Abstract
Background: Acute myeloid leukemia (AML) comprises a heterogeneous collection of morphologically related diseases. The identification and study of somatic mutations in AML using next-generation sequencing has led to insights into the pathogenesis of AML. Constellations of specific mutations may correlate with clinical phenotype and reveal the most relevant cooperating molecular pathways. Understanding AML biological pathways will contribute to more personalized, directed therapies and improve outcomes. Most sequencing efforts have focused on select groups of AML patients or select gene panels. Characterization of mutations identified by whole exome sequencing for AML patients from a variety of clinical scenarios has not been the subject of one study.
Methods: Whole exome sequencing (WES) was performed, in addition to complete clinical molecular diagnostics, on leukemia samples from a diverse group of 141 consecutive AML patients who underwent complete clinical evaluation at a single quaternary leukemia care institution (MD Anderson Cancer Center, MDACC). The cohort included adults of all ages, treatment histories (treated/untreated), and ontogenies (de novo/secondary/therapy-related). To facilitate the identification of driver mutations from WES data, an automatic pipeline was developed to filter and annotate raw variants, and then select non-polymorphism, protein-coding changing mutations. A relational mutation/clinical database was created to analyze gene mutations and gene categories in the context of well-annotated mutations and complete clinical indices including treatments and outcomes.
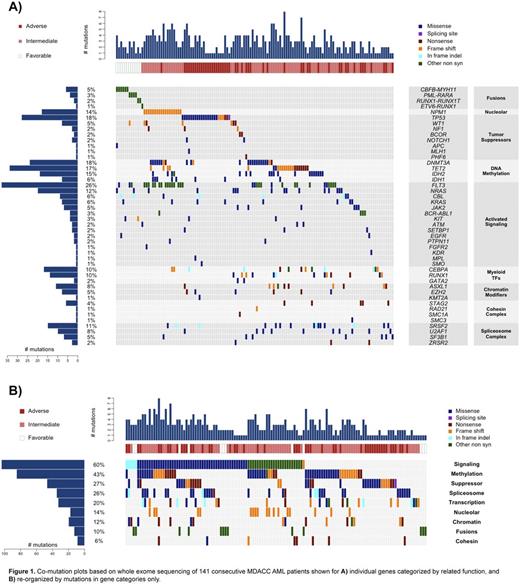
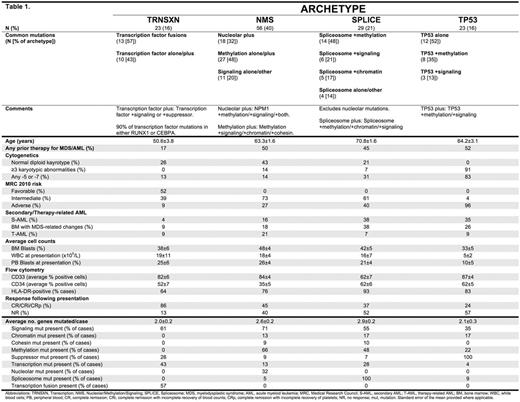
Results: We characterized the landscape of driver mutations in an unselected group of AML patients, and classified mutations by 9 predefined functional categories of known aberrant pathways implicated in AML (Figure 1). The frequency and distribution of gene mutations and gene categories were determined and compared to TCGA AML whole exome sequencing mutation data, derived from a de novo, untreated AML cohort. Forty-six separate genes/fusions were mutated in the MDACC cohort, including 326 mutations. The MDACC cohort included 23 cases (16%) with a TP53 mutation. Overall, a mean of 2.3+/-1.4 mutations-of-interest were identified per patient. For the MDACC dataset, the likelihood of co-occurrence and mutual exclusivity was determined for each gene (mutated in greater than 2 cases, n=34) and each gene category (n=9) versus every other gene, gene category, as well as each individual annotated clinical characteristic (n=30). 156 statistically significant co-occurring pairs and 71 statistically significant mutually exclusive pairs were found. We analyzed hierarchically clustered, strength-of-association correlograms to identify likely mutational frameworks with similarly associated clinical/molecular characteristics. We identified patterns of predominant genetic alterations that defined 4 distinct AML archetypes: 1) Transcription factor [TRNSXN] (16% of cases), 2) Nucleolar/Methylation/Signaling [NMS] (40%), 3) Spliceosome [SPLICE] (21%), and 4) TP53-mutated [TP53] (16%). Ten cases had no identifiable mutation-of-interest, compared to 4 cases without mutation in TCGA data. The defining mutational characteristics of each AML archetype, and select associated characteristics are shown in Table 1.
Conclusions: Analysis of whole exome sequencing data along with clinical information from all-comer AML patients identifies archetypal classes of the disease, each with particular tendencies for specific mutational, clinical, and outcome characteristics. Our approach demonstrates that genomic characterization of mutations in a variety of diverse clinical scenarios has the potential to identify representative mechanisms of AML development across the spectrum of disease presentations.
Daver:Otsuka: Consultancy, Honoraria; Sunesis: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Karyopharm: Honoraria, Research Funding; Ariad: Research Funding; Kiromic: Research Funding; BMS: Research Funding. Jabbour:ARIAD: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Novartis: Research Funding; BMS: Consultancy. DiNardo:Agios: Other: advisory board, Research Funding; Celgene: Research Funding; Novartis: Other: advisory board, Research Funding; Daiichi Sankyo: Other: advisory board, Research Funding; Abbvie: Research Funding. Konopleva:Reata Pharmaceuticals: Equity Ownership; Abbvie: Consultancy, Research Funding; Genentech: Consultancy, Research Funding; Stemline: Consultancy, Research Funding; Eli Lilly: Research Funding; Cellectis: Research Funding; Calithera: Research Funding. Cortes:ARIAD: Consultancy, Research Funding; BMS: Consultancy, Research Funding; Novartis: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Teva: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal