Key Points
Data from Dnmt3a−/− mice implicate Dot1l as a critical mediator of the malignant gene expression program of Dnmt3a-mediated leukemia.
Pharmacologic inhibition of DOT1L exerts potent antileukemic activity in DNMT3A-mutant human acute myeloid leukemia in vitro and in vivo.
Abstract
Mutations in DNA methyltransferase 3A (DNMT3A) are common in acute myeloid leukemia and portend a poor prognosis; thus, new therapeutic strategies are needed. The likely mechanism by which DNMT3A loss contributes to leukemogenesis is altered DNA methylation and the attendant gene expression changes; however, our current understanding is incomplete. We observed that murine hematopoietic stem cells (HSCs) in which Dnmt3a had been conditionally deleted markedly overexpress the histone 3 lysine 79 (H3K79) methyltransferase, Dot1l. We demonstrate that Dnmt3a−/− HSCs have increased H3K79 methylation relative to wild-type (WT) HSCs, with the greatest increases noted at DNA methylation canyons, which are regions highly enriched for genes dysregulated in leukemia and prone to DNA methylation loss with Dnmt3a deletion. These findings led us to explore DOT1L as a therapeutic target for the treatment of DNMT3A-mutant AML. We show that pharmacologic inhibition of DOT1L resulted in decreased expression of oncogenic canyon–associated genes and led to dose- and time-dependent inhibition of proliferation, induction of apoptosis, cell-cycle arrest, and terminal differentiation in DNMT3A-mutant cell lines in vitro. We show in vivo efficacy of the DOT1L inhibitor EPZ5676 in a nude rat xenograft model of DNMT3A-mutant AML. DOT1L inhibition was also effective against primary patient DNMT3A-mutant AML samples, reducing colony-forming capacity (CFC) and inducing terminal differentiation in vitro. These studies suggest that DOT1L may play a critical role in DNMT3A-mutant leukemia. With pharmacologic inhibitors of DOT1L already in clinical trials, DOT1L could be an immediately actionable therapeutic target for the treatment of this poor prognosis disease.
Introduction
Mutations of the de novo DNA methyltransferase DNA methyltransferase 3A (DNMT3A) occur in approximately 20% of all adult patients with acute myeloid leukemia (AML). Studies indicate that patients with DNMT3A mutations suffer particularly poor prognoses, indicating novel therapies are needed.1-4 DNMT3A mutations in AML are almost exclusively heterozygous, and approximately 60% affect the arginine at amino acid position 882 (R882) in the methyltransferase domain. R882-mutant DNMT3A is a hypomorphic protein that also inhibits the remaining WT DNMT3A, dramatically reducing cellular DNA methyltransferase activity5,6 ; however, the exact mechanisms by which DNMT3A loss contributes to leukemogenesis are poorly understood. DNA methylation profiling of DNMT3A-mutant AML samples revealed loci with decreased methylation, but, surprisingly, also a small subset of loci with increased methylation.1,4,7,8 These data suggest the pathologic changes in DNA methylation are mediated by additional, unknown factors.
Conditional ablation of Dnmt3a in the murine hematopoietic system results in a dramatic expansion of hematopoietic stem cells (HSCs), a progressive block in differentiation,9 and priming for malignant transformation.10,11 Whole genome bisulfite sequencing of Dnmt3a−/− HSCs revealed that the borders of expansive undermethylated regions, termed methylation canyons, are hotspots for DNA methylation loss, which leads to expansion of the canyon.12 Canyons that expand with Dnmt3a deletion are highly enriched for genes dysregulated in human leukemia, including HOX genes,12 suggesting these sites are important in leukemogenesis. Analysis of The Cancer Genome Atlas data confirmed many of these sites have methylation loss in DNMT3A-mutant AML8,12 ; and many canyon-associated genes, including HOX genes, are significantly changed in DNMT3A-mutant AML.4,8,12
In addition to the DNA methylation changes in Dnmt3a−/− HSCs, chromatin immunoprecipitation (ChIP)-sequencing (seq) and RNA-seq data revealed evidence of perturbations of histone modifications. Given the known functional interaction between DNA methylation and histone modifications, these alterations were intriguing.13-16 The observed overexpression of the histone 3, lysine 79 (H3K79) methyltransferase, Dot1l (disrupter of telomere silencing 1-like), was especially interesting because it plays a critical role in leukemia with MLL rearrangements.17-20 Pharmacologic inhibition of DOT1L has shown promising preclinical activity in MLL-rearranged leukemia and is now being tested in adult and pediatric clinical trials.21-25 MLL rearrangements rarely cooccur with DNMT3A mutations in AML.3,4,7 The essential mutual exclusivity7 of these lesions and the overexpression of Dot1l in our murine model led us to hypothesize that MLL rearrangements and DNMT3A mutations are distinct epigenetic aberrations that converge on a common mechanism, resulting in dysregulated gene expression mediated by H3K79 methylation. We therefore explored the role of DOT1L in DNMT3A-mediated leukemia and evaluated DOT1L as a therapeutic target for the treatment of this disease with a poor prognosis.
Methods
Murine model
Animal procedures were approved by the Animal Care and Use Committee of Baylor College of Medicine. For ChIP-seq experiments, C57Bl/6 CD45.2 Dnmt3afl/fl and Dnmt3aWT/WT were crossed to Rosa26-Cre ERT2.26 Dnmt3afl/fl-Rosa26-Cre ERT2 and Dnmt3aWT/WT-Rosa26-Cre ERT2 mice were treated with 5 daily intraperitoneal injections of tamoxifen (1 mg/0.1 mL corn oil per mouse per day) to induce deletion of the floxed Dnmt3a allele. This typically results in a >80% biallelic deletion as determined by genotyping of individual MethoCult colonies from the bone marrow of Dnmt3afl/fl-Rosa26-Cre ERT2 after tamoxifen treatment. Eight weeks later, bone marrow was harvested and transplanted (1 × 106 per mouse) into lethally irradiated syngeneic CD45.1 recipients. Additional details of murine experiments are provided in the supplemental Methods, available on the Blood Web site.
ChIP-seq
Four months after transplantation, recipient mice were euthanized, and pooled bone marrow HSCs from Dnmt3a−/− and control mice were purified using c-Kit magnetic enrichment (AutoMACS; Miltenyi Biotec) followed by gating on live cells and sorting for lineage−, Sca1+, CD48−, and CD150+ cells (FACSAria; BD Biosciences; antibodies from Becton Dickinson). ChIP-seq was performed on purified HSCs after chromatin crosslinking with 1% formalin followed by cell lysis in sodium dodecyl sulfate buffer. DNA was fragmented by sonication and ChIP performed using anti-H3K79me2 (ab3594; Abcam). Eluted DNA was used to prepare a library (Illumina ChIP-seq kit) and then sequenced on an Illumina HiSequation (100-base paired-end). Raw reads were quality trimmed (Trimgalore) and mapped (mm9) (Bowtie 2.0.6). See the supplemental Methods for details of analyses performed. Reviewers may access a track hub for H3K79me2 in the mouse genome (mm9) at: http://genome.ucsc.edu/cgi-bin/hgTracks?db5mm9&hubUrl5http://dldcc-web.brc.bcm.edu/lilab/benji/RaRau/k79.hub.txt.
Cell culture and reagents
Human leukemia cell lines OCI AML3 and OCI AML2 were provided by Mark Minden (Ontario Cancer Institute). Cells were grown in RPMI-1640 (Invitrogen) plus 10% fetal bovine serum, 1% l-glutamine, and 1% pen strep at 37°C in 5% carbon dioxide. Cell lines were validated by the short tandem repeat method. For in vitro experiments, we used DOT1L inhibitors SYC-52221 and EPZ00477723 (Epizyme, Inc). For in vivo studies, EPZ-567622 (Epizyme, Inc.) was used.
Cell proliferation, viability, and colony formation assays
Exponentially growing cells were plated in triplicate in 24-well plates (2 × 105/mL; final volume 1 mL). For dose-dependent assays, cells were incubated in increasing concentrations of SYC52221 or EPZ00477723 or dimethyl sulfoxide (DMSO) control. For time-dependent assays, cells were incubated in 3 μM EPZ00477723 or DMSO control. Every 2 to 3 days, media and compound were replaced and cells were split to 2 × 105/mL. At each replating, the viable cell number was determined (trypan blue). Total cell number is expressed as split-adjusted viable cells per milliliter. Analysis of apoptosis, cell-cycle, and cellular differentiation and gene expression changes were performed as described in the supplemental Methods. Viably frozen primary AML samples (obtained from MD Anderson Cancer Center or Texas Children’s Cancer Center under institutional review board–approved protocols) were thawed quickly and placed in fresh RPMI 1640 plus 10% fetal bovine serum, 1% l-glutamine, and 1% pen strep. After recovery for 2 to 3 hours, viable cells were counted and plated in triplicate (5000 viable cells/plate) in methylcellulose media (H4034; StemCell Technologies) with 3 μM EPZ00477 or DMSO vehicle control. After 12 to 14 days, plates were scored for colony number and morphology. Cells were isolated from plates; stained for CD45, CD14, CD13; and analyzed by flow cytometry. Cell morphology was examined by hematoxylin and eosin staining of cytospins (Cytopro).
Nude rat xenografts
In vivo studies were conducted after review by the Animal Care and Use Committee at Charles River Discovery Research Services (Durham, NC). OCI-AML3 cells were implanted subcutaneously into the right flank of female athymic nude rats (Hsd:RH-Foxn1rnu, Harlan Laboratories, Inc.). EPZ-5676 was delivered by continuous IV infusion via a catheter surgically implanted in the femoral vein of each rat. Animals were separated into either an efficacy or pharmacokinetic/pharmacodynamic cohort. Both cohorts were dosed by continuous IV infusion with 35 or 70 mg/kg per day of EPZ-5676. A control group received continuous IV infusion of the vehicle 5% hydroxypropyl-β-cyclodextrin in saline. Efficacy was determined after 21 days of drug treatment followed by a 7-day drug holiday. Animals assessed for pharmacokinetics/pharmacodynamics were dosed for 14 days and euthanized following the completion of infusion. Rats were weighed and tumors calipered twice weekly. At the completion of the study, animals were euthanized and tumor tissue collected in a ribonuclease-free environment, bisected, snap frozen in liquid nitrogen, pulverized, and stored at −80°C. Effects on H3K79 methylation, enzyme-linked immunosorbent assay, and gene expression by quantitative reverse transcription polymerase chain reaction (qRT-PCR) were performed (supplemental Methods).
Statistics
Student t test and 1-way analysis of variance were used for statistical comparisons where appropriate.
Results
Dot1l messenger RNA expression and H3K79 methylation are increased in Dnmt3a−/− HSCs
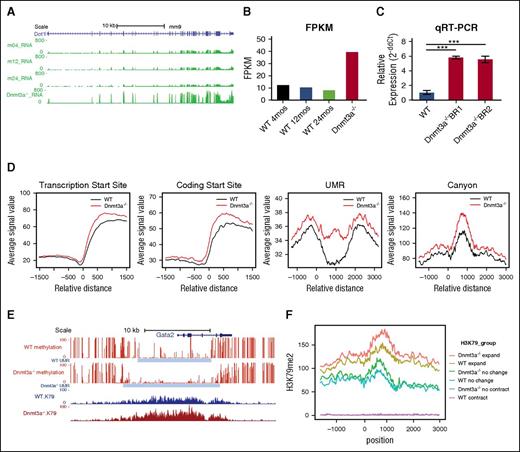
Reanalysis of previously performed RNA-seq of Dnmt3a−/− HSCs27 (Hoechst side population-KSL CD150+ after Mx1-Cre–mediated deletion and serial transplantation) revealed that Dot1l was overexpressed in the Dnmt3a−/− relative to WT HSCs isolated from mice of various ages (Figure 1A-B). Dot1l overexpression was confirmed by qRT-PCR of 2 biologic replicates of purified Dnmt3a−/− and WT HSCs (Figure 1C). In addition, modest reduction of DNA methylation and increased H3K79me2 density at the Dot1l promoter suggest that increased expression of Dot1l in this model may be attributable to altered epigenetic regulation (supplemental Figure 1A).
Dnmt3a−/−HSCs are characterized by increased Dot1l expression and increased H3K79 methylation. (A) RNA-seq signal density tracks of messenger RNA expression of Dot1l in murine Dnmt3a−/− HSCs purified as Hoechst side population-KSL and CD150+. Dnmt3a−/− HSCs were purified after Mx-Cre–mediated deletion and serial transplantation as have previously reported9,27 compared with HSCs from WT HSCs from 4-, 12-, and 24-month-old mice (m04_RNA, m12_RNA, and m24_RNA, respectively). (B) Average FPKM (fragments per kilobase of transcript per million mapped reads) value of Dot1L in WT vs Dnmt3a−/− HSCs (2 independently obtained biological replicates of each cohort represented). mos, months. (C) Dot1l expression determined by qRT-PCR relative to glyceraldehyde-3-phosphate dehydrogenase expression in WT HSCs (12 months of age) compared with 2 biologic replicates of Dnmt3a−/− HSCs purified after Mx-Cre–mediated deletion and serial transplantation (calculated by 2-ΔΔCt equation). Assay performed in triplicate. Error bars represent standard deviation. (D) ChIP-seq of H3K79me2 of Dnmt3afl/fl-Rosa26-Cre-ERT2 and Dnmt3awt/wt-Rosa26-Cre-ERT2 HSCs isolated from primarily transplanted mice after tamoxifen-induced deletion. Average normalized signal density of H3K79me2 at transcription start sites, protein coding start sites, undermethylated regions (UMR) and DNA methylation canyons in Dnmt3a−/− HSCs (red) and WT HSCs (black). (D) Representative DNA methylation canyon that expands with Dnmt3a deletion (DNA methylation, red; canyon extend, gray) and associated H3K79me2 in WT HSCs (blue) and Dnmt3a−/− HSCs (dark red). (E) Average normalized H3K79me2 signal at DNA methylation canyons that expand with Dnmt3a deletion (Dnmt3a−/− expand, red; WT expand, gold), canyons that do not change with Dnmt3a deletion (Dnmt3a−/− no change, green; WT no change, teal), and canyons that contract with Dnmt3a deletion (Dnmt3a−/− contract, blue; WT contract, purple). P value was determined using unpaired 2-way Student t test. ***P < .001.
Dnmt3a−/−HSCs are characterized by increased Dot1l expression and increased H3K79 methylation. (A) RNA-seq signal density tracks of messenger RNA expression of Dot1l in murine Dnmt3a−/− HSCs purified as Hoechst side population-KSL and CD150+. Dnmt3a−/− HSCs were purified after Mx-Cre–mediated deletion and serial transplantation as have previously reported9,27 compared with HSCs from WT HSCs from 4-, 12-, and 24-month-old mice (m04_RNA, m12_RNA, and m24_RNA, respectively). (B) Average FPKM (fragments per kilobase of transcript per million mapped reads) value of Dot1L in WT vs Dnmt3a−/− HSCs (2 independently obtained biological replicates of each cohort represented). mos, months. (C) Dot1l expression determined by qRT-PCR relative to glyceraldehyde-3-phosphate dehydrogenase expression in WT HSCs (12 months of age) compared with 2 biologic replicates of Dnmt3a−/− HSCs purified after Mx-Cre–mediated deletion and serial transplantation (calculated by 2-ΔΔCt equation). Assay performed in triplicate. Error bars represent standard deviation. (D) ChIP-seq of H3K79me2 of Dnmt3afl/fl-Rosa26-Cre-ERT2 and Dnmt3awt/wt-Rosa26-Cre-ERT2 HSCs isolated from primarily transplanted mice after tamoxifen-induced deletion. Average normalized signal density of H3K79me2 at transcription start sites, protein coding start sites, undermethylated regions (UMR) and DNA methylation canyons in Dnmt3a−/− HSCs (red) and WT HSCs (black). (D) Representative DNA methylation canyon that expands with Dnmt3a deletion (DNA methylation, red; canyon extend, gray) and associated H3K79me2 in WT HSCs (blue) and Dnmt3a−/− HSCs (dark red). (E) Average normalized H3K79me2 signal at DNA methylation canyons that expand with Dnmt3a deletion (Dnmt3a−/− expand, red; WT expand, gold), canyons that do not change with Dnmt3a deletion (Dnmt3a−/− no change, green; WT no change, teal), and canyons that contract with Dnmt3a deletion (Dnmt3a−/− contract, blue; WT contract, purple). P value was determined using unpaired 2-way Student t test. ***P < .001.
Given the aberrant expression of this histone methyltransferase, we examined whether Dot1L-induced H3K79 methylation was also altered in Dnmt3a−/− HSCs compared with WT controls and if these alterations were associated with altered DNA methylation. We previously reported that the edges of large undermethylated regions, termed DNA methylation canyons, are hotspots for DNA methylation loss in Dnmt3a−/− HSCs. However, only a portion of these canyons lose methylation and expand with Dnmt3a loss, and a close association between canyon DNA methylation changes and the associated histone marks was identified.12 Expanding canyons are characterized by the presence of the activating H3K4 tri-methyl (me3) mark and absence of the repressive histone mark H3K27me3,12 suggesting that Dnmt3a is particularly important in maintaining DNA methylation specifically at canyons with activating histone marks and active gene transcription. We speculated that H3K79me may be another key component of this activating histone signature. To determine if DOT1L-induced H3K79me was altered in Dnmt3a−/− HSCs, we performed ChIP-seq for H3K79me2 on Dnmt3afl/fl-Rosa26-Cre ERT2 and Dnmt3awt/wt-Rosa26-Cre ERT2 HSCs isolated from transplanted mice after tamoxifen-induced deletion. The H3K79me2 data were aligned with existing whole genome DNA methylation data from Dnmt3a−/− HSCs,12 revealing that levels of H3K79me2 were markedly increased at transcription start sites, protein coding start sites, and at undermethylated regions) (Figure 1D). Importantly, a substantial increase in signal intensity was noted, particularly at DNA methylation canyons (Figure 1D; supplemental Figure 1B-D). We then looked at the association between H3K79me2 and canyon dynamics with Dnmt3a loss. We found that H3K79me2 is found at very low levels in canyons that show little change or increased methylation after Dnmt3a loss; however, H3K79me2 densely coats canyons that expand when Dnmt3a is ablated, such as the canyon associated with the Gata2 gene (Figure 1E-F). This strong correlation between H3K79me and altered DNA methylation suggests a functional interaction.
DOT1L-induced H3K79 methylation is increased in DNMT3A-mutant AML
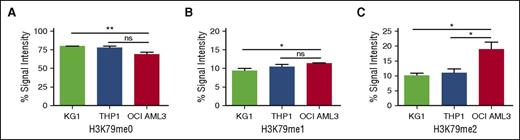
Based on our murine findings, we postulated that DOT1L might play a role in human DNMT3A-mutant AML. To explore this hypothesis, we examined the relative methylation of H3K79 of the DNMT3A-mutant human cell line OCI AML3, which harbors the most common and well-characterized type of DNMT3A mutation, the dominant-negative acting R882 mutation5,6 compared with the MLLr cell line, THP1, and KG-1 cells that have WT DNMT3A and MLL. Mass spectrometry demonstrated that OCI AML3 DNMT3A-mutant cells had decreased unmethylated H3K79 and increased H3K79me2 in the compared with DNMT3A WT cells (Figure 2A-C; supplemental Figure 2). These results are consistent with the increased H3K79me density observed in our murine Dnmt3a−/− model, despite the fact that DOT1L expression at the messenger RNA level was not increased in this cell line relative to DNMT3A WT AML cell lines (not shown).
DOT1L-induced H3K79 methylation is increased in DNMT3A-mutant human AML. Relative level of (A) unmethylated H3K79, (B) monomethylated H3K79, and (C) dimethylated H3K79 measured by mass spectrometry in 3 AML cell lines (KG1, THP1, and OCI AML3). Data are from a single biological replicate and 3 to 4 technical replicates. Error bars show standard deviation. P value was determined using unpaired 2-way Student t test. **P < .01; *P < .05. ns, not statistically significant.
DOT1L-induced H3K79 methylation is increased in DNMT3A-mutant human AML. Relative level of (A) unmethylated H3K79, (B) monomethylated H3K79, and (C) dimethylated H3K79 measured by mass spectrometry in 3 AML cell lines (KG1, THP1, and OCI AML3). Data are from a single biological replicate and 3 to 4 technical replicates. Error bars show standard deviation. P value was determined using unpaired 2-way Student t test. **P < .01; *P < .05. ns, not statistically significant.
Pharmacologic DOT1L inhibition reduces cellular H3K79me and decreases the proliferation of DNMT3A-mutant AML cells in a dose- and time-dependent fashion
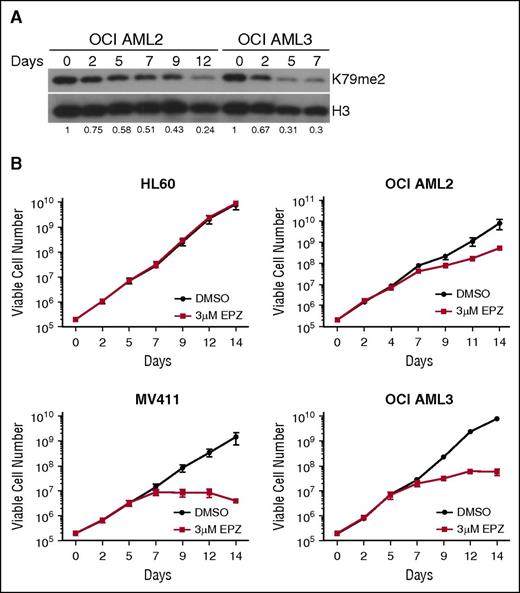
To explore H3K79 methylation as a potential therapeutic target in DNMT3A-mutant AML, we tested the efficacy of pharmacologic DOT1L inhibition in vitro using 2 specific DOT1L inhibitors with comparable potency and specificity: SYC-52221 and EPZ004777.23 We treated the only known human cell lines with DNMT3A mutations, R882 mutant OCI AML3, and OCI AML2 cells which have a functionally uncharacterized non-R882 mutation (and a possible cryptic MLL rearrangement28 ), with SYC-522 or EPZ004777. DOT1L inhibitor treatment resulted in a dose- and time-dependent reduction in H3K79me2 in both cell lines (supplemental Figure 3; Figure 3A, respectively).
Pharmacologic DOT1L inhibition reduces cellular H3K79me2 and suppresses proliferation of DNMT3A-mutant human AML cell lines. (A) Immunoblot analysis of cellular H3K79me2 in OCI AML2 and OCI AML3 cells after treatment with 3 μM EPZ004777. Fraction of H3K79me2/total H3 relative to DMSO-treated control is indicated below each lane. (B) Growth curves of HL60, MV411, OCI AML2, and OCI AML3 cells treated with 3 μM EPZ004777 or vehicle control for 14 days. Numbers are plotted on logarithmic scale. Assays were done in triplicate. Error bars represent standard deviation.
Pharmacologic DOT1L inhibition reduces cellular H3K79me2 and suppresses proliferation of DNMT3A-mutant human AML cell lines. (A) Immunoblot analysis of cellular H3K79me2 in OCI AML2 and OCI AML3 cells after treatment with 3 μM EPZ004777. Fraction of H3K79me2/total H3 relative to DMSO-treated control is indicated below each lane. (B) Growth curves of HL60, MV411, OCI AML2, and OCI AML3 cells treated with 3 μM EPZ004777 or vehicle control for 14 days. Numbers are plotted on logarithmic scale. Assays were done in triplicate. Error bars represent standard deviation.
Both compounds inhibited growth in a dose-dependent fashion, with more pronounced effects in the OCI AML3 cells compared with the OCI AML2 cells (supplemental Figures 4 and 5). To fully analyze the time-dependent impact of DOT1L inhibitor treatment on DNMT3A-mutant AML cells, we performed proliferation assays for 14 days, treating the DNMT3A-mutant cell lines with 3 μM EPZ004777 or vehicle control. We also included the DOT1L inhibitor-sensitive MLLr cell line, MV411, as a positive control, and the MLL- and DNMT3A-WT cell line, HL60, as a negative control. There was no impact on the growth of the HL60 cells and modest slowing of growth of the OCI AML2 cells, whereas the proliferation of the OCI AML3 cells was profoundly inhibited from around 7 days of treatment, comparable to effects in the MV411 cell line (Figure 3B). These results indicate that inhibition of DOT1L effectively suppresses the growth of DNMT3A-mutant cells.
Treatment with pharmacologic inhibitors of DOT1L induces apoptosis, cell-cycle arrest, and terminal differentiation of DNMT3A-mutant AML cell lines
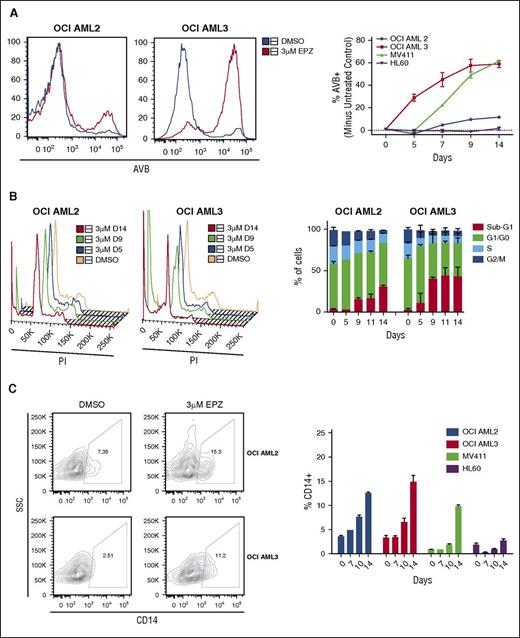
We next sought to determine the specific mechanism of DOT1L inhibitor–induced cytotoxicity in DNMT3A-mutant AML cells. By annexin-V binding (AVB) flow cytometry assay, DOT1L inhibition with either SYC-522 or EPZ004777 led to a dose-dependent induction of apoptosis in both OCI AML2 and OCI AML3 cells, though higher doses were required in the OCI AML2 cells to achieve significant apoptosis (supplemental Figures 5B and 6). Treatment with 3 μM EPZ004777 led to substantial induction of apoptosis in the OCI AML3 cells in a time-dependent fashion beginning around 5 days of treatment, earlier than observed in the MV411 cell line (Figure 4A). The HL60 cells had no induction of apoptosis, and the OCI AML2 cells experienced minimal induction of apoptosis (Figure 4A). We also examined the impact of DOT1L inhibitor treatment on cell-cycle kinetics by flow cytometry for DNA content. Both OCI AML2 and OCI AML3 cells experienced cell-cycle arrest with increased percentages of cells in sub-G1 and decreased percentages in S and G2/M phase in a dose- and time-dependent fashion, with the greatest percentage in the OCI AML3 cells (supplemental Figure 6B; Figure 4B, respectively). Additionally, both OCI AML2 and OCI AML3 cell lines showed evidence of induced differentiation with increased expression of the mature monocyte marker CD14, equivalent to the effects of DOT1L inhibition seen in the MV411 cells (Figure 4C).
EPZ004777 treatment induces apoptosis, cell-cycle arrest, and terminal differentiation in DNMT3A-mutant human AML cells. HL60, MV411, OCI AML2, and OCI AML3 cells were treated with 3 μM EPZ004777 or DMSO vehicle control for 14 days. Cells were replated at a constant concentration in fresh drug-containing media every 2 to 3 days. (A) Representative flow cytometry histograms of AVB for OCI AML2 and OCI AML3 cells on day 14 of treatment with DMSO vehicle control (blue) or EPZ004777 (red) (left). Quantification of % of EPZ004777-treated cells that are AVB+ minus % AVB+ vehicle control–treated cells (right). (B) Representative flow cytometry plots of propidium iodide (PI) DNA content cell-cycle analysis for OCI AML2 cells and OCI AML3 cells (left) with quantification of experiments (right) (C) Representative flow plots of CD14 cell surface expression of OCI AML 2 and OCI AML 3 cells treated with DMSO vehicle control or EPZ004777 for 14 days after gating out PI+ dead cells (left) and quantification of percentage of CD14+ cells treated with 3 μM EPZ004777 or vehicle control at specified time points (right). All assays were done in triplicate. Error bars represent standard deviation. SSC, side scatter.
EPZ004777 treatment induces apoptosis, cell-cycle arrest, and terminal differentiation in DNMT3A-mutant human AML cells. HL60, MV411, OCI AML2, and OCI AML3 cells were treated with 3 μM EPZ004777 or DMSO vehicle control for 14 days. Cells were replated at a constant concentration in fresh drug-containing media every 2 to 3 days. (A) Representative flow cytometry histograms of AVB for OCI AML2 and OCI AML3 cells on day 14 of treatment with DMSO vehicle control (blue) or EPZ004777 (red) (left). Quantification of % of EPZ004777-treated cells that are AVB+ minus % AVB+ vehicle control–treated cells (right). (B) Representative flow cytometry plots of propidium iodide (PI) DNA content cell-cycle analysis for OCI AML2 cells and OCI AML3 cells (left) with quantification of experiments (right) (C) Representative flow plots of CD14 cell surface expression of OCI AML 2 and OCI AML 3 cells treated with DMSO vehicle control or EPZ004777 for 14 days after gating out PI+ dead cells (left) and quantification of percentage of CD14+ cells treated with 3 μM EPZ004777 or vehicle control at specified time points (right). All assays were done in triplicate. Error bars represent standard deviation. SSC, side scatter.
Gene expression analysis after pharmacologic DOT1L inhibition
To assess the effects of DOT1L inhibition on gene expression, RNA-seq was performed on OCI AML2 and OCI AML3 cells after treatment with EPZ004777. We specifically probed canyon-associated genes and found that almost all differentially expressed genes had a significantly reduced expression with treatment (Figure 5A). Although a small subset of canyon-associated genes increased in expression, these canyons characteristically lacked H3K79me2 in our murine model. To validate the RNA-seq results, qRT-PCR was performed for specific HOX genes and MEIS1, confirming that expression of MEIS1 and HOX cluster genes was suppressed in both the OCI AML2 and OCI AML3 cells after treatment (Figure 5B). HOX B cluster genes were preferentially suppressed in the OCI AML3 cells, consistent with the data from patient cohorts showing specific overexpression of HOX B cluster genes in DNMT3A-mutant leukemia4,8 (Figure 5B). Additionally, Ingenuity Pathway Analysis of differentially expressed genes showed enrichment of genes involved in cell death, cell-cycle arrest and differentiation (Figure 5C), consistent with the phenotypic consequences of DOT1L inhibitor treatment reported previously. Gene Set Enrichment Analysis (GSEA) of genes upregulated after EPZ treatment demonstrated significant enrichment for genes associated with G1-S transition, including upregulation of cyclin-dependent kinase inhibitor CDKN1A. Strong positive enrichment was further observed for genes involved in myeloid differentiation, including increased expression of lysozyme and myeloperoxidase (Figure 5D-E). There was also significant overlap between upregulated genes in both cell lines after EPZ treatment and genes upregulated by knock-down of HOXA9 in the MLLr AML cell line MOLM14.29 Given the known correlation between suppression of HOXA9 expression and therapeutic effect of DOT1L inhibition in MLLr cells, this overlap highlights the likely important role of programs downstream of HOXA9 in both MLLr leukemia and DNMT3A-mutant leukemia (Figure 5E). This is confounded by a possible MLL fusion in the OCI AML2 cells; however, the overlap between this data set and the gene expression changes in the OCI AML3 cells point toward mechanistic overlap between MLLr leukemia and AML harboring R882 DNMT3A mutations.
Gene expression changes following DOT1L inhibitor treatment of DNMT3A-mutant AML cell lines. (A) Gene expression heat map (gene scaled and centered log2 counts per million) of top differentially expressed canyon-associated genes in OCI AML2 and OCI AML3 cell lines after 14 days of treatment with DOT1L inhibitor. (B) Relative expression determined by qRT-PCR of canyon-associated leukemogenic genes HOXA9, MEIS1, HOXB3, HOXB8, and the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in OCI AML 2 and OCI AML3 cells treated with 3 μM EPZ004777 or vehicle control for 14 days. qRT-PCR done in triplicate. Error bars represent standard deviation. (C) Ingenuity Pathway Analysis functional enrichment analysis of genes differentially expressed (false discovery rate [FDR] q value < 0.1) in AML cells after treatment with EPZ004777. P val, P value; prolif, proliferation. (D) GSEA of AML cells treated with EPZ004777. (E) Gene expression heat maps (gene scaled and centered log2 counts per million) of representative, core-enriched gene sets from GSEA. DN, downregulated; GO, gene ontogeny; inflamm, inflammatory; leuk, leukocyte; mDC, myeloid dendritic cell; meth, DNA methylation; Q-val, Q value; resp, response; signal size, enrichment signal strength of the leading edge subset of a gene set; trans, transition; UP, upregulated.
Gene expression changes following DOT1L inhibitor treatment of DNMT3A-mutant AML cell lines. (A) Gene expression heat map (gene scaled and centered log2 counts per million) of top differentially expressed canyon-associated genes in OCI AML2 and OCI AML3 cell lines after 14 days of treatment with DOT1L inhibitor. (B) Relative expression determined by qRT-PCR of canyon-associated leukemogenic genes HOXA9, MEIS1, HOXB3, HOXB8, and the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in OCI AML 2 and OCI AML3 cells treated with 3 μM EPZ004777 or vehicle control for 14 days. qRT-PCR done in triplicate. Error bars represent standard deviation. (C) Ingenuity Pathway Analysis functional enrichment analysis of genes differentially expressed (false discovery rate [FDR] q value < 0.1) in AML cells after treatment with EPZ004777. P val, P value; prolif, proliferation. (D) GSEA of AML cells treated with EPZ004777. (E) Gene expression heat maps (gene scaled and centered log2 counts per million) of representative, core-enriched gene sets from GSEA. DN, downregulated; GO, gene ontogeny; inflamm, inflammatory; leuk, leukocyte; mDC, myeloid dendritic cell; meth, DNA methylation; Q-val, Q value; resp, response; signal size, enrichment signal strength of the leading edge subset of a gene set; trans, transition; UP, upregulated.
DOT1L inhibition suppresses tumor growth in a nude rat AML xenograft
To test the in vivo efficacy of DOT1L inhibition on human DNMT3A-mutant AML, we used a nude rat xenograft model in which OCI AML3 cells were injected subcutaneously, forming a leukemic tumor. After tumor engraftment, the rats (n = 8 per treatment cohort) were treated with continuous IV infusion of vehicle control or EPZ-5676, a DOT1L inhibitor currently being tested in phase 1 clinical trials that is structurally similar to EPZ004777 but with improved pharmacokinetic properties and increased potency.22 Rats were treated at doses of either 35 mg/kg per day or 70 mg/kg per day via continuous IV infusion for 21 days followed by a 7-day drug holiday. Both doses of EPZ-5676 led to significant reduction of H3K79me2 (Figure 6A) and decreased MEIS1 and HOXB3 expression, consistent with in vitro results (Figure 6B). Tumor growth was inhibited in a dose-dependent fashion reaching statistical significance in the 70 mg/kg per day cohort (Figure 6C). These results indicate that DOT1L inhibition is effective at suppressing tumor growth in an in vivo nude rate xenograft model of DNMT3A-mutant AML. Ideally, treatment of patient-derived xenograft models would be performed to confirm these results; however, given the need for administration of the drug by continuous infusion for prolonged periods, such studies are currently not feasible.
In vivo efficacy of pharmacologic DOT1L inhibition in a rat xenograft model of DNMT3A-mutant AML. (A) H3K79me2 levels in acid-extracted histones as measured by enzyme-linked immunosorbent assay in OCI AML3 subcutaneous tumors and bone marrow from vehicle control–treated animals or animals treated with 35 or 70 mg/kg per day EPZ-5676 administered via continuous IV infusion. H3K79me2 levels were normalized to those of total histone H3 in the same sample and are plotted as a percent of the mean H3K79me2 level in tissue from the vehicle-treated group, which is set at 100%. Horizontal lines represent the mean percent H3K79me2 values for each group (N = 5 animals per cohort). (B) Relative expression of MEIS1 and HOXB3 in OCI AML3 subcutaneous tumors in vehicle control–treated mice and mice treated with 35 or 70 mg/kg per day EPZ-5676 for 21 days plotted as a percent of the mean transcript level in tumors from the vehicle-treated group, which is set at 100%. Horizontal lines represent the mean percent transcript level in each group (N = 5 animals per cohort). (C) Volume of OCI AML3 subcutaneous tumors over time in vehicle control–treated animals and animals treated with 35 or 70 mg/kg per day EPZ-5676 administered via continuous IV infusion for 21 days (N = 8 animals per cohort). Error bars represent standard deviation. CI, continuous infusion.
In vivo efficacy of pharmacologic DOT1L inhibition in a rat xenograft model of DNMT3A-mutant AML. (A) H3K79me2 levels in acid-extracted histones as measured by enzyme-linked immunosorbent assay in OCI AML3 subcutaneous tumors and bone marrow from vehicle control–treated animals or animals treated with 35 or 70 mg/kg per day EPZ-5676 administered via continuous IV infusion. H3K79me2 levels were normalized to those of total histone H3 in the same sample and are plotted as a percent of the mean H3K79me2 level in tissue from the vehicle-treated group, which is set at 100%. Horizontal lines represent the mean percent H3K79me2 values for each group (N = 5 animals per cohort). (B) Relative expression of MEIS1 and HOXB3 in OCI AML3 subcutaneous tumors in vehicle control–treated mice and mice treated with 35 or 70 mg/kg per day EPZ-5676 for 21 days plotted as a percent of the mean transcript level in tumors from the vehicle-treated group, which is set at 100%. Horizontal lines represent the mean percent transcript level in each group (N = 5 animals per cohort). (C) Volume of OCI AML3 subcutaneous tumors over time in vehicle control–treated animals and animals treated with 35 or 70 mg/kg per day EPZ-5676 administered via continuous IV infusion for 21 days (N = 8 animals per cohort). Error bars represent standard deviation. CI, continuous infusion.
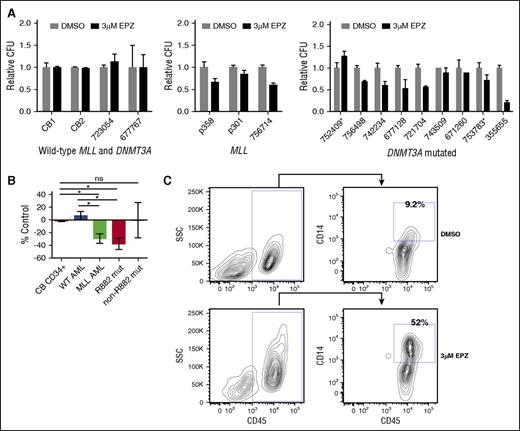
DOT1L inhibitor treatment selectively reduces the CFC and induces differentiation of primary patient samples with DNMT3A mutations
Although our therapeutic experiments using human AML cell lines allowed us to explore the efficacy of DOT1L inhibition in DNMT3A-mutant leukemia, given the potential confounding variables inherent to cell lines, we sought to test our hypothesis using primary patient samples. Viably frozen human primary samples were plated in methylcellulose media plus 3 μM EPZ00477 or DMSO vehicle control (patient sample characteristics provided in supplemental Table 2). Samples with MLL aberrations and most samples with DNMT3A mutations had reduced CFC, whereas normal human cord blood CD34+ cells and primary AML patient samples lacking MLL aberration and DNMT3A mutation had no change with treatment (Figure 7A). Given potential functional differences of R882 vs non-R882 mutations, we examined DNMT3A-mutant samples by mutation type. The average CFC of R882-mutant samples was significantly reduced compared with AML samples WT for both DNMT3A and MLL and normal CD34+ cord blood cells, similar to the average reduced CFC of MLL-aberrant samples (Figure 7B). Only 2 samples with non-R882 DNMT3A mutation were analyzed; therefore, no definitive conclusions about the responsiveness of this genotype can be reached.
DOT1L inhibitor treatment selectively inhibits in vitro growth and induces terminal differentiation of primary patient samples with DNMT3A mutations. (A) Relative colony-forming units (CFU) of normal cord blood CD34+ cells and primary AML samples wild-type for both DNMT3A and MLL (left), primary AML samples with MLL anomalies (middle), and primary AML samples with DNMT3A mutations (right). *Non-R882 DNMT3A mutation treated with DMSO vehicle control or 3 μM EPZ004777. Assays performed in triplicate; error bars represent standard error of the mean. (B) Average change in CFC of primary patient samples treated with 3 μM EPZ004777 compared with DMSO vehicle–treated control. Error bars represent standard error of the mean. (C) Flow cytometry analysis of CD14 expression of primary AML cells with DNMT3A mutation isolated from plates after treatment with DMSO vehicle control or 3 μM EPZ004777. P value was determined using unpaired 2-way Student t test. *P < .05. ns, not statistically significant.
DOT1L inhibitor treatment selectively inhibits in vitro growth and induces terminal differentiation of primary patient samples with DNMT3A mutations. (A) Relative colony-forming units (CFU) of normal cord blood CD34+ cells and primary AML samples wild-type for both DNMT3A and MLL (left), primary AML samples with MLL anomalies (middle), and primary AML samples with DNMT3A mutations (right). *Non-R882 DNMT3A mutation treated with DMSO vehicle control or 3 μM EPZ004777. Assays performed in triplicate; error bars represent standard error of the mean. (B) Average change in CFC of primary patient samples treated with 3 μM EPZ004777 compared with DMSO vehicle–treated control. Error bars represent standard error of the mean. (C) Flow cytometry analysis of CD14 expression of primary AML cells with DNMT3A mutation isolated from plates after treatment with DMSO vehicle control or 3 μM EPZ004777. P value was determined using unpaired 2-way Student t test. *P < .05. ns, not statistically significant.
Treatment with EPZ004777 also induced differentiation of DNMT3A-mutant samples evidenced by increased expression of the mature monocyte marker CD14 compared with vehicle-treated controls, including 1 sample that did not have a significant reduction in CFC (Pt 743509) (Figure 7C; supplemental Figure 7A). Histologic evaluation of the isolated cells also showed evidence of differentiation with a reduced nuclear to cytoplasmic ratio, increased granules in the cytoplasm, and condensation of the nuclei (supplemental Figure 7B). These results indicate that pharmacologic DOT1L inhibition reduces cellular proliferation and promotes differentiation of primary AML patient samples with DNMT3A mutations.
Discussion
DOT1L plays a critical role in leukemia with certain MLL rearrangements17-20,30 and is being explored as a therapeutic target for patients with these genetic alterations.21-25 Here, we report a possible role for DOT1L in leukemias with mutations of DNMT3A. Given the high prevalence of DNMT3A mutations across a variety of hematologic malignancies and that in most clinical studies DNMT3A mutations are associated with a particularly poor prognosis,1,3,4,31-33 identifying a novel therapeutic target is of substantial clinical impact. Our data suggest that DOT1L may be an immediately actionable target in DNMT3A-mutated AML.
In many MLLr leukemias, DOT1L is believed to contribute to leukemogenesis via the fusion partners, including AF9, AF10, and ENL, which normally interact with DOT1L.17,18,20 The fusions lead to aberrant recruitment of DOT1L and the H3K79me mark to the promoters of MLL target genes such as MEIS1 and HOX cluster genes, ultimately leading to their constitutive expression. However, recent evidence indicates that DOT1L and its cofactor AF10 may play a critical role in regulating HOX gene expression via H3K79me2 in a subset of additional leukemias without such MLL fusions, including AMLs with partial tandem duplication of MLL, NUP98-NSD1 fusions, and IDH mutations.18,30,34,35 We have identified DNMT3A-mutant AML as an additional distinct subset of leukemia in which DOT1L may contribute to leukemogenesis via MLL fusion-independent mechanisms.
Using our murine Dnmt3a−/− model, we identified a potential functional interaction between H3K79me2 and altered DNA methylation that may explain the site specification in DNMT3A-mutant AML. In murine HSCs, H3K79me2 is highly enriched in DNA methylation canyons, and is most prominent in canyons that expand with Dnmt3a loss. Expanding canyons coated by H3K79me2 are highly enriched for genes aberrantly expressed in hematologic malignancies, including HOX cluster gens.12 We previously reported that these sites are also characterized by the activating histone mark H3K4 me3 and lack of the repressive H3K27 me3 mark.12 We expect H3K79me2 is 1 component of an activating histone signature that dictates where in the genome DNMT3A is most critical for maintaining DNA methylation. When DNMT3A function is lost by deletion in a murine model or mutation in human AML, DNA methylation at these sites is eroded. We have further shown that in the Dnmt3a−/− HSC hypomethylated canyons gain additional H3K79 methylation. These observed perturbations of both DNA methylation and covalent histone modifications including H3K79 methylation may contribute to the aberrant expression of the associated genes, including HOX cluster genes. Indeed, when we treated DNMT3A-mutant AML cell lines with DOT1L inhibitor, we observed a reduction in the expression of genes associated with expanding H3K79me2-coated canyons including HOX and MEIS1 genes, potentially indicating a direct inhibitory effect. Supportive of this possibility is the finding that, in the R882-mutant cell line, the expression of HOX B cluster genes is affected to a greater extent than HOX A cluster genes, contrary to what is seen in MLLr leukemia cell lines. This is consistent with a more extensive loss of DNA methylation in the Hox B cluster–associated DNA methylation canyon compared with the canyon associated with the Hox A cluster in our murine Dnmt3a−/− HSCs12 and more pronounced overexpression of HOX B cluster genes in DNMT3A-mutant AML compared with HOX A cluster genes.4 However, as recent work has demonstrated, HOX gene expression in AML might simply reflect the HOX expression pattern of the HSC/progenitor cell from which the AML arose36 ; therefore, it is possible that the decreased expression of HOX genes observed in response to DOT1L inhibitor therapy reflects differentiation of the cells rather than a direct cause of DOT1L inhibition on gene expression. Furthermore, although in our murine model, Dot1l was overexpressed in Dnmt3a−/− HSCs relative to WT, we did not observe overexpression of DOT1L in human DNMT3A-mutant AML cell lines relative to DNMT3A WT cell lines nor in DNMT3A-mutant patient samples relative to WT in The Cancer Genome Atlas RNA-seq data. Thus, it is possible that DNMT3A-mutant AML may exploit DOT1L by alternative mechanisms such as aberrant recruitment. It is also possible that the therapeutic responses observed in DNMT3A-mutant cell lines and primary patient samples after treatment with DOT1L inhibitor could be attributable to other yet-unidentified factors. Additional work to fully define the mechanistic role of DOT1L in DNMT3A-mutant leukemia is ongoing.
We hypothesized that DOT1L-induced H3K79me might be a therapeutic target in human DNMT3A-mediated hematologic malignancies. Our results support this hypothesis, with pharmacologic DOT1L inhibitors leading to inhibition of cellular proliferation, induction of apoptosis, cell-cycle arrest, and terminal differentiation in DNMT3A-mutant cell lines and primary patient samples. In particular, the effects observed in the R882 DNMT3A-mutant cell line and patient samples were comparable to DOT1L inhibitor-sensitive MLL-rearranged cell lines and patient samples. We observed much less effect in the non-R882 mutant OCI AML2 cells, potentially suggesting there may be differences in response based on biological differences between types of DNMT3A mutations. However, our ability to evaluate the effect of DOT1L inhibitor therapy on non-R882 mutant AML is limited by a lack of available cell lines without additional confounding mutations and a paucity of patient samples with any given non-R882 DNMT3A mutation because no other true mutational hotspots exist.
Although our data indicate that sensitivity of the cell lines to DOT1L inhibition can be conferred by mutant DNMT3A, we cannot exclude contributions from other genetic or epigenetic aberrations. Whereas 1 study suggested that OCI AML2 cells harbor a MLLr,28 they lack cytogenetic evidence of 11q23 anomaly and exhibited only modest antiproliferative response to EPZ treatment. The OCI AML3 cell line, in addition to the DNMT3A mutation, also harbors a nucleophosmin (NPM1) mutation. Both DNMT3A and NPM1 mutations have been associated with overexpression of HOX cluster genes and altered DNA methylation4,6,8,37-39 ; however, as 2 of the most commonly cooccurring mutations in human AML,7 it is difficult to ascertain if these biologic features are attributable to the DNMT3A mutation or the NPM1 mutation—or are secondary to the combined effect of these likely cooperative events. Therefore, although the cell line experiments provided important evidence that DNMT3A-mutant AML might be sensitive to DOT1L inhibition, validation in human primary AML samples was essential. Using several DNMT3A-mutant samples specifically lacking other known confounding mutations, we observed near-universal reductions in CFC and induction of differentiation in the DNMT3A mutant primary AML patient samples consistent with the antiproliferative and pro-differentiation effects observed in our cell lines experiments. Furthermore, our initial data indicating a link between aberrant DNA methylation and H3K79 methylation come from our Dnmt3a−/− murine model, which only differ from the WT control by lack of Dnmt3a. Together, these observations suggest that the majority of therapeutic effect noted with DOT1L inhibitor therapy is attributable to the biology of the DNMT3A mutation.
In summary, based on novel observations from our Dnmt3a−/− murine model, we hypothesized that DOT1L may play a role in DNMT3A-mutant human leukemia and therefore may represent a therapeutic target. Our in vitro and in vivo work with both cell lines and primary patient samples support this hypothesis and provide the preclinical rationale for possible clinical investigation of pharmacologic DOT1L inhibitors for DNMT3A-muant leukemia. Ongoing adult and pediatric phase 1 clinical trials of the DOT1L inhibitor, EPZ-5676, for patients with MLL-rearranged hematologic malignancies could greatly facilitate the translation of these findings into clinical investigations for patients with DNMT3A mutations in the near term. Ultimately, if our results are validated, DOT1L inhibitors could be incorporated into the multiagent therapeutic regimen for the treatment of this relatively refractory group of patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Goodell Laboratory for helpful discussions; Y. Zheng, A. Guzman, and R. Gupta for technical support; C. Gillespie for critical review of the manuscript; M. Redell and M. Minden for providing cell lines; and J. Matthews and S. Piece for assistance with patient samples.
This work was supported by The Faust Foundation (R.E.R.); Edward P. Evans Foundation, Samuel Waxman Cancer Research Foundation, Henry Malvin Helis Foundation, and Lester and Sue Smith Foundation (M.A.G.); and the National Institutes of Health (NIH), National Cancer Institute (grants CA090433-11 [R.E.R.] and CA183252, CA183252, and CA126752), National Institute of Diabetes and Digestive and Kidney Diseases (grants DK092883 and DK084259), National Human Genome Research Institute (grants R01HG007538 and HG007538); and the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH, National Institute of Allergy and Infectious Diseases (grant nos. P30 AI036211 and P30 CA125123), National Institute of General Medical Sciences (S10 RR024574), and the expert assistance of J. Sederstrom and A. White.
National Institutes of Health
Authorship
Contribution: R.E.R. designed and performed experiments, analyzed results, made figures, and wrote the paper; B.A.R. analyzed the data and made figures; M.L., M.J., A.R., J.H.R., C.T.C., S.R.D., S.S., and T.C. designed and performed experiments; L.D. and Y.S. synthesized and provided DOT1L inhibitor compounds; M.A. and S.M.K. provided patient samples; W.L. helped analyze the data; and M.A.G. designed the experiments and wrote the paper. All authors commented on the edited manuscript.
Conflict-of-interest disclosure: C.T.C. and S.R.D. are employees of Epizyme, Inc. The remaining authors declare no competing financial interests.
Correspondence: Margaret A. Goodell, Department of Pediatrics and Stem Cells and Regenerative Medicine Center, Baylor College of Medicine, One Baylor Plaza, N1030, Houston, TX 77030; e-mail: goodell@bcm.edu; and Rachel E. Rau, Department of Pediatrics, Baylor College of Medicine, 1102 Bates St, Suite 1025, Houston, TX 77030; e-mail: rerau@bcm.edu.

![Figure 5. Gene expression changes following DOT1L inhibitor treatment of DNMT3A-mutant AML cell lines. (A) Gene expression heat map (gene scaled and centered log2 counts per million) of top differentially expressed canyon-associated genes in OCI AML2 and OCI AML3 cell lines after 14 days of treatment with DOT1L inhibitor. (B) Relative expression determined by qRT-PCR of canyon-associated leukemogenic genes HOXA9, MEIS1, HOXB3, HOXB8, and the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in OCI AML 2 and OCI AML3 cells treated with 3 μM EPZ004777 or vehicle control for 14 days. qRT-PCR done in triplicate. Error bars represent standard deviation. (C) Ingenuity Pathway Analysis functional enrichment analysis of genes differentially expressed (false discovery rate [FDR] q value < 0.1) in AML cells after treatment with EPZ004777. P val, P value; prolif, proliferation. (D) GSEA of AML cells treated with EPZ004777. (E) Gene expression heat maps (gene scaled and centered log2 counts per million) of representative, core-enriched gene sets from GSEA. DN, downregulated; GO, gene ontogeny; inflamm, inflammatory; leuk, leukocyte; mDC, myeloid dendritic cell; meth, DNA methylation; Q-val, Q value; resp, response; signal size, enrichment signal strength of the leading edge subset of a gene set; trans, transition; UP, upregulated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/7/10.1182_blood-2015-11-684225/6/m_971f5.jpeg?Expires=1769797130&Signature=Tg59Z1l0kFzrZxyeIermKdkj0legjskz1JdzzMRW1yAsGH2g1sYhL28hzk1cIB4ujhgQhvRIFe3PRp2VXMLGuBnzqpI6OebOgzLFuZeFP-KN4lOa70vRCDI28Sxnw9kFJ2mPQf32o~PUCdgDR5NNX-0JJNXzi4leLmgvcJm8cIElv8C~ARwHXBQSCLohpCqv79wsjT-NqMwiez095sk1qLhjWQHtW-n1Fv31Ji1-zxXy-NOiSt4k95knoUsfKJxbycvg3wbs12wzXY0PBOXaMIePdGd5pz9oImA8kRZ5ljNfFjiwj~9sLBYh~tOpjz49QygXIUxarpqF2ydInAxUUw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal