CD45RAlo, FOXP3lo) was spread over population B as well as outside the Treg area. Figures are overlays of manually gated Treg populations on viSNE plots of total CD4+ T cells from an IST responder AA patient.
Although viSNE clustered the Treg population in 1 area, density plots revealed a heterogeneous distribution of cells. Two subpopulations within Tregs with a different frequency between AA and HDs were identified and designated as Treg A and Treg B (Figure 2C-D; supplemental Figure 1B-C). To confirm the presence of these 2 subpopulations and eliminate any bias, dimensionality reduction and automated unsupervised clustering methodology was applied independently by the bioinformatician; this confirmed the presence of 2 subpopulations within Tregs (Figure 2C; supplemental Figure 1B-C). These 2 subpopulations showed distinct markers in AA Tregs (16 patients, 12 IST responders and 4 nonresponders) as well as 5 HDs. Although both subpopulations were CD25hi, FOXP3hi, and CD127lo compared with total CD4+ T cells, subpopulation B was additionally characterized by a lower expression of CD45RA (P < .0001), CD7 (P < .001), and CD27 (P < .05) and a higher expression of CCR4 (P < .0001), CCR6 (P < .0001), CD25 (P < .0001), CD28 (P < .01), CD45RO (P < .0001), CD95 (P < .0001), CXCR3 (P < .05), FOXP3 (P < .0001), and HLA-DR (P < .0001) compared with subpopulation A (Kruskal-Wallis 1-way analysis of variance by ranks).
In addition to total Tregs, the percentage of B Tregs was significantly lower in AA patients compared with HDs (40.8% ± 13.7% vs 72.2% ± 15%, P = .008).
Treg composition predicts response to IST at the time of AA diagnosis
When patients were stratified into IST responders (n = 12) and nonresponders (n = 4), although the overall frequency of Treg B was lower in both responder and non-responding patients at the time of diagnosis compared with HDs (n = 5) (48.8% ± 6.1% and 28.9% ± 2.7% vs 72.2% ± 6.7%, P = .005, P < .0001), nonresponders had significantly higher Treg A and lower Treg B cells compared with responders (63.5% ± 4.5 vs 38.8% ± 5.0%, P < .005 for Treg A, 28.9% ± 2.7 vs 48.8% ± 6.1, P < .05 for Treg B) (Figure 2E, Table 2).
Treg A and B markers and summary of changes in AA
| Treg subpopulation . | Markers . | Healthy donors . | IST responder AA pretreatment . | IST nonresponder AA pretreatment . | IST responder AA posttreatment . | IST nonresponder AA posttreatment . |
|---|---|---|---|---|---|---|
| Treg A | CD45RA↑ CD7↑ CD27↑ CCR4↓ CCR6↓ CD25↓* CD28↓ CD45RO↓CD95↓ CXCR3↓ FOXP3↓* HLA-DR↓ | Minor population | ↑38.8% ± 5.0% | ↑↑ 63.5% ± 4.5% | ↓19.26% ± 2.43% | NSC |
| 20.3% ± 6.6% | 73.9% ± 6.8% | |||||
| Treg A (TNF-a+) | TNF-α + IL-10↓ | R | NSC | NSC | ↓0.32% ± 0.12% | NSC |
| CD279↓ | 1.2% ± 0.3% | 0.75% ± 0.14% | 0.79% ± 0.13% | 1.81% ± 0.19% | ||
| HLA-DR↓ CD38↓ | ||||||
| Treg B | CD45RA↓ CD7↓ CD27↓ CCR4↑ CCR6↑ CD25↑† CD28↑ CD45RO↑CD95↑ CXCR3↑ FOXP3↑† | Major population 72.2% ± 6.7% | ↓48.8% ± 6.1% | ↓↓ 28.9% ± 2.7% | NSC 59.98% ± 3.18% | NSC 21.8% ± 4.3% |
| HLA-DR↑ |
| Treg subpopulation . | Markers . | Healthy donors . | IST responder AA pretreatment . | IST nonresponder AA pretreatment . | IST responder AA posttreatment . | IST nonresponder AA posttreatment . |
|---|---|---|---|---|---|---|
| Treg A | CD45RA↑ CD7↑ CD27↑ CCR4↓ CCR6↓ CD25↓* CD28↓ CD45RO↓CD95↓ CXCR3↓ FOXP3↓* HLA-DR↓ | Minor population | ↑38.8% ± 5.0% | ↑↑ 63.5% ± 4.5% | ↓19.26% ± 2.43% | NSC |
| 20.3% ± 6.6% | 73.9% ± 6.8% | |||||
| Treg A (TNF-a+) | TNF-α + IL-10↓ | R | NSC | NSC | ↓0.32% ± 0.12% | NSC |
| CD279↓ | 1.2% ± 0.3% | 0.75% ± 0.14% | 0.79% ± 0.13% | 1.81% ± 0.19% | ||
| HLA-DR↓ CD38↓ | ||||||
| Treg B | CD45RA↓ CD7↓ CD27↓ CCR4↑ CCR6↑ CD25↑† CD28↑ CD45RO↑CD95↑ CXCR3↑ FOXP3↑† | Major population 72.2% ± 6.7% | ↓48.8% ± 6.1% | ↓↓ 28.9% ± 2.7% | NSC 59.98% ± 3.18% | NSC 21.8% ± 4.3% |
| HLA-DR↑ |
Surface and intracellular markers that are significantly higher or lower in Treg subpopulations as well as their frequencies in HD, IST responder, and nonresponder AA patients before and after IST.
NSC, no significant change; R, reference value.
Lower expression compared with Treg B subpopulation.
Higher expression compared with Treg A subpopulation.
To investigate the overlap between the Treg subpopulations with Treg subpopulations identified by Miyara et al,23 Tregs were gated based on CD45RA and FOXP3 expression. Although subpopulation A and B mainly overlapped with subpopulations I (CD45RAhi, FOXP3lo) and II (CD45RAlo, FOXP3hi), respectively, subpopulation III (CD45RAlo, FOXP3lo) was spread over both population A and B areas, and some cells clustered outside Treg area (Figure 2F).
Following IST response, the frequency of population A was significantly reduced in responders (from 38.8% ± 5.0% to 19.2% ± 2.4, P < .01), but was not significant in nonresponder patients. Treg B frequency was significantly higher in responders compared with nonresponders (59.9% ± 3.4% vs 21.8% ± 4.3, P < .0001) and closer to HDs (Table 2). Treg population A or B was not significantly different between patients with severe/very severe (n = 11) and nonsevere AA (n = 5).
The cytokine profile of Tregs following stimulation with PMA, ionomycin, and brefeldin A was investigated. Treg subpopulations A and B were identified within Tregs (supplemental Figure 3). Stimulated Treg clusters expressed higher CD25 (P < .001) and FOXP3 (P = .001) and lower CD127 (P = .001) compared with total CD4+ T cells. The majority of CD4+ T cells with pro-inflammatory cytokine properties clustered outside the “Treg area”; thus, Treg clusters expressed negligible amounts of pro-inflammatory cytokines interferon-γ (IFN-γ), and IL-17 (Figure 3A). Tregs expressed significantly higher IL-10 compared with total CD4+ T cells (P = .002); however, the IL-10 expression was not significantly different between the 2 Treg subpopulations. Although tumor necrosis factor-α (TNF-α)–expressing cells were clustered within non-Treg subpopulations, a cluster of CD4+ TNF-α+ T cells clustered within the Treg A subpopulation (Figure 3B). These TNF-α+ cells expressed significantly higher CD127 (P < .001), and lower IL-10 (P < .001), CD279 (P < .001), HLA-DR (P = .003), CD38 (P < .001), CD25 (P < .001), and FOXP3 (P < .001) compared with total Tregs. Although at the time of diagnosis, the frequency of TNF-α+ cells was not significantly different between IST responders and nonresponders, IST responders had significantly lower TNF-α+ cells compared with nonresponders following response to IST (0.32% vs 1.81%, P = .008) (Figure 3C).
PMA/ionomycin-stimulated Treg and Tconsubpopulations. PBMCs from 16 patients and 5 HDs were stimulated for 4 hours with PMA, a calcium ionophore (ionomycin), and protein transport inhibitor (brefeldin A) and stained with a panel of antibodies based on 29 surface markers, transcription factors, and cytokines (supplemental Table 1). Treg subpopulations A and B were identified within Tregs after stimulation with Treg profiles that distinguished responders from nonresponders to IST (supplemental Figure 3). (A) Following 4 hours stimulation with PMA and ionomycin in the presence of brefeldin, PBMCs were stained for surface and intracellular markers (supplemental Table 1, panel 2) followed by mass cytometry and viSNE on CD4+ T cells. Cytokine-secreting CD4+ T cells, including IFN-γ, IL-2–, IL-17–, and IL-4–secreting cells, localized distinctly with minimal overlap and outside “Treg’s area.” Both Treg A and Treg B populations show higher expression of IL-10 compared with “non-Tregs” and TNF-α–secreting Tregs, but there was no significant difference between Tregs A and B in terms of IL-10 expression. (B) Unlike the rest of cytokine-secreting CD4+ T cells, TNF-α–secreting cells were spread over several areas, including the Treg A subpopulation (red arrow). (C) SPADE analysis of Tregs A and B following 4 hours’ stimulation with PMA/ionomycin and intracellular staining (supplemental Table 1, panel 2). Although the TNF-α–secreting cells within the Treg A subpopulation reduces after IST in responder AA patients, there is no similar reduction in nonresponder patients (patient AA-11 is an IST nonresponder and AA-6 is a responder patient). (D-E) SPADE clustering based on CD45RA, CD45RO, CD27, and CD62L. The Tcon subpopulations were defined as naïve (CD45RA+ CD45RO- CD27hi), memory (CD45RA− CD45RO+ CD27lo), central memory (CM; CD45RA− CD45RO+ CD27lo CD62Lhi), effector memory (EM; CD45RA− CD45RO+ CD27lo CD62Llo), effector (CD45RA− CD45RO+ CD27lo), and terminal effectors (Tee; CD45RA+ CD45RO− CD27lo). Tcon with effector phenotype expresses higher CD161 in nonresponder patients at the time of diagnosis compared with responder AA patients (patient AA-11 is an IST nonresponder and AA-6 is a responder patient). The frequencies of these subpopulations were not significantly different between IST responder and IST nonresponder patients at the time of diagnosis. The naïve Tcon from IST nonresponder patients were expressing slightly higher CCR4 compared with responder patients (supplemental Table 2).
PMA/ionomycin-stimulated Treg and Tconsubpopulations. PBMCs from 16 patients and 5 HDs were stimulated for 4 hours with PMA, a calcium ionophore (ionomycin), and protein transport inhibitor (brefeldin A) and stained with a panel of antibodies based on 29 surface markers, transcription factors, and cytokines (supplemental Table 1). Treg subpopulations A and B were identified within Tregs after stimulation with Treg profiles that distinguished responders from nonresponders to IST (supplemental Figure 3). (A) Following 4 hours stimulation with PMA and ionomycin in the presence of brefeldin, PBMCs were stained for surface and intracellular markers (supplemental Table 1, panel 2) followed by mass cytometry and viSNE on CD4+ T cells. Cytokine-secreting CD4+ T cells, including IFN-γ, IL-2–, IL-17–, and IL-4–secreting cells, localized distinctly with minimal overlap and outside “Treg’s area.” Both Treg A and Treg B populations show higher expression of IL-10 compared with “non-Tregs” and TNF-α–secreting Tregs, but there was no significant difference between Tregs A and B in terms of IL-10 expression. (B) Unlike the rest of cytokine-secreting CD4+ T cells, TNF-α–secreting cells were spread over several areas, including the Treg A subpopulation (red arrow). (C) SPADE analysis of Tregs A and B following 4 hours’ stimulation with PMA/ionomycin and intracellular staining (supplemental Table 1, panel 2). Although the TNF-α–secreting cells within the Treg A subpopulation reduces after IST in responder AA patients, there is no similar reduction in nonresponder patients (patient AA-11 is an IST nonresponder and AA-6 is a responder patient). (D-E) SPADE clustering based on CD45RA, CD45RO, CD27, and CD62L. The Tcon subpopulations were defined as naïve (CD45RA+ CD45RO- CD27hi), memory (CD45RA− CD45RO+ CD27lo), central memory (CM; CD45RA− CD45RO+ CD27lo CD62Lhi), effector memory (EM; CD45RA− CD45RO+ CD27lo CD62Llo), effector (CD45RA− CD45RO+ CD27lo), and terminal effectors (Tee; CD45RA+ CD45RO− CD27lo). Tcon with effector phenotype expresses higher CD161 in nonresponder patients at the time of diagnosis compared with responder AA patients (patient AA-11 is an IST nonresponder and AA-6 is a responder patient). The frequencies of these subpopulations were not significantly different between IST responder and IST nonresponder patients at the time of diagnosis. The naïve Tcon from IST nonresponder patients were expressing slightly higher CCR4 compared with responder patients (supplemental Table 2).
Validation by conventional polychromatic flow cytometry.
Conventional flow cytometry was performed on peripheral blood from a separate validation cohort of 15 AA patients (11 IST responders and 4 nonresponders) at the time of diagnosis to confirm that the identified markers were sufficient to detect Treg subpopulations and whether the CyTOF-identified combination of markers was still predictive for IST response. PBMCs were stained with anti-CD4, CD25, CD127, FOXP3, CD95, CCR4, and CD45RA and, similar to the initial cohort, Treg A cells were significantly higher in nonresponders compared with responders (37.82% ± 5.11% vs 18.33 ± 2.31, P = .006), whereas Treg B cells were significantly higher in responders (69.97% ± 3.02% vs 49.34% ± 4.92%, P = .01) (supplemental Figure 4).
Conventional CD4+ T cells
Our marker panels also identified and clustered conventional CD4+ Tcon. The Treg population was first gated out on a viSNE plot and SPADE clustering based on CD45RA, CD45RO, CD27, and CD62L performed. Tcon subpopulations were defined as naïve (CD45RA+ CD45RO− CD27hi), memory (CD45RA− CD45RO+ CD27lo), central memory (CD45RA− CD45RO+ CD27lo CD62Lhi), effector memory (CD45RA− CD45RO+ CD27lo CD62Llo), effector (CD45RA− CD45RO+ CD27lo), and terminal effectors (CD45RA+ CD45RO− CD27lo). Subset frequencies were not significantly different between IST responders (n = 12) and nonresponders (n = 4) at the time of diagnosis. However, effector CD4+ T cells (Te) expressed a significantly higher CD161 level in nonresponders compared with responders (P < .01) (Figure 3D-E; supplemental Table 2).
Function, ontogeny, and in vitro expansion of Treg subsets
To assess the function of the Treg subpopulations, HD CD4+ Tregs were sorted based on CD25, CD127, and CD95 expression, which showed the highest expression of differences between subpopulations A and B on viSNE clusters, excluding intracellular markers to avoid fixation and permeabilization of the cells. Sorted cells were CD4+CD25hi CD127lo CD95−, CD45RAhi, CCR4lo (subpopulation A) and CD4+CD25hi CD127lo CD95+, CD45RAlo, and CCR4hi (subpopulation B). To confirm these markers were enough to identify Treg subpopulations, viSNE runs were performed based on these markers and both subpopulations were identified (supplemental Figure 5). Treg B cells were significantly more functional compared with those of subpopulation A in suppression of both IFN-γ and TNF-α secretion by Tcon (P < .05, Figure 4A).
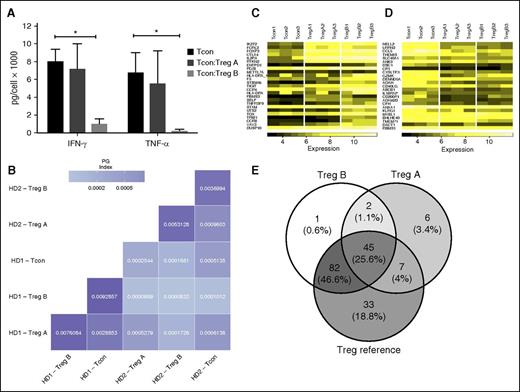
Function and ontogeny of Treg subpopulations. (A) Suppression of Tcon cytokine secretion by Treg subpopulations: CD4+CD25hi CD127lo CD45−RAhi, CD95−, CCR4lo (subpopulation A), CD4+CD25hi CD127lo CD45-RAlo CD95+ CCR4hi (subpopulation B) and CD4+CD25low Tcon were sorted (FacsAria) and cultured for 5 days with anti-CD3/CD28 beads (Tcon:Treg:bead = 20:20:1). After 5 days of culture, the supernatant was analyzed with ProcartaPlex 6 Plex (eBioscience) according to the manufacturer’s instructions. The cytokine concentrations were corrected for the cell number. Treg B cells were able to significantly reduce both IFN-γ and TNF-α secretion by Tcon (P < .05) in a 1:1 coculture. Average of 3 replicates, Student t test, *P < .05. Error bars are standard error of mean. (B) Pairwise comparison of TCR repertoire overlaps. The color shading reflects the numerical value of the (PG indices. The TCR sequences shared between Tcon, Treg A, and Treg B were very low with a PG index <0.001 in all comparisons. (C-D) GEP of Treg A and Treg B subpopulations compared with Tcon: We have used the published GEP profile of Tregs (Ferraro et al.) as the reference list, and all 3 T-cell populations were compared with the list, which is sorted based on highly expressed genes in human Treg. Both Treg A and Treg B subpopulations were enriched with Treg-related genes, in particular IKZF2, FCRL3, FOXP3, CTLA4, and IL-2R. However, the genes that are expressed at a lower level in human Tregs were enriched in Tcon, but in none of the Treg subpopulations. (D) The frequency of common genes between Treg A, Treg B, and referenced published genes (Ferraro et al.) are demonstrated (E), which includes genes that are highly expressed by human Tregs.
Function and ontogeny of Treg subpopulations. (A) Suppression of Tcon cytokine secretion by Treg subpopulations: CD4+CD25hi CD127lo CD45−RAhi, CD95−, CCR4lo (subpopulation A), CD4+CD25hi CD127lo CD45-RAlo CD95+ CCR4hi (subpopulation B) and CD4+CD25low Tcon were sorted (FacsAria) and cultured for 5 days with anti-CD3/CD28 beads (Tcon:Treg:bead = 20:20:1). After 5 days of culture, the supernatant was analyzed with ProcartaPlex 6 Plex (eBioscience) according to the manufacturer’s instructions. The cytokine concentrations were corrected for the cell number. Treg B cells were able to significantly reduce both IFN-γ and TNF-α secretion by Tcon (P < .05) in a 1:1 coculture. Average of 3 replicates, Student t test, *P < .05. Error bars are standard error of mean. (B) Pairwise comparison of TCR repertoire overlaps. The color shading reflects the numerical value of the (PG indices. The TCR sequences shared between Tcon, Treg A, and Treg B were very low with a PG index <0.001 in all comparisons. (C-D) GEP of Treg A and Treg B subpopulations compared with Tcon: We have used the published GEP profile of Tregs (Ferraro et al.) as the reference list, and all 3 T-cell populations were compared with the list, which is sorted based on highly expressed genes in human Treg. Both Treg A and Treg B subpopulations were enriched with Treg-related genes, in particular IKZF2, FCRL3, FOXP3, CTLA4, and IL-2R. However, the genes that are expressed at a lower level in human Tregs were enriched in Tcon, but in none of the Treg subpopulations. (D) The frequency of common genes between Treg A, Treg B, and referenced published genes (Ferraro et al.) are demonstrated (E), which includes genes that are highly expressed by human Tregs.
Genomic DNA from sorted Tregs A and B was used for TCR Vβ chain CDR3 high-throughput sequencing.19,20 On average, subpopulations A and B had 33 773 and 26 090 unique TCR sequences, respectively, and 233 sequences were common to both (r = 0.008) (supplemental Figure 6). Tcon also shared 137 and 383 sequences with subpopulations A and B, respectively (r = 0.154 and r = 0.092, respectively). In assessing a naturally, highly diverse TCR subpopulations, undersampling can introduce bias.18,24 We therefore used PG index for pairwise comparison of TCR repertoire overlap.18 Overlap between Treg A, B, and Tcon was small, suggesting these subpopulations were distinct (Figure 4B).
GEP analysis of Treg subpopulations
Whole GEP data25,26 showed both Treg A and Treg B had different GEP compared with Tcon. Nevertheless, when principal component analysis was performed, Treg B and Tcon had the most differences, whereas the Treg A subpopulation showed a transcriptional profile in between Treg B and Tcon (supplemental Figure 7).
Comparing GEPs of Treg subpopulations to Tcon using the human Treg’s gene signature27 showed both Treg A and B subpopulations were significantly enriched in genes upregulated in human Tregs, including IL-2RA, FOXP3, IKZF2, TIGIT, and CTLA4 (false discovery rate [FDR] < 0.0001) for both Treg subpopulations compared with Tcon. Treg B cells were enriched with Treg-related memory/activation genes compared with Treg A, including JAKMIP1, CCR8, TRIB1, and GZMK (FDR < 0.0001, Figure 4C-E). Thus, although both Treg subpopulations were enriched with Treg-associated genes, Tregs B were characterized by an activation gene signature in agreement with mass cytometry findings.
Gene set enrichment functional analysis22 highlighted several gene sets as significantly overexpressed in the Treg B subpopulation, including G2M checkpoint (FDR < 0.0001), mitosis (FDR = 0.015), M phase of mitotic cell cycle (FDR = 0.018) IL-2–STAT5 signaling (FDR = 0.023), and immune response genes (FDR = 0.032) (Table 3). Protein interaction networks of proteins encoded by messenger RNA (mRNA) that are enriched in Treg B subpopulation cells were also mapped (supplemental Figure 8A-B). Ontology analyses of the functions of these protein complexes (supplemental Table 3, supplemental Figure 9) showed they are involved in mitotic functions, DNA replication, and cell-cycle–dependent transcription. MKI67 mRNA (encodes nuclear Ki67 protein) was enriched in Treg B subpopulation cells. This is important because the Ki67 protein is often used as a marker of proliferating cells.28,29
Genes that are upregulated in the Treg population B compared with population A
| Genes . | FDR q-value . | Normalized enrichment score . | Significant genes . |
|---|---|---|---|
| G2M checkpoint | <0.0001 | 2.0188198 | CASC5, NUSAP1, CENPE, TPX2, TOP2A, KIF11, BARD1, EZH2, SLC7A5, HN1, GINS2, CKS2, BUB1, SMC4, STIL, BRCA2, CHEK1, SAP30, E2F3, MKI67, CDKN2C, NEK2, KIF15, KPNA2, KIF23, ZAK, POLQ, WHSC1, TFDP1, UCK2, E2F2, CDKN3, CDC7, E2F1, AURKA, CDC20, EFNA5, KIF4A, EXO1, CDC25A, PRC1, KIF5B |
| Mitosis | 0.014934987 | 2.020 | NCAPH, NUSAP1, NDC80, CENPE, BUB1B, TPX2, KIF11, TTN, BUB1, NEK2, SMC4, PCBP4, PAM |
| M phase of mitotic cell cycle | 0.01755665 | 2.0006573 | NCAPH, NUSAP1, NDC80, CENPE, BUB1B, TPX2, KIF11, TTN, BUB1, NEK2, SMC4, PCBP4, PAM |
| IL2-STAT5 signaling | 0.02301426 | 1.5519865 | SYT11, CCR4, TNFRSF9, CST7, ADAM19, IL1R2, ANXA4, PHLDA1, SLC1A5, IL18R1, FGL2, TNFRSF18, TNFRSF4, GALM, CXCL10, BATF, SPP1, TNFSF10, PHTF2, CD86, AHNAK, IL10, LIF, TLR7, F2RL2, CD79B, CTLA4, FURIN, TNFRSF1B, CAPG, ALCAM, CSF1, CASP3, CSF2, MUC1, MYO1E RORA, ITGAV, PRNP, ICOS, UCK2, CYFIP1, SOCS1 |
| Immune response genes | 0.031976275 | 1.9183398 | IL7, CCR2, CCR8, CCR4, IL1R2, CCR6, CST7, CCL20, GZMA, CADM1, AIM2, CIITA, CCR5, CTSC, IL12A, NCF4, IL4, CCR9, GEM, IL32, TNFRSF4, LAX1, DEFB4A, FCGR3B, TLR7, CD74, APOA4, CCL5, APOBEC3G, CD79B, CTLA4 |
| Genes . | FDR q-value . | Normalized enrichment score . | Significant genes . |
|---|---|---|---|
| G2M checkpoint | <0.0001 | 2.0188198 | CASC5, NUSAP1, CENPE, TPX2, TOP2A, KIF11, BARD1, EZH2, SLC7A5, HN1, GINS2, CKS2, BUB1, SMC4, STIL, BRCA2, CHEK1, SAP30, E2F3, MKI67, CDKN2C, NEK2, KIF15, KPNA2, KIF23, ZAK, POLQ, WHSC1, TFDP1, UCK2, E2F2, CDKN3, CDC7, E2F1, AURKA, CDC20, EFNA5, KIF4A, EXO1, CDC25A, PRC1, KIF5B |
| Mitosis | 0.014934987 | 2.020 | NCAPH, NUSAP1, NDC80, CENPE, BUB1B, TPX2, KIF11, TTN, BUB1, NEK2, SMC4, PCBP4, PAM |
| M phase of mitotic cell cycle | 0.01755665 | 2.0006573 | NCAPH, NUSAP1, NDC80, CENPE, BUB1B, TPX2, KIF11, TTN, BUB1, NEK2, SMC4, PCBP4, PAM |
| IL2-STAT5 signaling | 0.02301426 | 1.5519865 | SYT11, CCR4, TNFRSF9, CST7, ADAM19, IL1R2, ANXA4, PHLDA1, SLC1A5, IL18R1, FGL2, TNFRSF18, TNFRSF4, GALM, CXCL10, BATF, SPP1, TNFSF10, PHTF2, CD86, AHNAK, IL10, LIF, TLR7, F2RL2, CD79B, CTLA4, FURIN, TNFRSF1B, CAPG, ALCAM, CSF1, CASP3, CSF2, MUC1, MYO1E RORA, ITGAV, PRNP, ICOS, UCK2, CYFIP1, SOCS1 |
| Immune response genes | 0.031976275 | 1.9183398 | IL7, CCR2, CCR8, CCR4, IL1R2, CCR6, CST7, CCL20, GZMA, CADM1, AIM2, CIITA, CCR5, CTSC, IL12A, NCF4, IL4, CCR9, GEM, IL32, TNFRSF4, LAX1, DEFB4A, FCGR3B, TLR7, CD74, APOA4, CCL5, APOBEC3G, CD79B, CTLA4 |
The most significant gene sets that are upregulated in the Treg B subpopulation compared with Treg A are listed. Leading-edge analysis revealed several genes that significantly contribute in each gene set and are enriched in Treg B compared with Treg A (marked as significant genes).
Expandability of Tregs in Treg-promoting culture.
One of the aims of this study was to investigate the potential expandability of Tregs in AA. We first tested the IL-2 sensitivity of AA total Tregs based on STAT5 phosphorylation.30 Freshly isolated PBMCs from 2 HDs and 6 AA patients (3 IST responders and 3 nonresponders, at diagnosis) were cultured in the presence of IL-2 at different concentrations (from 0.1 to 1000 IU/mL). Treg p-STAT5 expression significantly increased after 15 minutes’ culture with IL-2 (0.5 IU/mL) (median fluorescence intensities, 92.6 ± 35.1 pre-IL-2 vs 787 ± 109.6 post-IL-2, P < .001), confirming their responsiveness to IL-2. There was no difference between HDs and AA Tregs in response to IL-2 (Figure 5A).
(A) Tregs from both AA patients and HDs were sensitive to IL-2 (at 0.5 and 1 U/mL concentrations) as evident by STAT5 phosphorylation after 15 and 30 minutes. Flow cytometry results are representative of 8 experiments (2 HDs and 6 AA patients). (B) Expanded Tregs were >90% FOXP3+ following expansion. (C) There was no significant difference between AA and HD Tregs in terms of fold-change increase after 4 weeks’ expansion (represents median-fold increase of 3 HDs and 3 AA). Error bars are standard error of mean. (D) Expanded Tregs were able to suppress Tcon proliferation in both autologous and allogeneic settings (crisscross assay) (1:1 Treg/Tcon ratio after 96 hours’ culture). It is noticeable that in this experiments, TconAA were proliferating slightly more than TconHD in the presence of expanded Tregs (6.43% vs 3.86%); however, this difference was not statistically significant when all replicates were compared. Tcon was stained by violet proliferation dye (VPD). TconAA, conventional T cells from AA patients; TconHD, conventional T cells from healthy donors; TreAA, expanded Tregs from AA patients; TreHD, expanded Tregs from healthy donors.
(A) Tregs from both AA patients and HDs were sensitive to IL-2 (at 0.5 and 1 U/mL concentrations) as evident by STAT5 phosphorylation after 15 and 30 minutes. Flow cytometry results are representative of 8 experiments (2 HDs and 6 AA patients). (B) Expanded Tregs were >90% FOXP3+ following expansion. (C) There was no significant difference between AA and HD Tregs in terms of fold-change increase after 4 weeks’ expansion (represents median-fold increase of 3 HDs and 3 AA). Error bars are standard error of mean. (D) Expanded Tregs were able to suppress Tcon proliferation in both autologous and allogeneic settings (crisscross assay) (1:1 Treg/Tcon ratio after 96 hours’ culture). It is noticeable that in this experiments, TconAA were proliferating slightly more than TconHD in the presence of expanded Tregs (6.43% vs 3.86%); however, this difference was not statistically significant when all replicates were compared. Tcon was stained by violet proliferation dye (VPD). TconAA, conventional T cells from AA patients; TconHD, conventional T cells from healthy donors; TreAA, expanded Tregs from AA patients; TreHD, expanded Tregs from healthy donors.
To test their in vitro expandability, Tregs were obtained from 6 AA patients (3 IST responders and 3 nonresponders, at diagnosis) and 8 HDs and were cultured and stimulated with anti-CD3/CD28 beads (1:1 ratio) and high-dose IL-2 (1000 IU/mL) for 4 weeks with added all-trans-retinoic acid 2 μM and rapamycin 100 nM.17 AA Tregs expanded in a comparable rate to HD Tregs, with a median 33-fold increase (range, 29-149) compared with a 21-fold increase (range, 8-36) in HD. Expanded Tregs demonstrated more than 90% FOXP3+ expression in both AA and HDs (Figure 5 B-C).
Expanded Tregs were gradually (within 5 days) deprived of IL-2 following expansion and p-STAT5 evaluated in both AA and HDs Tregs to assess IL-2 dependency. Although expanded AA Tregs showed slightly lower p-STAT5 level following IL-2 deprivation, they responded to low-dose IL-2 and p-STAT5 and returned to a similar level as in HDs (data not shown).
Expanded Tregs function, methylation status of TSDR, and TCR diversity.
To assess the suppressive activity of expanded Tregs, Tcon were stained with a fluorescent proliferation dye (CellTrace Violet, Life Technologies) and cocultured with autologous expanded Tregs (1:1 ratio) for 5 days in the presence of anti-CD3/CD28 beads (Tcon:Treg:beads = 20:20:1). Expanded AA Tregs suppressed proliferation of CD4+ Tcon (an average reduction from 43% to 5%, P = .009) in both autologous and allogeneic conditions (Figure 5D). The suppressive activity of AA expanded Tregs was not significantly different from HD expanded Tregs.
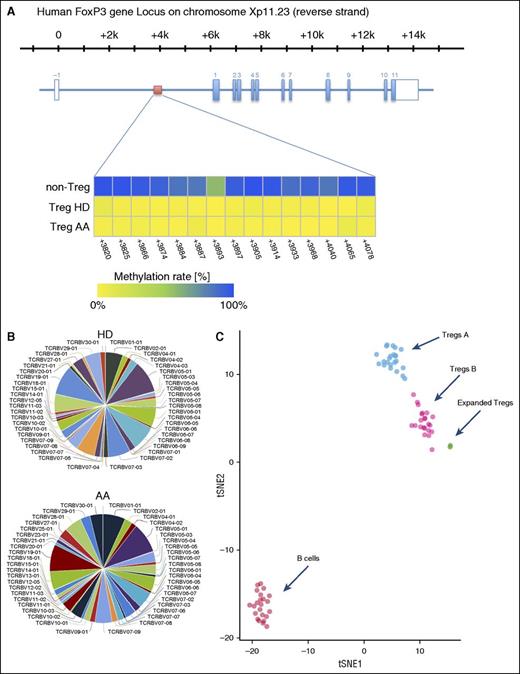
To assess the stability of expanded Tregs, we investigated the methylation status of 15 cytosine guanine dinucleotide sites within the FOXP3 Treg-specific demethylated region (TSDR)31 by amplicon sequencing of bisulfite treated DNA on an Illumina MiSeq sequencing platform. TSDR cytosine guanine dinucleotide sites in expanded HDs and AA Tregs were >98% unmethylated, confirming the stability of expanded Tregs (Figure 6A).
(A) Overview of the human FOXP3 gene locus with the exon/intron structure in blue and the TSDR region in red. The lower panel shows the methylation statues of TSDR in HD and AA expanded Tregs compared with non-Tregs. (B) Expanded Tregs were polyclonal in both AA and HD patients. The normalized Shannon entropy of expanded Treg repertoire was used to calculate the degree of clonality, which, on average, was 0.12. (C) Because we did not succeed in isolating enough Treg A and Treg B cells from AA patients for individual in vitro expansion because of the low number of Tregs in these patients, the composition of in vitro expanded Tregs were assessed by calculating the Euclidian distance between the mean expression for each parameter in Treg A and B, which was calculated in 24 Treg A and B (5 HD and 19 AA samples) and 5 (2 AA and 3 HDs) in vitro expanded Tregs. B cells were used as an irrelevant control. The following parameters, which showed the highest differences between subpopulations, were used for the Euclidian distance calculation: FOXP3, CD25, CD127, CD45RA, HLA-DR, CCR6, CCR4, CD69, CD27, CXCR3, CD45RO, CD4, CD20, CD95, CD161, CD28, CD152, CD7, CD279, and CD19. t-SNE1 and t-SNE2 were used for distance calculation (see also supplemental Figure 10). viSNE plot of expression centroids for all Treg cell subpopulations, B cells, and expanded Tregs across all samples. Treg A and Treg B cells were automatically gated from 24 individual samples (19 samples from AA patients, 5 HDs) using the automated clustering algorithm FLOCK on a subset of 700 000 cells proportionally selected from all samples. B cells were gated from the same 24 samples in Cytobank. Expanded Tregs were from 3 HDs and 2 AA patients. Expression centroids were computed for each cell population and used as input for the dimensionality algorithm t-SNE as implemented in the tool cyt (see “Methods” for more details). Each dot in the plot represents 1 particular cell population in a particular sample. Expression values were transformed using the asinh function in a cofactor of 5. Using a 1-tailed Welch’s 2-sample t test, we can reject the null hypothesis that the distance between Treg A and expanded Tregs is lower than in Treg B (P < 2.2 × 10−16), which suggests that expanded Tregs are more similar to subpopulation B than A.
(A) Overview of the human FOXP3 gene locus with the exon/intron structure in blue and the TSDR region in red. The lower panel shows the methylation statues of TSDR in HD and AA expanded Tregs compared with non-Tregs. (B) Expanded Tregs were polyclonal in both AA and HD patients. The normalized Shannon entropy of expanded Treg repertoire was used to calculate the degree of clonality, which, on average, was 0.12. (C) Because we did not succeed in isolating enough Treg A and Treg B cells from AA patients for individual in vitro expansion because of the low number of Tregs in these patients, the composition of in vitro expanded Tregs were assessed by calculating the Euclidian distance between the mean expression for each parameter in Treg A and B, which was calculated in 24 Treg A and B (5 HD and 19 AA samples) and 5 (2 AA and 3 HDs) in vitro expanded Tregs. B cells were used as an irrelevant control. The following parameters, which showed the highest differences between subpopulations, were used for the Euclidian distance calculation: FOXP3, CD25, CD127, CD45RA, HLA-DR, CCR6, CCR4, CD69, CD27, CXCR3, CD45RO, CD4, CD20, CD95, CD161, CD28, CD152, CD7, CD279, and CD19. t-SNE1 and t-SNE2 were used for distance calculation (see also supplemental Figure 10). viSNE plot of expression centroids for all Treg cell subpopulations, B cells, and expanded Tregs across all samples. Treg A and Treg B cells were automatically gated from 24 individual samples (19 samples from AA patients, 5 HDs) using the automated clustering algorithm FLOCK on a subset of 700 000 cells proportionally selected from all samples. B cells were gated from the same 24 samples in Cytobank. Expanded Tregs were from 3 HDs and 2 AA patients. Expression centroids were computed for each cell population and used as input for the dimensionality algorithm t-SNE as implemented in the tool cyt (see “Methods” for more details). Each dot in the plot represents 1 particular cell population in a particular sample. Expression values were transformed using the asinh function in a cofactor of 5. Using a 1-tailed Welch’s 2-sample t test, we can reject the null hypothesis that the distance between Treg A and expanded Tregs is lower than in Treg B (P < 2.2 × 10−16), which suggests that expanded Tregs are more similar to subpopulation B than A.
TCR Vβ CDR3 high-throughput sequencing of expanded Tregs was used to investigate their clonality. The normalized Shannon entropy of expanded Treg repertoire was used to calculate the degree of clonality that, on average, was 0.12 (4 expanded Tregs, 1 being the most clonal and 0 with the most TCR diversity). Both AA and HD expanded Tregs showed a comparable level of TCR Vβ CDR3 diversity, as defined in Figure 6B and the supplemental Methods.
The (dis)similarity of in vitro expanded Tregs with Treg A and Treg B was investigated next. As the isolated Tregs were stimulated and treated in a Treg skewing environment, the expression level of some markers such as CD25 or FOXP3 were high in expanded Tregs, making it difficult to cluster expanded Tregs with Treg A or B of untouched Tregs for comparison. To overcome this technical issue, we used an alternative analysis approach based on distance calculation and relative expression of markers. The subpopulations of Tregs expanded in vitro were assessed using the Euclidian distance between the mean expression for each parameter in Tregs A and B (Figure 6C; supplemental Figure 10).
Using a 1-tail Welch’s 2-sample t test, we rejected the null hypothesis that the distance between Treg A and expanded Tregs is lower than in Treg B (P < 2.2 × 10−16), suggesting that expanded Tregs were more similar to subpopulation B than A.
Discussion
Although the importance of Tregs in the pathophysiology of autoimmune diseases is well established, the definition and significance of Treg subpopulations is less clear. Identification of Treg subsets is challenging in autoimmune diseases because the number of Tregs is usually low and Tregs may express aberrant markers; in addition, gating strategies for Treg subpopulations are often subjective. Biomarkers that, first, identify AA patients from HDs and, second, identify at time of diagnosis who are less likely to respond to IST, have as yet not been identified. It is now possible to comprehensively characterize rare, complex populations of cells with minimal bias15,32 using CyTOF to measure the expression level of more than 40 parameters at the single-cell level.32-34 The complexity of Treg subsets in HDs has been previously demonstrated by mass cytometry on sorted Tregs35 ; however, their biological importance has not been investigated. In the current study, by using this multidimensional phenotyping and unbiased approach, 2 distinct Treg subpopulations were characterized in HD and AA patients and the changes in these subsets predicted response to IST at diagnosis of AA. We sorted these cells based on their immunological markers and confirmed their dissimilar TCRs, gene expression signatures, and functions. Our analytical strategy eliminated the unavoidable subjectivity of Treg subpopulation definition based on 2-dimensional gating without unnecessary overclustering.
Within the CD25hi, FOXP3hi, and CD127lo Treg population, AA Tregs expressed CD27hi, CD45RAlo, CD45ROhi, CD95hi, CD7lo, CD28hi, and CCR4hi compared with the total CD4+ T cell. We have identified 2 well-defined subpopulations within this Treg population (Treg A and B). Although total Treg numbers were reduced in AA, Treg A was significantly higher in AA patients compared with HDs. In contrast, the number of Treg B subpopulation was significantly lower in AA patients compared with HDs (Table 2). Subpopulation B was characterized by a lower expression of CD45RA, CD7, and CD27 and a higher expression of CCR4, CCR6, CD25, CD28, CD45RO, CD95, CXCR3, FOXP3, and HLA-DR. The most significantly different markers were CD95, CCR4, and CD45RO.36-38 The identified Treg subpopulations were compared with established Treg subpopulation definitions.23 Although Tregs A and B overlap with Treg subpopulations I and II, respectively, our approach demarcates those Treg III cells that are closer to Tregs and combine them with subpopulation A or B based on their phenotype and eliminate cells that are closer to Tcon and less likely to be regulatory. There is an unmet need for more robust predictive factors for response to IST at the time of AA diagnosis. Known predictive factors for response to IST include less severe disease, young age, and absolute reticulocyte and lymphocyte counts of ≥25 and ≥1.0 × 109/L, respectively.39 Short telomeres in children, but not in adults, also predict response to IST.40,41 The presence of a PIGA mutation predicts for response to IST, as does a somatic BCOR/BCORL mutation. In contrast, somatic mutation of DNMT3A or ASXL1 is associated with worse outcomes following IST.42 We have identified an immune signature that predicts for response to IST for an individual patient at time of diagnosis of AA. Nonresponders to IST were more likely to have higher Treg A compared with nonresponders, whereas responders had higher Treg B numbers compared with HDs (Figure 7). Treg B subpopulation characterized by a more “activated/memory” phenotype. We also tested this combination of identified markers in a separate validation cohort of AA patients using conventional flow cytometry and confirmed the predictive value of this combination for response to IST.
Graphical abstract. AA patients with higher number of Treg B cells before IST are more likely to respond to therapy. Following response to IST, responder patients have a higher number of Treg B cells compared with nonresponders. Treg B cells are enriched with cell-cycle–related proteins and are more likely to enter the cell cycle compared with Treg A subpopulations. In vitro expanded Tregs are also phenotypically closer to Treg B than Treg A. Tconv, conventional CD4+ T cells.
Graphical abstract. AA patients with higher number of Treg B cells before IST are more likely to respond to therapy. Following response to IST, responder patients have a higher number of Treg B cells compared with nonresponders. Treg B cells are enriched with cell-cycle–related proteins and are more likely to enter the cell cycle compared with Treg A subpopulations. In vitro expanded Tregs are also phenotypically closer to Treg B than Treg A. Tconv, conventional CD4+ T cells.
To explore the potential therapeutic application of expanded Tregs in the treatment of AA, we examined the characteristics and in vitro expandability of the Treg subpopulations in depth. Both AA and HDs Tregs were sensitive to IL-2 as assessed by STAT5 phosphorylation. The expansion rate of AA Tregs (3 of them from IST nonresponders) was not different from HDs after 4 weeks’ culture. Expanded Tregs were functional in both autologous and allogeneic settings, and TSDR of expanded Tregs indicated a stable phenotype. Treg B population was more functional in suppressing IFN-γ and TNF-α secretion by Tcon compared with Treg A cells. Although both Treg subpopulations were significantly enriched with Treg-specific genes such as IL-2RA, FOXP3, IKZF2, TIGIT, and CTLA4 compared with Tcon, nevertheless, TCR sequence overlaps between Treg A, Treg B, and Tcon were minimal, suggesting possible distinct developmental origins. Functional GEP analysis revealed marked enrichment of Treg B subpopulation with G2M checkpoint and mitosis-related genes, suggesting that Tregs B are more prone to enter cell cycle. In studies of human primary lymphocytes and CD34+ cells, we showed that quiescent cells do not contain many proteins required for cell proliferation (eg, DNA synthesis, mitosis) or molecules that regulate cell cycle.43-47 Expression of mRNA-encoding proteins involved in mitosis, DNA replication, and other proliferation functions suggest that the Treg B subpopulation is more likely to be proliferating, primed to proliferate, or has recently exited the cell cycle. The limitation of this study was the very low number of Tregs in AA patients, which made it technically difficult to individually assess the IL-2 sensitivity and in vitro expandability of patients’ Treg subpopulations. Nonetheless, the observation that IST-responder patients had a significantly higher frequency of Treg B following IST therapy compared with IST nonresponders would support the hypothesis that Treg B cells are more likely to proliferate and perhaps better control the immune response in AA, particularly following treatments with antithymocyte globulin–based immunosuppressive therapy, when the number of T cells is reduced and the ability of Tregs to proliferate would be crucial to reinstate a balanced immune response (Figure 7).
In vitro expansion of Tregs for clinical use is an emerging cellular therapy in graft-versus-host disease, type I diabetes, and organ transplant rejection48-52 ; nevertheless, the quality control of expanded Tregs is a time-consuming procedure. Our data comparing expanded Tregs with preexpansion Treg subpopulations could provide a robust and quick screening for quality control of cell therapy products and may serve as a predictive tool for expandability of Tregs in AA and other autoimmune diseases.
In summary, we have shown for the first time that a novel strategy for multidimensional deep phenotyping can reliably identify an immune signature for AA based on Treg subpopulations. This approach also identifies an immune signature that predicts for response to IST at time of diagnosis of AA and that may allow a more patient-specific approach to future treatment decision-making in severe AA. Our findings also pave the way for future novel therapeutic approaches such as expanded autologous Tregs and low-dose IL-2 in AA.
Global gene expression (GEP) data of this manuscript is deposited at gene expression omnibus (accession number GSE78004).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank King’s College London Genomic Centre for performing gene expression experiments; in particular, Matthew Arno. The authors also thank the hematology department tissue bank for processing patient samples and are grateful to Giovanna Lombardi, Cristiano Scotta, Behdad Afzali, and Claudia Kemper for their scientific input.
This work is supported by financial support from the Department of Health via the National Institute for Health Research comprehensive Biomedical Research Centre Award to Guy's & St Thomas' National Health Service Foundation Trust in partnership with King's College London and King's College Hospital National Health Service Foundation Trust. This work is also supported by a research program grant from Bloodwise UK (grant number 14017) and a grant from The Aplastic Anemia and MDS International Foundation, USA (research grant awarded to S.K.).
Authorship
Contribution: S.K. designed, supervised, and performed experiments; analyzed and interpreted data; and wrote the paper. B.C. and T.S. designed and performed experiments, analyzed and interpreted data, and wrote the paper. P.P.A. validated metal-tagged antibodies. M.M.L. analyzed and interpreted the global gene expression (GEP) data and contributed in writing the paper. D.M. provided clinical data and contributed in writing the paper. K.E.D. contributed in data analysis. A.K. provided clinical data. C.B. performed experiments. X.F. provided clinical samples and data. A.S. analyzed data. S.A.M. performed experiments. R.M. provided bioinformatics and statistical analysis. E.d.R. supervised the bioinformatics and statistical analysis. S.H. provided mass cytometry data quality control and contributed in writing the paper; R.E. provided mass cytometry data acquisition and quality control. N.P. provided bioinformatics and statistical analysis. G.A.M.P. provided GEP data analysis. S.S.C. provided cell-cycle–related bioinformatics data analysis. N.S.B.T. provided cell-cycle–related data analysis and contributed in writing the paper; F.F. contributed in writing the paper. J.M.I. contributed in data analysis, interpreting data, and in writing the paper. N.S.Y. provided clinical samples and data and contributed in writing the paper. J.C.W.M. provided clinical samples and data, supervised the project, interpreted the results, and contributed in writing the paper. G.J.M. supervised the project, interpreted the results, and wrote the paper.
Conflict-of-interest disclosure: J.M.I. is a cofounder and board member at Cytobank Inc. The remaining authors declare no competing financial interests.
Correspondence: Ghulam J. Mufti, Department of Haematological Medicine, King’s College London, Rayne Institute, 123 Coldharbour Ln, London SE5 9NU, United Kingdom; e-mail: ghulam.mufti@kcl.ac.uk.
References
Author notes
S.K., B.C., and T.S. contributed equally to this study.