Key Points
Inhibition of HDAC reverses epigenetic silencing to upregulate miRs that target BTK and suppress its downstream prosurvival signaling.
We identified a rationale for the dual targeting of BTK when combined with ibrutinib and a strategy to eliminate the C481S-mutant BTK clone.
Abstract
Bruton’s tyrosine kinase (BTK) is a critical mediator of survival in B-cell neoplasms. Although BTK inhibitors have transformed therapy in chronic lymphocytic leukemia (CLL), patients with high-risk genetics are at risk for relapse and have a poor prognosis. Identification of novel therapeutic strategies for this group of patients is an urgent unmet clinical need, and therapies that target BTK via alternative mechanisms may fill this niche. Herein, we identify a set of microRNAs (miRs) that target BTK in primary CLL cells and show that the histone deacetylase (HDAC) repressor complex is recruited to these miR promoters to silence their expression. Targeting the HDACs by using either RNA interference against HDAC1 in CLL or a small molecule inhibitor (HDACi) in CLL and mantle cell lymphoma restored the expression of the BTK-targeting miRs with loss of BTK protein and downstream signaling and consequent cell death. We have also made the novel and clinically relevant discovery that inhibition of HDAC induces the BTK-targeting miRs in ibrutinib-sensitive and resistant CLL to effectively reduce both wild-type and C481S-mutant BTK. This finding identifies a novel strategy that may be promising as a therapeutic modality to eliminate the C481S-mutant BTK clone that drives resistance to ibrutinib and provides the rationale for a combination strategy that includes ibrutinib to dually target BTK to suppress its prosurvival signaling.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by the constitutive activation of Bruton’s tyrosine kinase (BTK),1,2 which plays a critical role in supporting neoplastic cell survival via its signaling intermediaries phospholipase Cγ2 (PLCγ2), extracellular signal-regulated kinase (ERK), and AKT.3 Ibrutinib, a potent, orally bioavailable, irreversible inhibitor that covalently binds BTK at the cysteine-481 (C481) residue showed remarkable clinical activity in CLL and other B-cell malignancies.4-6 However, in a subset of patients with CLL, disease relapse was linked to acquired mutations in BTK7,8 at the C481 site, which converted ibrutinib from an irreversible to a less potent reversible inhibitor or to gain-of-function mutations in its immediate downstream target PLCγ2.8,9 Clinically, progression of disease while receiving ibrutinib therapy is associated with a poor outcome.10-13
The histone deacetylases (HDACs) function in multicomponent repressor complexes that include HDAC1, HDAC2, and the lysine demethylase KDM1.14 They mediate the deacetylation and demethylation of lysine residues (H3K9/14 and H3K4me3) on histones to silence genes,15,16 and in CLL, they have been shown to silence the expression of key microRNA genes (miRNAs or miRs) and miRs such as miR-15a, miR-16, miR-29b, and miR-106b.17,18 However, the role of miRs in targeting BTK, their expression levels in CLL, and the role of HDACs in dysregulating the BTK-targeting miRs are unknown.
Broad-spectrum small molecule inhibitors of HDACs such as vorinostat and panobinostat are approved for the treatment of lymphomas19 and multiple myeloma,11 and abexinostat has shown clinical activity in follicular lymphoma and CLL.7,20 Panobinostat potently inhibits multiple HDACs (HDAC1, HDAC2, HDAC4, HDAC5, and HDAC6) with Ki values between 0.6 and 1 nM,21 whereas abexinostat selectively targets HDAC1 with a Ki of 7 nM, is modestly effective against HDAC2, HDAC 3, and HDAC 6 (Ki, 8 to 20 nM), and has low potency toward HDAC8 (Ki, 280 nM).22,23
The goal of this study was to identify mechanisms that contributed to the overexpression of BTK and to devise a therapeutic strategy aimed at targeting BTK in B-cell malignancies. We identified a key set of miRs that target BTK in primary CLL cells that undergo HDAC-directed silencing in CLL. Inhibition of HDACs by using either RNA interference (RNAi) or a small molecule inhibitor (HDACi) in CLL and mantle cell lymphoma (MCL) restored the expression of the BTK-targeting miRs, with loss of BTK protein and downstream signaling and consequent cell death. Interestingly, BTK-targeting miRs that become upregulated in response to an HDACi effectively target both wild-type (wt) and C481S-mutant BTK. Our studies identify inhibition of HDACs as a novel and promising therapeutic strategy to eliminate the C481S-mutant BTK clone that drives resistance to ibrutinib and provides a rationale for using a combination strategy that includes ibrutinib to dually target BTK to suppress its prosurvival signaling.
Materials and methods
Materials and patient samples
Abexinostat was provided by Pharmacyclics (Sunnyvale, CA), and panobinostat was purchased from Fisher Scientific (Waltham, MA). MEC2 cells were purchased from the Leibniz-Institute DSMZ (Braunschweig, Germany). Blood was obtained from patients with CLL or MCL or from healthy donors after they provided informed consent under an institutional review board–approved protocol. Peripheral blood mononuclear cells were isolated and used or purified for the CD19+ fraction (Stem Cell Technologies). Goat α-human immunoglobulin M (IgM [Fc5μ]) was purchased from Jackson ImmunoResearch (West Grove, PA).
Immunoblotting
Immunoblotting was performed by using antibodies against phospho-BTK (Tyr223), BTK, phospho-AKT (Ser473), AKT, phospho-ERK (Thr202/204), ERK1/2, phospho-PLCγ2 (Tyr759), PLCγ2, H3K9Ac, and total H3 from Cell Signaling Technologies (Beverly, MA); HDAC1, HDAC2, H3K9/14Ac, KDM1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from Millipore (Billerica, MA); BTK from Santa Cruz Biotechnologies (Dallas, TX); and IgG from Jackson ImmunoResearch. Immunoblots were visualized by using the LI-COR Odyssey quantitative infrared imaging system (LI-COR Biosciences, Lincoln, NE).17
Chromatin and protein immunoprecipitation
DNA from HDAC1-, HDAC2-, KDM1-, H3K4me2/3-, or H3K9/14Ac-bound chromatin was subjected to next-generation sequencing (supplemental Data, available on the Blood Web site) or polymerase chain reaction (PCR) by using probes specific for or nonspecific for the miR-147b, miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p promoters. In parallel, CLL nuclear lysates were immunoprecipitated with HDAC1, KDM1, and IgG for 1 hour, linked to protein A agarose beads, washed, and assayed for the presence of HDAC1, HDAC2, and KDM1 by immunoblotting.
Real-time PCR
Total RNA was used to quantitate the expression of miR-147b, miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p (Applied Biosystems) after normalizing to small nuclear RNA48 (snRNA48). Similarly, the levels of HDAC1 and BTK messenger RNA (mRNA) and GAPDH (normalizer) were measured by using one-step real-time PCR procedure probes from Applied Biosystems.
miR, anti-miR, and small interfering RNA transfection
Pre-miR-147b, pre-miR-210, pre-miR-425, pre-miR-1253, pre-miR-4269, pre-miR-4667-3p, anti-miR-210, anti-miR-425, and pre-miR negative control #1 were obtained from Life Technologies. Validated stealth small interfering RNAs against HDAC1 and negative controls were obtained from Thermo Fisher Scientific (Waltham, MA). Oligonucleotides were transfected into primary CLL or Mec2 cells at 150 nM by using the Amaxa Nucleofector System (Solution V, U-13; Lonza, Basel, Switzerland)24 according to manufacturer’s instructions for 48 to 72 hours, after which cells were harvested for miRs, mRNA, and protein analysis.
Luciferase assay
The psiCHECK-2 vector with BTK wt insert (full-length 3′UTR) or mutated BTK 3′UTR (containing deletion of the miR-147b, miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p target sites) were co-transfected into Mec2 cells with each of the following: miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p, or an irrelevant miR (miR-181). Cells were lysed 48 hours after transfection, and the luciferase activity (dual luciferase assay; Promega, Madison, WI) was measured at 48 hours with data expressed as the mean ± standard error of the mean from triplicate determinations from 3 independent transfections.
Cytotoxicity assays
CLL cells were exposed to either 0.4 µM abexinostat, or to 1 µM ibrutinib (for 2 hours with washout) or to a combination of ibrutinib (for 2 hours with washout) with abexinostat for 48 hours before being assayed for annexin V positivity as described.18
In vivo treatment with ibrutinib and abexinostat
C57BL/6 mice were engrafted with spleen lymphocytes derived from an Eμ-TCL1 (TCL1) transgenic mouse. Mice were observed for leukemia development (defined as ≥10% CD5+/CD19+ cells in the leukocyte population). Once they were leukemic, they were randomly assigned to a control group (vehicle [2-hydroxypropyl-β-cyclodextrin]/vehicle and 50 mmol/L sodium lactate [pH 4.2]), ibrutinib alone (∼30 mg/kg/day via drinking water), abexinostat (50 mg/kg for 3 days of each week via tail vein injection), or ibrutinib (given continuously) combined with abexinostat (given 3 times each week). Animals were euthanized at 2 weeks for pharmacodynamic evaluations.
Statistical analysis
Quantitative data are shown as the median or mean ± standard error of the mean for 3 or more independent experiments. Paired Student t tests or nonparametric Wilcoxon signed rank tests were used to compare expression between conditions. A two-sample t test was used to compare miR expression in B cells from young TCL1 mice with no evidence of disease with B cells from leukemia mice with CLL. Mixed effects models were used in experiments with multiple conditions and multiple time points to account for correlations among observations from the same subject; an interaction test was used to directly test for synergy.25 For miR expression data, ΔCT values rather than fold changes were used to reduce skewing and stabilize variances. P values for multiple comparisons were adjusted by using Hochberg’s procedure. All analyses were performed by using SAS/STAT software, Version 9.4 (SAS Institute, Cary, NC).
Results
BTK is a direct target of a set of miRs in primary CLL
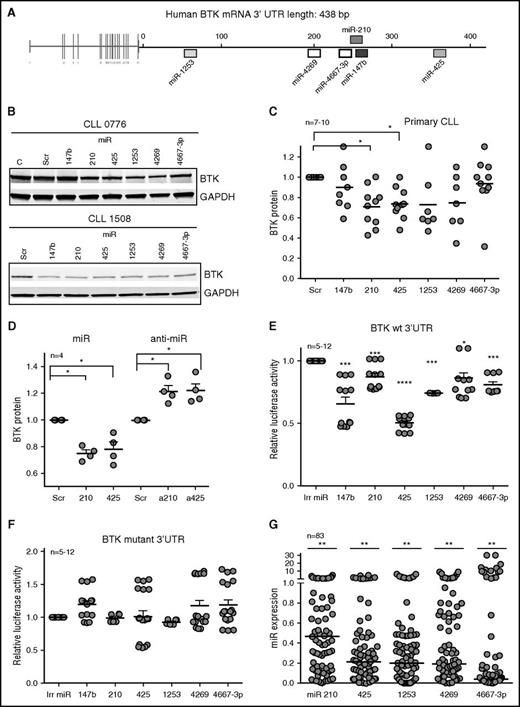
Of 8 miRs (miR-147b, miR-210, miR-425, miR-590, miR-622, miR-1253, miR-4269, and miR-4667-3p) predicted to target BTK with high specificity by 3 or more algorithms,26 5 miRs (miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p) decreased BTK protein levels by 20% to 50% compared with cells expressing the scrambled oligonucleotides or an irrelevant miR not predicted to target BTK (miR-4667-5p) in Mec2 cells (Figure 1A; supplemental Figure 1A-B). Next, to determine whether these miRs target BTK in primary tumor cells, we expressed the 5 BTK-targeting miRs validated in Mec2 cells as well as miR-147b (because the miR-147b and miR-210 target sequences differ by 1 base pair on the BTK 3′UTR; supplemental Figure 1A) in primary CLL samples (Figure 1B). Ectopic expression of each miR decreased BTK protein levels by 10% to 40% in individual CLL samples in comparison with cells expressing scrambled oligonucleotides (Figure 1B-C). Specifically, miR-210 and miR-425 significantly decreased BTK across samples (P < .05), whereas miR-147b, miR-1253, miR-4269, and miR-4667-3p reduced BTK in some CLL samples but not all.
An miR signature targets BTK in CLL. (A) Putative binding sites of 6 miRs in the 3′UTR of BTK. (B) Primary CLL cells were transiently transfected with scrambled (Scr) oligonucleotides or miR-147b, miR-210, miR-425, miR-1253, miR-4269, or miR-4667-3p for 48 hours and immunoblotted for BTK and GAPDH. The figure is representative of 7 to 10 independent CLL samples which are quantitated in (C) (P < .05 for miR-210 and miR-425, Wilcoxon signed rank test). (D) Mec2 cells transfected with mimics or inhibitors of miR-210 and miR-425 and analyzed for the levels of BTK and GAPDH (P < .05, one-sample Student t test; two-tailed P). (E-F) Reporter gene analyses using BTK 3′UTR constructs. Mec1 cells were transfected with luciferase reporter constructs expressing wt BTK or BTK with the binding sites for miR-147, miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p mutated. Cells were then transfected with an irrelevant miR (miR-181) or miR-147b, miR-210, miR-425, miR-1253, miR-4269, or miR-4667-3p, and the luciferase counts were quantitated. Data represent mean ± standard error of the mean (SEM) of 12 independent transfections (P < .05 for miR-4269 and P < .001 for miR-147b, miR-210, miR-425, miR-1253, and miR-4667-3p; paired Student t test). (G) Expression analysis of miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p in 83 CLL samples expressed as a fraction of the levels for these miRs from CD19+CD5+ B cells from healthy donors (set at 1) (P < .01 average reduction across all miRs, mixed effects model). *P ≤ .05; **P ≤ .01; ***P ≤ .001; **** P ≤ .0001.
An miR signature targets BTK in CLL. (A) Putative binding sites of 6 miRs in the 3′UTR of BTK. (B) Primary CLL cells were transiently transfected with scrambled (Scr) oligonucleotides or miR-147b, miR-210, miR-425, miR-1253, miR-4269, or miR-4667-3p for 48 hours and immunoblotted for BTK and GAPDH. The figure is representative of 7 to 10 independent CLL samples which are quantitated in (C) (P < .05 for miR-210 and miR-425, Wilcoxon signed rank test). (D) Mec2 cells transfected with mimics or inhibitors of miR-210 and miR-425 and analyzed for the levels of BTK and GAPDH (P < .05, one-sample Student t test; two-tailed P). (E-F) Reporter gene analyses using BTK 3′UTR constructs. Mec1 cells were transfected with luciferase reporter constructs expressing wt BTK or BTK with the binding sites for miR-147, miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p mutated. Cells were then transfected with an irrelevant miR (miR-181) or miR-147b, miR-210, miR-425, miR-1253, miR-4269, or miR-4667-3p, and the luciferase counts were quantitated. Data represent mean ± standard error of the mean (SEM) of 12 independent transfections (P < .05 for miR-4269 and P < .001 for miR-147b, miR-210, miR-425, miR-1253, and miR-4667-3p; paired Student t test). (G) Expression analysis of miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p in 83 CLL samples expressed as a fraction of the levels for these miRs from CD19+CD5+ B cells from healthy donors (set at 1) (P < .01 average reduction across all miRs, mixed effects model). *P ≤ .05; **P ≤ .01; ***P ≤ .001; **** P ≤ .0001.
Next, we expressed mimics and inhibitors of miR-210 and miR-425 (because those miRs decreased BTK across patient samples) in Mec2 cells (miR mimics were used as a positive control to show increased miR expression and BTK targeting) and demonstrated that antagonizing the action of miR-210 and miR-425 reduced the levels of endogenous miR-210 and miR-425 (supplemental Figure 1C) and increased the levels of BTK protein in Mec2 cells (P < .05; Figure 1D; supplemental Figure 1D), establishing a cause-and-effect relationship between these miRs and BTK expression. Because miRs modulate target proteins either by destabilizing their transcript or by repressing protein translation, we evaluated the action of miR-210 and miR-425 mimics or antago-miRs on BTK mRNA and found that overexpression of miR-210 or miR-425 or their antago-miRs in Mec2 cells (supplemental Figure 1E) or expression of miR-210 and miR-425 in primary CLL (supplemental Figure 1F) did not significantly alter the levels of BTK mRNA.
We then used a luciferase reporter system to investigate whether the 6 BTK-targeting miRs directly interacted with the BTK 3′UTR as a mechanism of gene silencing. Expression of each of these miRs significantly reduced luciferase expression (P < .05 or lower) (Figure 1E), demonstrating that these miRs directly interact with the BTK 3′UTR. Conversely, deletions in the seed interacting regions for these miRs in the BTK 3′UTR rescued the luciferase activity (Figure 1F). Taken together, these data show that BTK is a direct functional target of a discrete set of miRs that serve to regulate BTK expression in CLL.
BTK-targeting miRs are epigenetically silenced in CLL
We then quantitated the levels of all 6 BTK-targeting miRs in 83 primary CLL samples and established that the median expression of miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p was significantly lower in CLL cells compared with normal B cells (P < .01; Figure 1G). The levels of miR-147b were readily detected when overexpressed in CLL (supplemental Figure 1G) but showed low endogenous expression (at the limit of detection) in both CLL and normal B cells (supplemental Figure 1H). Furthermore, the downregulation of the BTK-targeting miRs was independent of cytogenetic group or IGHV mutational status in CLL (supplemental Figure 1I-L).
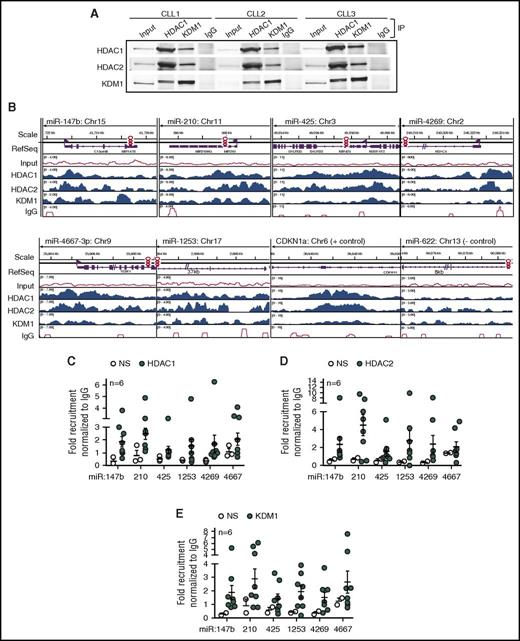
We have previously shown that HDACs silence the expression of specific miRs in CLL,17,18 so we hypothesized that an HDAC repressor complex would mediate the silencing of the BTK-targeting miRs. To determine key components of the HDAC repressor complex in primary CLL cells, we conducted reciprocal immunoprecipitations with HDAC1, KDM1, or IgG as a nonspecific control and determined that HDAC1 formed a complex with HDAC2 and KDM1 in the nuclei (Figure 2A). We then conducted genome-wide chromatin immunoprecipitation (ChIP) linked to high-throughput sequencing (ChIP-Seq) profiles in 3 CLL samples and determined that HDAC1, HDAC2, and KDM1 were recruited to the promoters for all 6 BTK-targeting miRs (Figure 2B). To show binding specificity, we conducted ChIP for HDAC1, HDAC2, and KDM1 in 6 CLL samples followed by quantitative PCR with primers either specific for the promoter regions for each miR or nonspecific to a region upstream of the defined promoter (supplemental Figure 2A) and confirmed that the HDAC repressor complex was recruited specifically to the promoters of the BTK-targeting miRs (Figure 2C-E). To further establish the link between HDAC recruitment and silencing of miR expression, we selected 3 samples with robust expression of the BTK-targeting miRs in which we would not expect recruitment of the repressor complex. As predicted, these samples showed no specific recruitment of HDAC1 or HDAC2 to the BTK-targeting miR promoter regions, whereas KDM1 was recruited only at low levels (supplemental Figure 2B-D). Taken together, our data indicate that the BTK-targeting miRs undergo HDAC-mediated silencing in primary CLL.
Recruitment of HDAC1, HDAC2, and KDM1 to the promoters of BTK-targeting miRs in CLL. (A) Co-immunoprecipitation (IP) of HDAC1, HDAC2, and KDM1 from the nuclei of 3 CLL samples in comparison with IgG. Figure is representative of 3 experiments. (B) Relative recruitment of HDAC1, HDAC2, and KDM1 in comparison with IgG at the miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p, CDKN1a (gene regulated by HDACs used as a positive control), and miR-622 (gene predicted to target BTK but not regulated by HDACs used as negative control) promoters in CLL samples with adverse cytogenetics. Data representative of 3 individual CLL samples analyzed by ChIP-Seq. (C-E) Recruitment of HDAC1, HDAC2, and KDM1 at miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p using primers specific or nonspecific (NS) to the promoters for these miRs. Data represent mean ± SEM of 6 CLL samples.
Recruitment of HDAC1, HDAC2, and KDM1 to the promoters of BTK-targeting miRs in CLL. (A) Co-immunoprecipitation (IP) of HDAC1, HDAC2, and KDM1 from the nuclei of 3 CLL samples in comparison with IgG. Figure is representative of 3 experiments. (B) Relative recruitment of HDAC1, HDAC2, and KDM1 in comparison with IgG at the miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p, CDKN1a (gene regulated by HDACs used as a positive control), and miR-622 (gene predicted to target BTK but not regulated by HDACs used as negative control) promoters in CLL samples with adverse cytogenetics. Data representative of 3 individual CLL samples analyzed by ChIP-Seq. (C-E) Recruitment of HDAC1, HDAC2, and KDM1 at miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p using primers specific or nonspecific (NS) to the promoters for these miRs. Data represent mean ± SEM of 6 CLL samples.
HDAC inhibition upregulates the expression of BTK-targeting miRs to reciprocally decrease BTK protein expression
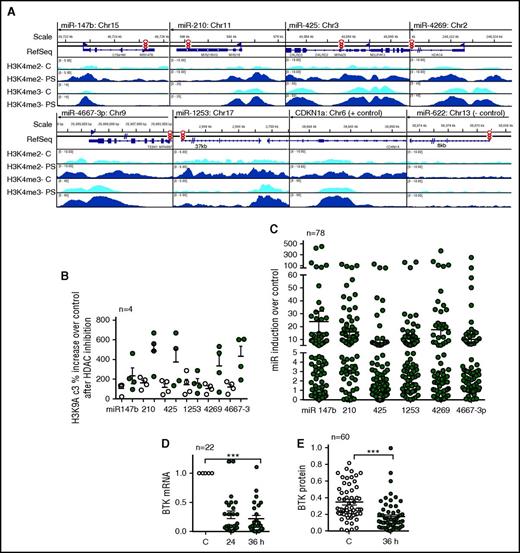
HDAC inhibition induces activating chromatin modifications such as H3K9/14Ac and H3K4me3 at gene promoters to induce transcription.15,17 We therefore treated primary CLL cells with the pan HDAC inhibitor panobinostat for 5 hours followed by ChIP-Seq using H3K4me3 antibody. HDAC inhibition resulted in a robust accumulation of the H3K4me3-activating chromatin modification at the promoters for each of the BTK-targeting miRs (Figure 3A). Because the accumulation of H3K4me3 is preceded by the accumulation of H3K9/14Ac3, we also conducted ChIP-quantitative PCR assays to determine the presence of H3K9/14Ac3 in the same samples and determined an increase in the levels of H3K9/14Ac3 at the promoters of each of the BTK-targeting miRs with the exception of miR-1253 after HDAC inhibition (Figure 3B). To establish that accumulation of these activating chromatin modifiers was linked with target gene expression, we measured the BTK-targeting miR expression in 78 CLL samples after treatment with abexinostat and determined that there was a significant increase in expression of each of the 6 BTK-targeting miRs (P < .001; Figure 3C), whereas the expression of snRNA48 (normalizing control) remained unchanged. miR induction occurred irrespective of cytogenetic abnormalities (supplemental Figure 3A,C,E,G,I). Panobinostat performed similarly to abexinostat, indicating that the effect is a class effect (supplemental Figure 3J). Importantly, induction of the BTK-targeting miRs was linked to reciprocal decreases in BTK mRNA (P < .001; Figure 3D) and protein expression (P < .001; Figure 3E; supplemental Figure 3B,D,F,H).
HDAC inhibition increases H3K4me3 at the promoters of the BTK-targeting miRs, induces their expression, and results in a reciprocal decrease in BTK protein in CLL. (A) Accumulation of the transcriptionally permissive marks H3K4me2 and H3K4me3 after HDAC inhibition at the miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p, CNDN1a (positive control), and miR-622 promoters (negative control) in CLL samples with adverse cytogenetics. Data are representative of 3 individual CLL samples analyzed by ChIP-Seq. (B) Accumulation of the transcriptionally permissive mark H3K9Ac3 after HDAC inhibition at the miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p promoters using primers specific or nonspecific to the promoters for these miRs. Data represent mean ± SEM of 4 CLL samples. (C) Induction of miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p in 79 CLL samples after HDAC inhibition. Expression of the BTK-targeting miRs were normalized to the expression of snRNA48, which did not change after HDAC inhibition (P < .001, mixed effects model). (D) Effect of abexinostat for 24 or 36 hours on BTK mRNA. Data represent mean ± SEM of 22 CLL samples (P < .001, paired Student t test). (E) Effect of abexinostat for 36 hours on BTK protein levels. Data represent mean ± SEM of 60 CLL samples (P < .001, Wilcoxon signed rank test). ***P ≤ .001.
HDAC inhibition increases H3K4me3 at the promoters of the BTK-targeting miRs, induces their expression, and results in a reciprocal decrease in BTK protein in CLL. (A) Accumulation of the transcriptionally permissive marks H3K4me2 and H3K4me3 after HDAC inhibition at the miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p, CNDN1a (positive control), and miR-622 promoters (negative control) in CLL samples with adverse cytogenetics. Data are representative of 3 individual CLL samples analyzed by ChIP-Seq. (B) Accumulation of the transcriptionally permissive mark H3K9Ac3 after HDAC inhibition at the miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p promoters using primers specific or nonspecific to the promoters for these miRs. Data represent mean ± SEM of 4 CLL samples. (C) Induction of miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p in 79 CLL samples after HDAC inhibition. Expression of the BTK-targeting miRs were normalized to the expression of snRNA48, which did not change after HDAC inhibition (P < .001, mixed effects model). (D) Effect of abexinostat for 24 or 36 hours on BTK mRNA. Data represent mean ± SEM of 22 CLL samples (P < .001, paired Student t test). (E) Effect of abexinostat for 36 hours on BTK protein levels. Data represent mean ± SEM of 60 CLL samples (P < .001, Wilcoxon signed rank test). ***P ≤ .001.
To determine whether the HDACs suppressed the expression of the BTK-targeting miRs in other B-cell neoplasms in which BTK inhibitors are approved for clinical use,27 we exposed primary MCL samples to abexinostat and showed that, similar to CLL, exposure to an HDACi led to the upregulation of each of the BTK-targeting miRs with decreases in the levels of BTK protein and inhibition of BTK signaling (supplemental Figure 3K-L).
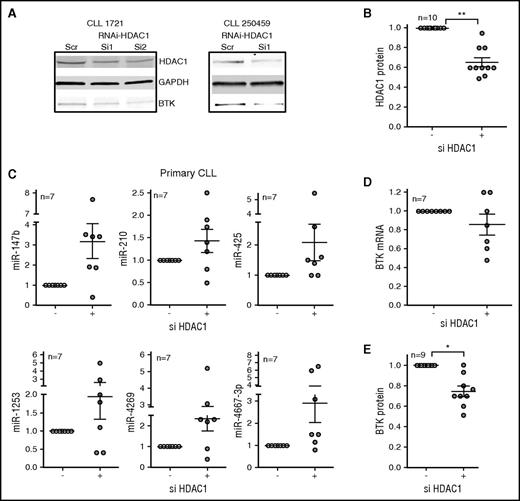
To determine that inhibition of HDAC1 is the primary mechanism of miR induction, we knocked down HDAC1 in primary CLL cells by using small interferring RNA oligonucleotides. Although the response was variable in individual primary CLL samples, overall, the knockdown of HDAC1 (Figure 4A-B) was associated with a modest induction of each of the BTK-targeting miRs (Figure 4C), which in turn was associated with moderate decreases in the levels of BTK mRNA (Figure 4D) and statistically significant decreases in BTK protein (P < .05; Figure 4A,E), establishing that specific loss of HDAC1 increases the expression of BTK-targeting miRs, thereby reducing BTK protein expression.
RNAi against HDAC1 induces BTK-targeting miRs to reduce BTK protein in primary CLL cells. (A-B) Primary CLL cells were nucleofected with small interferring RNA (siRNA) against HDAC1 for 48 hours before being harvested for RNA and protein. The top panel shows the effect of siRNA-HDAC1 on HDAC1 protein. GAPDH was used as a loading control (middle panel). The bottom panel shows the consequence of si-HDAC1 on BTK protein. The figure is representative of 10 independent transfections. (B) Quantitation of the effect of siHDAC1 on HDAC1 protein. Data represent mean ± SEM of 10 CLL samples (P < .01, paired Student t test). (C) The expression of miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p was quantitated in the primary CLL cells nucleofected in (A). Data represent mean ± SEM of 7 CLL samples. (D) The expression of BTK mRNA in the primary CLL cells nucleofected in (A). Data represent mean ± SEM of 7 CLL samples. (E) The expression of BTK protein (P < .05, Wilcoxon signed rank test) in the primary CLL cells nucleofected in (A). Data represent mean ± SEM of 9 CLL samples. *P ≤ .05; **P ≤ .01.
RNAi against HDAC1 induces BTK-targeting miRs to reduce BTK protein in primary CLL cells. (A-B) Primary CLL cells were nucleofected with small interferring RNA (siRNA) against HDAC1 for 48 hours before being harvested for RNA and protein. The top panel shows the effect of siRNA-HDAC1 on HDAC1 protein. GAPDH was used as a loading control (middle panel). The bottom panel shows the consequence of si-HDAC1 on BTK protein. The figure is representative of 10 independent transfections. (B) Quantitation of the effect of siHDAC1 on HDAC1 protein. Data represent mean ± SEM of 10 CLL samples (P < .01, paired Student t test). (C) The expression of miR-147b, miR-210, miR-425, miR-1253, miR-4269, miR-4667-3p was quantitated in the primary CLL cells nucleofected in (A). Data represent mean ± SEM of 7 CLL samples. (D) The expression of BTK mRNA in the primary CLL cells nucleofected in (A). Data represent mean ± SEM of 7 CLL samples. (E) The expression of BTK protein (P < .05, Wilcoxon signed rank test) in the primary CLL cells nucleofected in (A). Data represent mean ± SEM of 9 CLL samples. *P ≤ .05; **P ≤ .01.
HDAC inhibition effectively synergizes with ibrutinib to target BTK through independent mechanisms
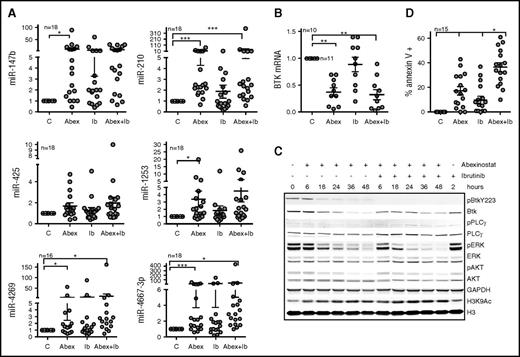
Survival is poor after relapse during ibrutinib therapy, so alternative therapies and combination strategies to prevent relapse are greatly needed10,13,28 for CLL patients. We sought to combine HDAC inhibitors with ibrutinib with the aim of targeting BTK by independent mechanisms and thus augmenting the activity of either agent alone. CLL cells were treated with abexinostat alone, ibrutinib alone, or a combination of abexinostat and ibrutinib for up to 48 hours. Although treatment with ibrutinib alone induced miR-4269 and miR-4667-3p (P < .05) expression (Figure 5A), treatment with abexinostat alone significantly upregulated each BTK-targeting miR with the exception of miR-425 (P < .05; Figure 5A); the combination of abexinostat and ibrutinib significantly induced miR-210, miR-4269, and miR-4667-3p (P < .05; Figure 5A). Upregulation of the BTK-targeting miRs was associated with consequent decreases in BTK mRNA (P < .01; Figure 5B). As predicted, abexinostat treatment significantly decreased BTK protein expression in a time-dependent manner, whereas ibrutinib treatment inhibited BTK phosphorylation while preserving total protein expression (Figure 5C). As a consequence of total protein reduction, the absolute level of phosphorylated BTK was lower in abexinostat- or combination-treated samples, and this was associated with loss of the immediate downstream effector phospho-PLCγ2 (p-PLCγ2) and with reductions in the distal downstream intermediaries p-ERK and p-AKT, thus indicating that loss of BTK protein is sufficient to abrogate BTK signaling. When cells were exposed to the combination of abexinostat and ibrutinib, p-BTK was inhibited at all the evaluated times (6 to 48 hours), whereas total BTK underwent a time-dependent decline indicating a dual targeting of BTK (Figure 5C). Similar to cells exposed to abexinostat alone, there were time-dependent losses in p-PLCγ2, p-ERK, and p-AKT, indicating the inhibition of BTK-mediated signaling (Figure 5C).
Effect of abexinostat combined with ibrutinib on BTK protein, signaling, and CLL survival. (A) Induction of miR-147b, miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p in CLL cells left untreated and exposed to abexinostat (Abex), ibrutinib (IB), or abexinostat and ibrutinib combined. Data represent mean ± SEM of 18 CLL samples. After abexinostat: P < .001 for miR-210 and miR-4667-3p; P < .05 for miR-147b, miR1253, and miR-4269; after abexinostat and ibrutinib combined: P = .001 for miR-210; P < .05 for miR-4269 and miR-4667-3p mixed effects model. (B) BTK mRNA in cells left untreated and exposed to abexinostat, ibrutinib, or abexinostat and ibrutinib combined. Data represent the mean ± SEM of 10 CLL samples (P < .01 for BTK expression after exposure to either abexinostat or abexinostat plus ibrutinib, mixed effects model). (C) Action of abexinostat alone or combined with ibrutinib on p-Y223-BTK, total BTK, p-PLCγ2, total PLCγ2, p-ERK, total ERK, p-AKT, total AKT, and H3K9Ac. GAPDH was assayed as a loading control, and H3 was assayed as a control for H3K9Ac. (D) Effect of abexinostat, ibrutinib, or combined abexinostat and ibrutinib on the induction of apoptosis (Annexin V–positive cells) in primary CLL samples. Data represent the mean ± SEM of 13 CLL samples (P < .05, interaction test of synergy from a mixed effects model). *P ≤ .05; **P ≤ .01; ***P ≤ .001.
Effect of abexinostat combined with ibrutinib on BTK protein, signaling, and CLL survival. (A) Induction of miR-147b, miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p in CLL cells left untreated and exposed to abexinostat (Abex), ibrutinib (IB), or abexinostat and ibrutinib combined. Data represent mean ± SEM of 18 CLL samples. After abexinostat: P < .001 for miR-210 and miR-4667-3p; P < .05 for miR-147b, miR1253, and miR-4269; after abexinostat and ibrutinib combined: P = .001 for miR-210; P < .05 for miR-4269 and miR-4667-3p mixed effects model. (B) BTK mRNA in cells left untreated and exposed to abexinostat, ibrutinib, or abexinostat and ibrutinib combined. Data represent the mean ± SEM of 10 CLL samples (P < .01 for BTK expression after exposure to either abexinostat or abexinostat plus ibrutinib, mixed effects model). (C) Action of abexinostat alone or combined with ibrutinib on p-Y223-BTK, total BTK, p-PLCγ2, total PLCγ2, p-ERK, total ERK, p-AKT, total AKT, and H3K9Ac. GAPDH was assayed as a loading control, and H3 was assayed as a control for H3K9Ac. (D) Effect of abexinostat, ibrutinib, or combined abexinostat and ibrutinib on the induction of apoptosis (Annexin V–positive cells) in primary CLL samples. Data represent the mean ± SEM of 13 CLL samples (P < .05, interaction test of synergy from a mixed effects model). *P ≤ .05; **P ≤ .01; ***P ≤ .001.
CLL cells respond to stimulation with IgM by activating BCR and BTK signaling to favor survival,29 which was measured by increased p-ERK and p-AKT compared with their levels in unstimulated cells (supplemental Figure 5B). Exposure to ibrutinib inhibited p-BTK and BTK signaling under both basal and IgM-stimulated conditions (supplemental Figure 5B). Significantly, exposure to abexinostat alone or in combination with ibrutinib led to an increase in the levels of BTK-targeting miRs (Figure 5A; supplemental Figure 5A) with corresponding decreases in total BTK and an inhibition of BTK signaling measured by p-ERK and p-AKT in unstimulated as well as IgM-stimulated cells (supplemental Figure 5B). Finally, abexinostat and ibrutinib combined induced synergistic cytotoxicity in primary CLL cells compared with either agent alone (P < .05; Figure 5D).
HDAC inhibition effectively targets the C481S-mutant BTK clone in ibrutinib-resistant CLL
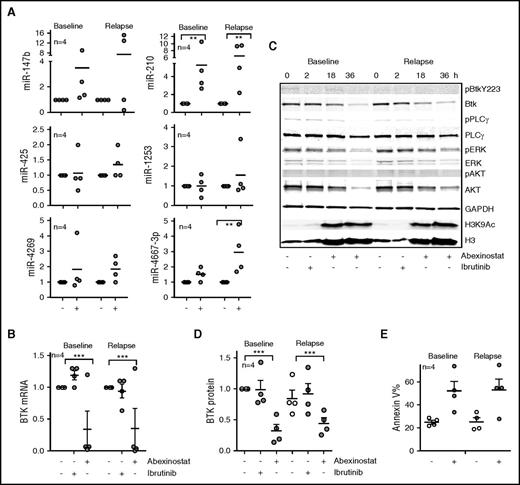
Because resistance to ibrutinib is largely associated with the acquisition of substitution mutations in BTK (C481S), we evaluated whether HDAC inhibition could target both wt and mutant BTK. We obtained sequential samples from 4 patients with CLL before ibrutinib therapy and after the acquisition of the C481S clone and development of ibrutinib resistance. In samples taken before exposure to ibrutinib, in vitro abexinostat led to a significant increase in miR-210 (P < .01; Figure 6A) with a resulting reduction in BTK gene expression (P < .01; Figure 6B). Notably, in the samples with C481S BTK, in vitro treatment with abexinostat induced both miR-210 and miR-4667-3p (P < .01 for both; Figure 6A) and showed an equivalent reduction in BTK mRNA (P < .01; Figure 6B). In parallel, signaling interrogation after ibrutinib treatment revealed loss of p-BTK in baseline samples (Figure 6C), whereas in the samples with BTK C481S, p-BTK levels were lower than at baseline, and administration of ibrutinib did not alter the phosphorylation of BTK or its downstream targets (Figure 6C). However, treatment with abexinostat depleted BTK protein (P < .001; Figure 6D), reduced the levels of p-PLCγ2, p-ERK, and p-AKT to attenuate BTK-mediated signaling (Figure 6C), and induced cell death (Figure 6E) in all samples regardless of BTK C481 mutational status or response to ibrutinib therapy.
HDAC inhibition effectively targets the C481S-mutant BTK clone in ibrutinib-resistant CLL. (A) Induction of miR-147b, miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p in response to abexinostat in paired CLL samples obtained at baseline or after acquisition of the C481S BTK clone while the patient was receiving ibrutinib therapy. Data represent mean ± SEM of 4 CLL samples (P < .01 for miR-210 before and after ibrutinib resistance had developed, and P < .01 for miR-4667-3p after ibrutinib resistance had developed; mixed effects model). (B) Effect of ibrutinib or abexinostat on BTK mRNA in paired CLL samples obtained at baseline or after acquisition of the C481S BTK clone while the patient received ibrutinib therapy. Data represent mean ± SEM of 4 CLL samples (P < .01; mixed effects model). (C) Effect of ibrutinib or abexinostat at 18 and 36 hours on BTK phosphorylation, protein, and signaling in paired CLL samples obtained at baseline or after acquisition of the C481S BTK clone while the patient received ibrutinib therapy. Levels of H3K4Ac3 were evaluated as a measure of HDAC inhibition, and H3 was measured as a control for H3K9Ac. (D) The decrease in BTK protein was quantitated; data represent mean ± SEM of 4 CLL samples (P < .001; mixed effects model). (E) Effect of abexinostat on CLL survival at 48 hours as measured by increase in percentage of annexin V–positive cells in paired CLL samples obtained at baseline or after acquisition of the C481S BTK clone while the patient received ibrutinib therapy. ***P ≤ .001.
HDAC inhibition effectively targets the C481S-mutant BTK clone in ibrutinib-resistant CLL. (A) Induction of miR-147b, miR-210, miR-425, miR-1253, miR-4269, and miR-4667-3p in response to abexinostat in paired CLL samples obtained at baseline or after acquisition of the C481S BTK clone while the patient was receiving ibrutinib therapy. Data represent mean ± SEM of 4 CLL samples (P < .01 for miR-210 before and after ibrutinib resistance had developed, and P < .01 for miR-4667-3p after ibrutinib resistance had developed; mixed effects model). (B) Effect of ibrutinib or abexinostat on BTK mRNA in paired CLL samples obtained at baseline or after acquisition of the C481S BTK clone while the patient received ibrutinib therapy. Data represent mean ± SEM of 4 CLL samples (P < .01; mixed effects model). (C) Effect of ibrutinib or abexinostat at 18 and 36 hours on BTK phosphorylation, protein, and signaling in paired CLL samples obtained at baseline or after acquisition of the C481S BTK clone while the patient received ibrutinib therapy. Levels of H3K4Ac3 were evaluated as a measure of HDAC inhibition, and H3 was measured as a control for H3K9Ac. (D) The decrease in BTK protein was quantitated; data represent mean ± SEM of 4 CLL samples (P < .001; mixed effects model). (E) Effect of abexinostat on CLL survival at 48 hours as measured by increase in percentage of annexin V–positive cells in paired CLL samples obtained at baseline or after acquisition of the C481S BTK clone while the patient received ibrutinib therapy. ***P ≤ .001.
Abexinostat and ibrutinib combined is superior to each agent alone in vivo
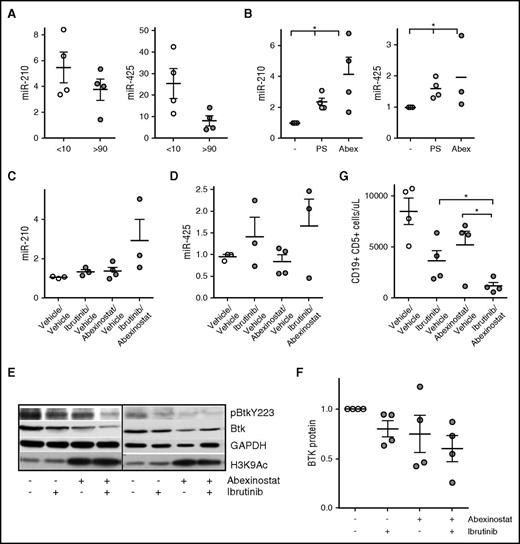
To determine whether the BTK-targeting miRs were epigenetically silenced in the TCL1 transgenic mouse model of progressive CLL,1,27,30 we compared the expression of miR-210, miR-425, and miR-147b (the 3 BTK-targeting miRs expressed in mice) in B cells from young TCL1 mice with no evidence of disease with expression in B cells from leukemia mice with CLL and found that levels of miR-210 and miR-425 were downregulated in mice with CLL; miR-425 showed a significant decrease (P < .05; Figure 7A). Similar to our findings in human CLL, the levels of miR-147b were low in B cells from both nonleukemic and CLL-bearing mice (supplemental Figure 4A). To determine whether HDACs mediated the selective downregulation of these miRs, we exposed cells obtained from mice with CLL to abexinostat and determined that each miR became induced after HDAC inhibition (P < .05; Figure 7B; supplemental Figure 4B). Next, to test the action of abexinostat combined with ibrutinib in vivo, we used an adoptive transfer model in which leukemic lymphocytes from TCL1 mice were engrafted into wt C57BL/6 mice. At the time of leukemia development, mice were randomly assigned to treatment with vehicle, ibrutinib administered in drinking water (0.016 mg/mL), abexinostat at 50 mg/kg by oral gavage, or combined ibrutinib and abexinostat. After 2 weeks of treatment, mice were euthanized, and CD19-selected leukemic cells were obtained from the spleen. Combined abexinostat and ibrutinib significantly induced miR-210 (P < .05; Figure 7C), whereas miR-425 and miR-147b were induced in some but not all mice (Figure 7D; supplemental Figure 4C). More importantly, similar to primary CLL cells, exposure to abexinostat and ibrutinib combined reduced p-BTK as well as total BTK protein, indicating dual targeting of BTK in vivo (Figure 7E-F). Finally, although treatment with either ibrutinib or abexinostat decreased leukemia cell counts in the peripheral blood compared with vehicle, abexinostat and ibrutinib combined reduced leukemic cell counts to a greater degree than either agent alone, again demonstrating that treatment with the combination was superior to treatment with each individual drug (P < .05; Figure 7G).
Role of HDACs in silencing the BTK-targeting miR in the TCL1 model of CLL and consequence of the combination of abexinostat and ibrutinib on BTK protein, signaling, and leukemia cell survival in vivo. (A) CLL cells were isolated from the spleen of TCL1 transgenic mice before (CD19+CD5+ cells <10%) and after developing CLL (CD19+CD5+ cells >90%). RNA was extracted and used to quantitate the expression of miR-210 and miR-425 (P < .05 for miR-425, two-sample Student t test). (B) CLL cells from TCL1 mice with leukemia (CD19+CD5+ cells >90%) were exposed to either panobinostat or abexinostat for 18 hours and then evaluated for the expression of miR-210 and miR-425 (P < .05 for both, paired Student t test). (C-D) Mice were randomly assigned to vehicle/vehicle (control), abexinostat/vehicle, ibrutinib/vehicle, or a combination of abexinostat and ibrutinib for 2 weeks. Spleen CLL cells were isolated and used to quantitate miR-210 and miR-425 (P < .05 for miR-210 with the combination; analysis of variance). (E) CLL cells from TCL1 mice with leukemia (CD19+CD5+ cells >90%) were left untreated or exposed to abexinostat, ibrutinib, or the combination before being assayed for p-BTK and BTK. GAPDH was used as a loading control, and H3K9/14 was assayed to measure HDAC inhibition. (F) BTK protein was quantified in CLL cells from (E) (P < .05 for the combination vs vehicle; paired Student t test). (G) Mice were randomly assigned to vehicle/vehicle (control), abexinostat/vehicle, ibrutinib/vehicle, or a combination of abexinostat and ibrutinib for 2 weeks. Spleen CLL cells were isolated from each group, and the number of CD19+CD5+ CLL cells was quantitated (P < .05 for the combination over treatment with each individual drug; mixed effects model). *P ≤ .05.
Role of HDACs in silencing the BTK-targeting miR in the TCL1 model of CLL and consequence of the combination of abexinostat and ibrutinib on BTK protein, signaling, and leukemia cell survival in vivo. (A) CLL cells were isolated from the spleen of TCL1 transgenic mice before (CD19+CD5+ cells <10%) and after developing CLL (CD19+CD5+ cells >90%). RNA was extracted and used to quantitate the expression of miR-210 and miR-425 (P < .05 for miR-425, two-sample Student t test). (B) CLL cells from TCL1 mice with leukemia (CD19+CD5+ cells >90%) were exposed to either panobinostat or abexinostat for 18 hours and then evaluated for the expression of miR-210 and miR-425 (P < .05 for both, paired Student t test). (C-D) Mice were randomly assigned to vehicle/vehicle (control), abexinostat/vehicle, ibrutinib/vehicle, or a combination of abexinostat and ibrutinib for 2 weeks. Spleen CLL cells were isolated and used to quantitate miR-210 and miR-425 (P < .05 for miR-210 with the combination; analysis of variance). (E) CLL cells from TCL1 mice with leukemia (CD19+CD5+ cells >90%) were left untreated or exposed to abexinostat, ibrutinib, or the combination before being assayed for p-BTK and BTK. GAPDH was used as a loading control, and H3K9/14 was assayed to measure HDAC inhibition. (F) BTK protein was quantified in CLL cells from (E) (P < .05 for the combination vs vehicle; paired Student t test). (G) Mice were randomly assigned to vehicle/vehicle (control), abexinostat/vehicle, ibrutinib/vehicle, or a combination of abexinostat and ibrutinib for 2 weeks. Spleen CLL cells were isolated from each group, and the number of CD19+CD5+ CLL cells was quantitated (P < .05 for the combination over treatment with each individual drug; mixed effects model). *P ≤ .05.
Discussion
We present a novel mechanism by which BTK is regulated through miR expression and can be targeted by HDAC inhibition. This study is the first to comprehensively elucidate the specificity, expression, and regulation of a panel of miRs that converge on BTK, an essential target in CLL and other B-cell neoplasms. In this study, we identified an miR signature that targeted BTK in which each of the miRs in our signature reduced BTK effectively in reporter assays, but their ability to reduce BTK varied when they were overexpressed in primary CLL cells, suggesting that additional divergent factors may modulate the miR-directed targeting of BTK in individual patient samples.7,20 By using miR-210 and miR-425 (the 2 miRs that consistently targeted BTK in all CLL samples), we were able to establish a causal relation between BTK and these miRs. We were unable to conduct similar evaluations for miR-147b, miR-1253, and miR-4269 because of the low levels of expression of these miRs in Mec2 cells.
The finding that the BTK-targeting miRs undergo epigenetic silencing offers a mechanistic explanation for the robust expression of BTK in CLL.31 In the large cohort of CLL samples tested, each of the 6 BTK-targeting miRs was significantly induced after HDAC inhibition, whereas in the smaller cohort of samples exposed to abexinostat combined with ibrutinib, induction of miR-210, miR-425, and miR-4667-3p was sufficient to reduce BTK. In addition, miR-210 was the only miR significantly induced in paired samples before and after ibrutinib resistance was acquired, suggesting that of the 6 BTK-targeting miRs, induction of miR-210, and to a lesser extent miR-425 and miR-4667-3p, may be critical in targeting BTK. HDAC inhibition reduced BTK mRNA and protein, whereas ectopic expression of the BTK-targeting miRs reduced BTK protein without affecting the transcript, suggesting that HDAC inhibition may also reduce BTK by more than 1 mechanism.32
The finding that HDAC inhibition robustly reduces BTK protein and signaling is clinically relevant because it points toward a rational combination strategy of ibrutinib with an HDAC inhibitor to dually target BTK and potentially preclude the acquisition of a mutation while the patient is receiving ibrutinib therapy. Most CLL cells express IgM receptors that, upon stimulation, activate signaling via the BCR, BTK, ERK, and AKT kinases to favor survival.29 Our results demonstrating the ability of HDAC inhibitors singly or in combination with ibrutinib to effectively target BTK protein and abrogate its downstream signaling under conditions of IgM stimulation highlight the utility of this combination to antagonize the BCR-mediated survival advantage. In addition, because HDAC inhibitors can effectively also target mutated BTK in vitro, trials of HDAC inhibitors in treating ibrutinib-resistant CLL would be a rational strategy for patients with ibrutinib-resistant disease because survival for this group of patients is extremely poor.10,12,13,28 Another helpful strategy would be to use HDAC inhibitors in combination with ibrutinib for patients who have molecular relapse (ie, increased cloning of C481S-mutated cells) without clinical relapse with the goal of prolonging sensitivity to ibrutinib. This strategy is attractive because it has the potential to limit exposure to HDAC inhibitors, which are known to be difficult for patients with CLL to tolerate.33,34 Finally, although our data here are largely specific to CLL, this strategy is potentially applicable to other B-cell malignancies in which response to ibrutinib is not as prolonged as in CLL.
In conclusion, our data uncovered a novel mechanism of BTK regulation. We also presented, for the first time, a dual epigenetic and kinase inhibition of an oncogenic pathway by inhibition of BTK and showed the therapeutic potential of this strategy in CLL. Our data showed that besides synergistic preclinical activity in CLL, this combination has the potential to circumvent ibrutinib resistance, an increasingly common unmet clinical need.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by grants from the American Cancer Society, CLL-Global Research Foundation, D. Warren Brown Foundation, Four Winds Foundation, Michael and Judy Thomas, and the National Institutes of Health, National Cancer Institute (K23-CA178183, R01-CA197870, P30-CA016058, and R35-CA197734).
Authorship
Contribution: A.B., L.R., and T.-H.L. performed experiments and analyzed and interpreted data; C.L., L.L.S., R.M., and S.R. performed experiments; D.E.-G. provided study materials and analyzed data; K.L., A.J.J., R.L., J.S.B., W.P., and J.C.B. analyzed and interpreted data; A.L. provided statistical analysis; J.A.W. and D.S. conceived and designed the study, analyzed and interpreted data, and wrote the manuscript; and all authors reviewed and approved the manuscript before submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Deepa Sampath, Division of Hematology, Department of Internal Medicine, The Ohio State University, 410 W. 12th Ave, Room 485, Columbus, OH 43210; e-mail: deepa.sampath@osumc.edu.
References
Author notes
A.B. and L.R. contributed equally to this work.
J.A.W. and D.S. contributed equally to this work.