Key Points
Notch1 mutations are selected in the murine T-ALL model despite genetic pan-Notch inhibition; other pathways do not easily substitute it.
Myc is the key Notch target responsible for Notch-selective pressure in T-ALL as it can substitute for Notch; by contrast, Akt cannot.
Abstract
Activating NOTCH1 mutations are frequent in human T-cell acute lymphoblastic leukemia (T-ALL) and Notch inhibitors (γ-secretase inhibitors [GSIs]) have produced responses in patients with relapsed, refractory disease. However, sustained responses, although reported, are uncommon, suggesting that other pathways can substitute for Notch in T-ALL. To address this possibility, we first generated KrasG12D transgenic mice with T-cell–specific expression of the pan-Notch inhibitor, dominant-negative Mastermind (DNMAML). These mice developed leukemia, but instead of accessing alternative oncogenic pathways, the tumor cells acquired Notch1 mutations and subsequently deleted DNMAML, reinforcing the notion that activated Notch1 is particularly transforming within the context of T-cell progenitors. We next took a candidate approach to identify oncogenic pathways downstream of Notch, focusing on Myc and Akt, which are Notch targets in T-cell progenitors. KrasG12D mice transduced with Myc developed T-ALLs that were GSI-insensitive and lacked Notch1 mutations. In contrast, KrasG12D mice transduced with myristoylated AKT developed GSI-sensitive T-ALLs that acquired Notch1 mutations. Thus, Myc can substitute for Notch1 in leukemogenesis, whereas Akt cannot. These findings in primary tumors extend recent work using human T-ALL cell lines and xenografts and suggest that the Notch/Myc signaling axis is of predominant importance in understanding both the selective pressure for Notch mutations in T-ALL and response and resistance of T-ALL to Notch pathway inhibitors.
Introduction
Notch1 belongs to a family of transmembrane receptors that regulate fate decisions and differentiation during normal development. In the absence of ligands, Notch receptors are held in an inactive state by the extracellular negative regulatory region (NRR), which consists of the Lin-12/Notch repeat domain and a heterodimerization domain (HD).1 Engagement of Notch by ligands expressed on neighboring cells triggers a conformational change in the NRR that permits successive cleavages by ADAM metalloproteases and γ-secretase that release the intracellular domain of Notch1 (ICN1), a process that can be inhibited by γ-secretase inhibitors (GSIs). Following γ-secretase cleavage, ICN translocates to the nucleus and binds the DNA-binding factor RBPJ, creating a composite surface that recruits a coactivator of the Mastermind (MAML) family to form a Notch transcription complex.2 Dominant-negative Mastermind (DNMAML), which consists of the RBPJ/ICN1-binding portion of MAML1, lacks transactivation function and inhibits target gene induction by activated Notch.3 DNMAML is well validated in cell-based assays and in mouse models as a potent and specific Notch inhibitor4-6 that replicates phenotypes produced by Notch or RBPJ deficiency.3-7
Supraphysiological Notch signaling is implicated in a wide variety of human cancers.8 Oncogenic activating NOTCH1 mutations occur most commonly in T-cell acute lymphoblastic leukemia (T-ALL), chronic lymphocytic leukemia, mantle cell lymphoma, adenoid cystic carcinoma, and breast cancer.9-14 Of these, the consequences of dysregulated Notch signaling are best understood in T-ALL. In contrast to wild-type NOTCH1, oncogenic NOTCH1 mutations in T-ALL often lead to ligand-independent signaling.9,15 Mutations that disrupt NRR function occur frequently in both human and murine T-ALL.9 In murine T-ALL, recombination activating gene (RAG)–mediated 5′ Notch1 deletions leading to expression of truncated transcripts that fail to encode the NRR predominate,16,17 whereas in human T-ALL, the NOTCH1 NRR often is destabilized by point substitutions or small in-frame indels that trigger ligand-independent γ-secretase cleavage.15,18 Also frequently seen in T-ALL in both species are frameshift or nonsense mutations that result in loss of a C-terminal degron domain containing a PEST (peptide sequence that is rich in proline, glutamic acid, serine, and threonine) sequence, leading to enhanced ICN1 stability.19-22 Notch activation in T-ALL induces sustained, high-level expression of Notch target genes, many of which are implicated in oncogenesis, including Myc, components of the phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway, Il7ra, and Hes1.23-30 The Myc and AKT pathways appear to be particularly important, as inhibiting them abrogates the growth of Notch-mutated T-ALL cell lines and the development of Notch-driven T-ALL in mouse models.23,29,31-36
Owing to the high prevalence of activating Notch mutations in T-ALL, Notch inhibitors, such as GSIs, may benefit patients with this disease.37,38 In phase 1 clinical trials, GSIs typically produce a decrease in circulating blasts39 and occasionally complete remissions37,38 ; however, responses are usually incomplete or transient. Chronic GSI exposure of human T-ALL cell lines results in outgrowth of resistant cells that no longer require activated NOTCH1, apparently because of epigenetic alterations that lead to Notch-independent MYC upregulation.35 However, cell lines do not replicate the genetic and epigenetic heterogeneity of primary leukemias.40 Work focused on testing the dependence of T-ALL on Notch signaling in vivo using mice bearing Notch-addicted primary T-ALLs or T-ALL xenografts showed that tumors initially regress but then recur for uncertain reasons,35,36,41 whereas other studies suggest that T-ALL requires persistent Notch signaling for tumor maintenance in vivo.37,38
Because of these uncertainties, we undertook studies designed to address the ability of other pathways or signals to substitute for Notch in the genesis of T-ALL. We created a “first hit” in murine thymocytes by expressing a KrasG12D oncogene and scored leukemogenesis in the presence or absence of a dominant-negative inhibitor of canonical Notch signaling, DNMAML. Like pharmacological Notch inhibitors, DNMAML presents a steep (but not insurmountable) barrier; because all components of the Notch signaling machinery are intact, cells can adapt to this strong negative selective pressure by restoring Notch signaling, or alternatively can reexpress key target genes through other mechanisms. Indeed, we observed that T-ALL still developed in DNMAML mice by suppressing DNMAML expression and reactivating the Notch pathway, indicating strong selection for Notch activation during development of Ras-driven T-ALL. We further show that the key Notch target responsible for this selective pressure is Myc, which can substitute for Notch; by contrast, activated Akt cannot. These studies highlight the importance of the Notch/Myc axis in providing critical signals that cannot be easily duplicated through other mechanisms.
Methods
Mice
LSL-DNMAML-GFP,42 LSL-KrasG12D,43 LCR,44 LC,4 and LCD4 mice are described; Table 1 shows additional mice used. For bone marrow (BM) transplantation (BMT) studies, 4- to 12-week-old LCR mice were donors and littermate LC mice were recipients. Donors were divided into pools that were age-matched for each experimental condition. Experiments were performed according to National Institutes of Health (NIH) guidelines with approved institutional animal care and use committee protocols.
Mouse strains used in the study
| Mouse identification . | Genotype . |
|---|---|
| LC | Lck-cre+ |
| LCR | Lck-cre+ LSL-KrasG12D |
| LCD | Lck-cre+ LSL-DNMAML-GFP+/− |
| LCDR | Lck-cre+ LSL-KrasG12D LSL-DNMAML-GFP+/− |
| LCDD | Lck-cre+ LSL-DNMAML-GFP+/+ |
| LCDDR | Lck-cre+ LSL-KrasG12D LSL-DNMAML-GFP+/+ |
| Mouse identification . | Genotype . |
|---|---|
| LC | Lck-cre+ |
| LCR | Lck-cre+ LSL-KrasG12D |
| LCD | Lck-cre+ LSL-DNMAML-GFP+/− |
| LCDR | Lck-cre+ LSL-KrasG12D LSL-DNMAML-GFP+/− |
| LCDD | Lck-cre+ LSL-DNMAML-GFP+/+ |
| LCDDR | Lck-cre+ LSL-KrasG12D LSL-DNMAML-GFP+/+ |
LSL-KrasG12D refers to a transgene containing a floxed-stop cassette upstream of the KrasG12D oncogene inserted into the endogenous Kras locus. LSL-DNMAML-GFP refers to a transgene containing a floxed-stop cassette upstream of the DNMAML peptide/GFP fusion protein inserted into the Rosa26 locus.
Constructs and retroviruses
Antibodies
Antibodies for flow cytometry and western blotting are provided in supplemental Table 1 (available on the Blood Web site).
Flow cytometry
Cells were stained on ice in phosphate-buffered saline containing 2% fetal bovine serum (FBS), 10 mN 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid, N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) (HEPES), and 0.02% NaN3 after blocking with rat and mouse immunoglobulin G (Sigma-Aldrich) and 2.4G2 cell supernatant. Acquisition was performed on a FACSCalibur (Becton Dickinson). Cell cycle and Annexin V analyses were performed by staining with 7-aminoactinomycin D (7-AAD) solution or allophycocyanin Annexin V antibody according to the manufacturer’s protocol (Becton Dickinson). A FACSAria (Becton Dickinson) or Mo-Flo (DAKO) was used to sort cells. Dead cells and doublets were excluded based on forward scatter (FSC) and side scatter (SSC) characteristics on the FACSCalibur/Mo-Flo and FSC width and SSC width characteristics and 4′,6-diamidino-2-phenylindole, dihydrochloride staining on the FACSAria. Data were analyzed with FlowJo (Tree Star).
BM transduction and transplantation
Lymphoma-initiating cell assay
LCDDR thymic lymphomas were sorted into green fluorescence protein–positive/negative (GFP+/GFP−) populations. Dead and doublet cells were excluded. Defined numbers of sorted leukemia cells mixed with 200 000 competitor C57BL/6 cells were injected into lethally irradiated syngeneic recipients. Mice were observed at least 6 months for T-ALL development. The progenitor frequency was calculated with L-Calc software (StemCell Technologies).
Cell culture
Primary tumor cells extracted from leukemic mice were grown in RPMI 1640 (Invitrogen) supplemented with 10% FBS (GIBCO), 2 mM l-glutamine, 2-mercaptoethanol (0.0005% [vol/vol]; Sigma-Aldrich), and antibiotics. Treatment of cell lines with GSI (JC-19), provided by Yueming Ling (Memorial Sloan Kettering Cancer Center [MSKCC], New York, NY), is described.44
Quantitative real-time polymerase chain reaction
Total RNA was prepared using TRIzol (Invitrogen). Random-primed total RNAs (2 µg) were reverse-transcribed with SuperScript II (Invitrogen). Mouse Deltex1 (Mm00492297_m1), Hes1 (Mm00468601_m1), and Myc (Mm00487803_m1) RNAs were quantified using primer/probe sets from TaqMan Gene Expression Assays (Applied Biosystems). Sequences of the 18s primer set are described.49 Transcripts were amplified with TaqMan Universal PCR Master Mix or Sybr Green PCR Master Mix on the ABI Prism 7900 sequence detection system (Applied Biosystems).
Statistical analysis
Comparison of survival curves using a log-rank (χ2) analysis and the Student t test analysis were done with the Prism 4.03 software package (GraphPad Software).
Genomic PCR analysis and sequencing
DNA was isolated from fresh or snap-frozen thymic lymphomas or mouse tail clippings. Polymerase chain reaction (PCR) was performed using the following primers: Rosa1 (AAAGTCGCTCTGAGTTGTTAT), Rosa2 (GCGAAGAGTTTGTCCTCAACC), and Rosa3 (GGAGCGGGAGAAATGGATATG). DNA sequencing for NRR and PEST domain mutations50 and RAG-mediated 5′ deletions in Notch116,17 is described.50
TAT-Cre
TAT-Cre protein production is described.7 Thymocytes (107) obtained from thymic lymphomas of LCDR mice or LSL-DNMAML-GFP controls were incubated in 1 mL of 100 μg/mL TAT-Cre in serum-free OPTI-MEM (Life Technologies) for 45 minutes at 37°C. Cells were then washed and cultured 20 hours in RPMI 1640 (Life Technologies) supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM l-glutamine, and 0.0005% (vol/vol) 2-mercaptoethanol.
Western blot analyses
Twenty-five micrograms of extract harvested from thymic lymphomas were run on a 9% acrylamide gel, transferred to a polyvinylidene difluoride membrane, and blocked with 5% milk. Blots were stained with the indicated antibodies (supplemental Table 1).
Results
Coexpressing KrasG12D and the pan-Notch inhibitor DNMAML-GFP
In our KrasG12D-initiated T-ALL mouse model,44 thymocyte-specific conditional KrasG12D expression is regulated by a proximal Lck-promoter Cre recombinase (“Lck-cre”) transgene, which becomes active prior to thymocyte β-selection.51 Cre activates the KrasG12D allele from its endogenous regulatory elements by excising an upstream floxed stop cassette.43 These mice, called “LCR,” develop fully penetrant T-ALL with a high frequency (>80%) of acquired Notch1 mutations.44 We also engineered LSL-DNMAML-GFP mice that conditionally express DNMAML-GFP, a pan-Notch inhibitor consisting of DNMAML fused to GFP.4 Cre activates DNMAML-GFP expression from the ROSA26 locus by excising an upstream floxed stop cassette. When LSL-DNMAML-GFP mice are bred to Lck-cre mice to generate LCD mice, DNMAML-GFP is expressed in the majority of T cells at the DN3 stage and all T cells by the DN4 stage of T-cell development.4 LCD mice,4 LckCre+Notch1f/f mice,52 and LckCre+Rbpjf/f mice53 all show an approximately fivefold to sixfold decrease in thymocytes compared with controls, and show similar perturbations in thymocyte subsets, consisting of slightly higher percentages of double-negative (DN) T cells and slightly lower percentages of double-positive (DP) T cells.4,52,53 Thus, based on its ability to phenocopy other loss-of-function models, DNMAML-GFP appears to be a potent Notch inhibitor.
In order to test the requirement for Notch in Ras-induced T-ALL, we bred LSL-KrasG12D mice, LSL-DNMAML mice, and Lck-Cre mice to generate 6 mouse strains (LC, LCR, LCD, LCDR, LCDD, and LCDDR; Table 1). LC mice contain the Lck-cre transgene. LCR mice contain both the Lck-cre transgene and the KrasG12D transgene. LCD and LCDD contain the Lck-cre transgene and a single copy (LCD) or 2 copies (LCDD) of the DNMAML-GFP transgene. LCDR and LCDDR contain the Lck-cre/KrasG12D transgenes and a single copy (LCDR) or 2 copies (LCDDR) of the DNMAML-GFP transgene.
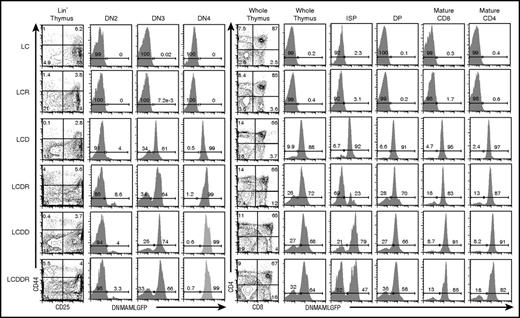
DNMAML-GFP expression was verified in all thymic T-cell subsets in these mouse strains (Figure 1). In Lck-cre transgenic mice, Cre recombinase is first expressed at the DN2 stage of thymocyte development. DNMAML-GFP expression was similar in LCD, LCDR, LCDD, and LCDDR mice (Figure 1; Maillard et al4 ). In each mouse strain, DNMAML-GFP is expressed by 3% to 9% of DN2 cells, 64% to 74% of DN3 cells, and >95% of DN4 cells. After the DN4 stage, the fraction of DNMAML-GFP dropped to lower levels (23%-79% in the immature single-positive [ISP]; 58%-70% in the DP) but recovered to >80% in the mature CD4 and CD8 compartments. Thus, DNMAML-GFP was expressed throughout thymocyte development in the LCDR and LCDDR mice. As expected, DNMAML-GFP was not expressed in LC or LCR mouse thymocytes (Figure 1). Consistent with Notch inhibition, the absolute number of thymocytes and each post-DN2 thymic T-cell subset (when the Lck-Cre transgene was expressed in the majority of cells) was decreased in the DNMAML-GFP–expressing cells, except in the DN4 LCDR+ and LCDDR+ populations and ISP LCDR− and LCDDR− populations (supplemental Figure 1). The basis for the increase in these populations is uncertain; one possibility is that it is related to emergence of transformed cells.
The pan-Notch inhibitor DNMAML-GFP is expressed in most T cells beginning at the DN3 stage. Flow cytometric analysis of DN T-cell development and DNMAML-GFP expression in DN2 (CD44+CD25+Lin−), DN3 (CD44−CD25+Lin−), and DN4 (CD44−CD25−Lin−) subsets at 5 to 7 weeks of age in the 6 different mouse strains used in the study (Table 1). Flow cytometric analysis of thymic development beyond the DN T-cell stage and expression of the DNMAML-GFP genetic inhibitor in ISP (CD8+TCRβ−), DP (CD4+CD8+), mature CD8 (CD8+TCRβ+), and mature CD4 (CD4+TCRβ+) subsets in the 6 different mouse strains used in the study. See supplemental Figure 1 for absolute numbers.
The pan-Notch inhibitor DNMAML-GFP is expressed in most T cells beginning at the DN3 stage. Flow cytometric analysis of DN T-cell development and DNMAML-GFP expression in DN2 (CD44+CD25+Lin−), DN3 (CD44−CD25+Lin−), and DN4 (CD44−CD25−Lin−) subsets at 5 to 7 weeks of age in the 6 different mouse strains used in the study (Table 1). Flow cytometric analysis of thymic development beyond the DN T-cell stage and expression of the DNMAML-GFP genetic inhibitor in ISP (CD8+TCRβ−), DP (CD4+CD8+), mature CD8 (CD8+TCRβ+), and mature CD4 (CD4+TCRβ+) subsets in the 6 different mouse strains used in the study. See supplemental Figure 1 for absolute numbers.
T-ALLs developing in heterozygous and homozygous DNMAML-GFP mice suppress DNMAML-GFP expression
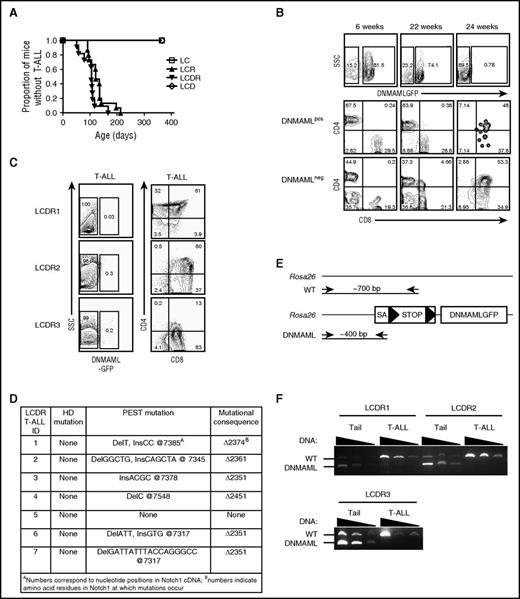
We compared the rate of T-ALL development in LCR mice (which express oncogenic KrasG12D) and LCDR mice (which express KrasG12D and DNMAML-GFP) (Figure 2A). As expected, LC and LCD control mice did not develop T-ALL after >1 year of observation. LCR mice developed T-ALL with 100% penetrance and median survival of ∼121 days. Unexpectedly, LCDR mice also developed T-ALL with 100% penetrance, with a median survival of ∼105 days. The MEK–extracellular signal-regulated kinase pathway was activated, suggesting that the KrasG12D transgene was functional (supplemental Figure 2). All tumors in LCDR mice lost DNMAML-GFP expression (Figure 2B-C). Figure 2B shows a representative LCDR mouse that was bled every 2 to 3 weeks to monitor T-ALL development. Initially, the percentage of peripheral blood T cells that expressed DNMAML-GFP was high (∼80% of Thy-1+ cells expressed GFP). By 22 weeks of age, an immature CD4+CD8+ DNMAML-GFP− blast population was first detected (∼4.66% of GFP−Thy-1+ cells). By 24 weeks of age, this aberrant DNMAML-GFP− T-ALL population dominated the blood, GFP+ cells were absent, and the mouse succumbed to T-ALL. All T-ALLs that developed in LCDR mice lacked DNMAML-GFP expression in the thymus and other organs, including spleen, BM, and lymph nodes (Figure 2C and not shown). These tumors were transplantable into secondary, sublethally irradiated recipients (not shown). Mice were sacrificed due to morbidity at an average age of 110 days (range, 80-167 days). At necropsy, the vast majority of these tumors (∼86%) contained PEST domain mutations in the endogenous Notch1 locus (Figure 2D). Past work showed that PEST domain mutations often are found together with 5′ Notch1 deletions that disrupt the Notch1 NRR in mouse T-ALL models (including LCR).16 Indeed, we noted previously that cell lines derived from LCDR1 and LCDR4 tumors (cell lines 385 and 330, respectively; Ashworth et al16 ) contain 5′ Notch1 deletions. Together, these data show that these tumors escaped Notch inhibition by losing DNMAML expression and activating Notch1 through mutation, emphasizing the strong positive selection for Notch1 activation in the Kras T-ALL model.
Escape from Notch suppression by heterozygous DNMAML-GFP occurs frequently during T-ALL development. (A) Survival curve showing the fraction of LC (N = 5 mice), LCR (N = 15), LCDR (N = 11), and LCD (N = 6) mice developing T-ALL over time. (B) Flow cytometric analysis of peripheral blood leukocytes showing expression of DNMAML-GFP of a representative LCDR mouse. Both DNMAML-GFP+ and DNMAML-GFP− (internal negative control) subsets were analyzed for immature T-cell blasts using CD4 and CD8 markers at 6 weeks, 22 weeks, and 24 weeks of age. (C) Flow cytometric analysis of thymic lymphomas of 3 representative LCDR mice with T-ALL showing expression of DNMAML-GFP, CD4, and CD8. (D) DNA sequence analysis of endogenous Notch1 in thymic lymphomas of 7 LCDR mice with T-ALL. Mutations in the HD and PEST domains are shown in addition to the mutational consequence in the protein. Mutations were detected upon morbidity at 110 days (total average) of age, 107 days (LCDR1), 105 days (LCDR2), 80 days (LCDR3), 167 days (LCDR4), and 90 days (LCDR6). (E) Genomic PCR strategy to determine the presence of DNMAML-GFP at the ROSA26 locus. The DNMAML-GFP (“DNMAML”) allele generates a ∼400-bp fragment whereas the WT allele generates a ∼700-bp fragment. (F) Genomic PCR analysis of 3 thymic lymphomas for the DNMAML-GFP and WT alleles at the ROSA26 locus. cDNA, complementary DNA; ID, identification; SA, splice acceptor.
Escape from Notch suppression by heterozygous DNMAML-GFP occurs frequently during T-ALL development. (A) Survival curve showing the fraction of LC (N = 5 mice), LCR (N = 15), LCDR (N = 11), and LCD (N = 6) mice developing T-ALL over time. (B) Flow cytometric analysis of peripheral blood leukocytes showing expression of DNMAML-GFP of a representative LCDR mouse. Both DNMAML-GFP+ and DNMAML-GFP− (internal negative control) subsets were analyzed for immature T-cell blasts using CD4 and CD8 markers at 6 weeks, 22 weeks, and 24 weeks of age. (C) Flow cytometric analysis of thymic lymphomas of 3 representative LCDR mice with T-ALL showing expression of DNMAML-GFP, CD4, and CD8. (D) DNA sequence analysis of endogenous Notch1 in thymic lymphomas of 7 LCDR mice with T-ALL. Mutations in the HD and PEST domains are shown in addition to the mutational consequence in the protein. Mutations were detected upon morbidity at 110 days (total average) of age, 107 days (LCDR1), 105 days (LCDR2), 80 days (LCDR3), 167 days (LCDR4), and 90 days (LCDR6). (E) Genomic PCR strategy to determine the presence of DNMAML-GFP at the ROSA26 locus. The DNMAML-GFP (“DNMAML”) allele generates a ∼400-bp fragment whereas the WT allele generates a ∼700-bp fragment. (F) Genomic PCR analysis of 3 thymic lymphomas for the DNMAML-GFP and WT alleles at the ROSA26 locus. cDNA, complementary DNA; ID, identification; SA, splice acceptor.
We next investigated the mechanism by which tumors lost DNMAML-GFP. Insufficient Cre recombinase levels were unlikely to account for the absence of DNMAML-GFP expression in LCDR tumors, as treating LCDR tumor cells with Tat-cre peptide7 failed to induce DNMAML-GFP expression, whereas thymocytes from a control mouse efficiently expressed DNMAML-GFP following Tat-cre peptide treatment (supplemental Figure 3). To determine whether the DNMAML-GFP transgene was deleted in LCDR tumors, we used a multiplex genomic PCR assay (Figure 2E). Amplification of LCDR mouse tail DNA generated PCR products of the expected size for the wild-type Rosa26 locus and the DNMAML-GFP transgenic locus. However, LCDR mouse tumor DNA generated only products consistent with the presence of the wild-type Rosa26 allele (Figure 2F). These data show that the LCDR tumors deleted the DNMAML-GFP locus, which is the likely mechanism by which these tumors escaped Notch inhibition.
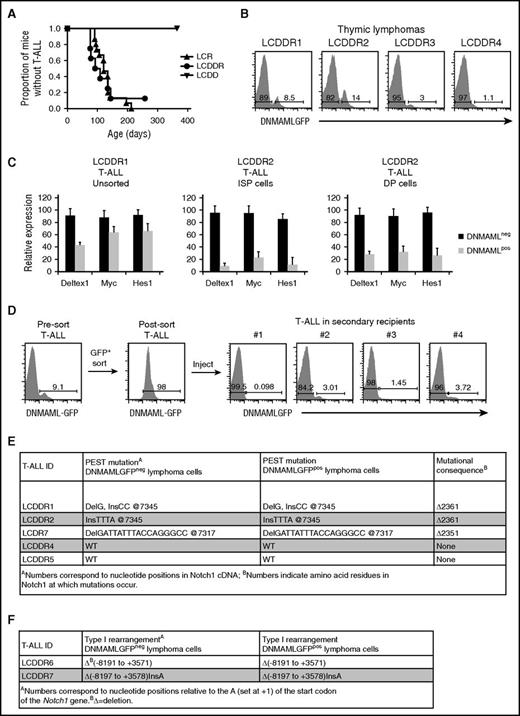
We hypothesized that 2 copies of the DNMAML-GFP allele (in the LCDDR mice) would inhibit leukemogenesis more efficiently than 1 copy (in the LCDR mice). However, the LCDDR mice developed T-ALL with nearly 100% penetrance and median survival of ∼101 days (Figure 3A). Survival of the LCDDR mice was similar to the LCDR and LCR mice. Like the LCDR tumors, DNMAML-GFP expression was lost in the LCDDR tumors (Figure 3B). Assuming that deletion also is the main mechanism of DNMAML-GFP loss in the homozygous model (as in the heterozygous model), it appears that deletion was incomplete in LCDDR tumors. Specifically, although DNMAML-GFP was lost in >99.5% of total cells in LCDR tumors (Figure 2C), small subsets of cells in LCDDR tumors (∼1%-15% of total cells) maintained DNMAML-GFP expression (Figure 3B).
Escape from Notch suppression by homozygous DNMAML-GFP occurs frequently during T-ALL development. (A) Survival curve showing the fraction of LCR (N = 15), LCDDR (N = 8), and LCDD (N = 6) mice that develop T-ALL over time. (B) Flow cytometric analysis of thymic lymphomas of 4 representative LCDDR mice with T-ALL showing loss of expression of DNMAML-GFP. (C) Quantitative real-time polymerase chain reaction analysis for expression of Notch1 target genes (Deltex1, Myc, and Hes1). For the LCDDR1 tumor, messenger RNA (mRNA) of DNMAML-GFP+ cells is compared with mRNA of DNMAML-GFP− cells. For the LCDDR2 tumor, mRNA of DNMAML-GFP+ cells is compared with mRNA of DNMAML-GFP− cells of sorted ISP cells (CD8+TCRβ−) or DP cells (CD4+CD8+). (D) Thymic lymphoma cells from LCDDR mice were enriched for cells expressing DNMAML-GFP and transferred to lethally irradiated recipients. DNMAML-GFP expression was analyzed by flow cytometry in presort T-ALL cells, postsort T-ALL cells, and T-ALL cells in 4 representative secondary recipients. (E-F) DNA sequence analysis of (E) exon 34 (where PEST mutations occur; 4 LCDDR mice and 1 LCDR mouse) or (F) the 5′ region of endogenous Notch1 (where type I rearrangements occur16 ; 2 LCDDR mice) in thymic lymphomas sorted for DNMAML-GFP+ and DNMAML-GFP− subpopulations.
Escape from Notch suppression by homozygous DNMAML-GFP occurs frequently during T-ALL development. (A) Survival curve showing the fraction of LCR (N = 15), LCDDR (N = 8), and LCDD (N = 6) mice that develop T-ALL over time. (B) Flow cytometric analysis of thymic lymphomas of 4 representative LCDDR mice with T-ALL showing loss of expression of DNMAML-GFP. (C) Quantitative real-time polymerase chain reaction analysis for expression of Notch1 target genes (Deltex1, Myc, and Hes1). For the LCDDR1 tumor, messenger RNA (mRNA) of DNMAML-GFP+ cells is compared with mRNA of DNMAML-GFP− cells. For the LCDDR2 tumor, mRNA of DNMAML-GFP+ cells is compared with mRNA of DNMAML-GFP− cells of sorted ISP cells (CD8+TCRβ−) or DP cells (CD4+CD8+). (D) Thymic lymphoma cells from LCDDR mice were enriched for cells expressing DNMAML-GFP and transferred to lethally irradiated recipients. DNMAML-GFP expression was analyzed by flow cytometry in presort T-ALL cells, postsort T-ALL cells, and T-ALL cells in 4 representative secondary recipients. (E-F) DNA sequence analysis of (E) exon 34 (where PEST mutations occur; 4 LCDDR mice and 1 LCDR mouse) or (F) the 5′ region of endogenous Notch1 (where type I rearrangements occur16 ; 2 LCDDR mice) in thymic lymphomas sorted for DNMAML-GFP+ and DNMAML-GFP− subpopulations.
This surprising finding led us to study whether DNMAML was functional in the small subset of GFP+LCDDR T-ALL cells. Sequencing showed that the DNMAML-GFP transgene was intact in the DNMAML-GFP+ tumor cells (not shown). To further prove that DNMAML-GFP inhibited Notch signaling, we compared the expression of the Notch1 target genes Dtx1, Myc, and Hes1 in DNMAML-GFP+ and DNMAML-GFP− T-ALL cells. Expression of these target genes was inhibited in the DNMAML-GFP+ tumor cells when compared with DNMAML-GFP− cells, a result that was even more evident in purified ISP and DP tumor cells (Figure 3C). Thus, DNMAML-GFP was functional in the small subset of DNMAML-GFP+ LCDDR tumor cells.
Our ability to detect and isolate rare T-ALL cells with functional DNMAML-GFP provided an opportunity to study the effect of DNMAML-GFP suppression in vivo. Our working model was that DNMAML-GFP− T-ALL cells arose from DNMAML-GFP+ cells. To test this idea, we purified DNMAML-GFP+ and DNMAML-GFP− T-ALL cells and transferred them to secondary recipients. DNMAML-GFP+ T-ALL cells generated DNMAML-GFP− T-ALLs in secondary recipients (Figure 3D). To further test the clonal relationships between these 2 populations, we purified the DNMAML-GFP+ and DNMAML-GFP− T-ALL cells from 6 LCDDR and 1 LCDR tumors, and sequenced the endogenous PEST or the 5′ regions of Notch1.16,44 Both the DNMAML-GFP+ and DNMAML-GFP− subsets harbored the same PEST mutations (3 tumors) (Figure 3E) or 5′ Notch1 rearrangements (2 tumors) (Figure 3F). Together, these data suggest a precursor-progeny relationship between the DNMAML-GFP+ and DNMAML-GFP− subsets, and support a model in which KrasG12D expressing DNMAML-GFP+ thymocytes acquire Notch1 mutations and then further amplify Notch1 signals by suppressing DNMAML-GFP.
Suppressing DNMAML-GFP enhances cell proliferation, survival, and LIC frequency
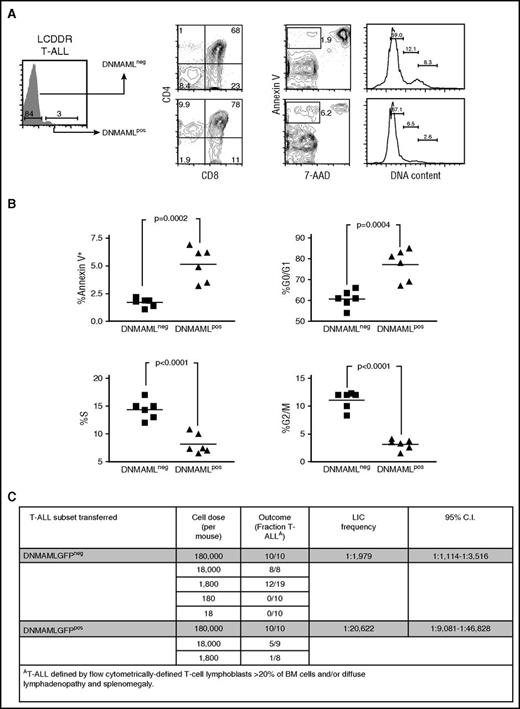
Previous studies established that Notch inhibition leads to G0/G1 cell cycle arrest and apoptosis in murine T-ALLs.54 The LCDDR tumors offered a unique opportunity to study this effect in vivo. The DNMAML-GFP− tumor subset had ∼2.5-fold fewer apoptotic cells and also had fewer cells in G0/G1 and more cells in G2/M/S phases of the cell cycle than the DNMAML-GFP+ subset (Figure 4A-B). These data show that once tumors suppress DNMAML-GFP, they are released from G0/G1 arrest and have a survival advantage.
Suppression of DNMAML-GFP improves survival, relieves cell cycle arrest, and increases LIC frequency. Sorted DNMAML-expressing thymic lymphoma cells from LCDDR mice were transferred into secondary recipients. T-ALLs that developed in these mice were subdivided into T-ALL cells that continued to express DNMAML-GFP (DNMAML+) and T-ALL cells that suppressed DNMAML-GFP (DNMAML−). These 2 populations were then analyzed for T-cell markers (CD4 and CD8), apoptotic cells (Annexin V+/7-AAD−), and cell cycle status using flow cytometry. A representative recipient mouse is shown in panel A and the rest of the cohort is shown in panel B in scatter plot format. P values (Student t test) were .0002 (% Annexin V+/7-AAD−), .0004 (% G0/G1), <.0001 (% S), and <.0001 (% G2M) and are shown on the plots. (C) Sorted DNMAML-GFP+ or DNMAML-GFP− thymic lymphoma cells were transferred at limiting dilution into lethally irradiated secondary recipient mice. The number of mice in each cohort that developed T-ALL over at least 6 months was used to determine the frequency of LIC activity by Poisson statistics. C.I., confidence interval.
Suppression of DNMAML-GFP improves survival, relieves cell cycle arrest, and increases LIC frequency. Sorted DNMAML-expressing thymic lymphoma cells from LCDDR mice were transferred into secondary recipients. T-ALLs that developed in these mice were subdivided into T-ALL cells that continued to express DNMAML-GFP (DNMAML+) and T-ALL cells that suppressed DNMAML-GFP (DNMAML−). These 2 populations were then analyzed for T-cell markers (CD4 and CD8), apoptotic cells (Annexin V+/7-AAD−), and cell cycle status using flow cytometry. A representative recipient mouse is shown in panel A and the rest of the cohort is shown in panel B in scatter plot format. P values (Student t test) were .0002 (% Annexin V+/7-AAD−), .0004 (% G0/G1), <.0001 (% S), and <.0001 (% G2M) and are shown on the plots. (C) Sorted DNMAML-GFP+ or DNMAML-GFP− thymic lymphoma cells were transferred at limiting dilution into lethally irradiated secondary recipient mice. The number of mice in each cohort that developed T-ALL over at least 6 months was used to determine the frequency of LIC activity by Poisson statistics. C.I., confidence interval.
Although inhibiting Notch can eradicate T-ALL blasts,37,38 it is questionable whether Notch inhibition targets lymphoma-initiating cells (LICs). This is particularly relevant because in vivo GSI treatment of murine T-ALL leads to only transient regression.35,36,41 Our ability to detect and isolate T-ALL cells with functional DNMAML-GFP in the LCDDR tumors provided an opportunity to test the effect of Notch inhibition on LICs. We purified DNMAML-GFP+ and DNMAML-GFP− cells and transferred these populations at varied doses into cohorts of lethally irradiated recipient mice (Figure 4C). DNMAML-GFP− T-ALL cells were more efficient than DNMAML-GFP+ T-ALL cells in transferring T-ALL, as the LIC frequency was >10 times higher in DNMAML-GFP− cells (∼1 in 1979) than in DNMAML-GFP+ cells (∼1 in 20 622). As expected, the T-ALLs that developed in mice receiving DNMAML-GFP+ cells lost DNMAML-GFP after transplantation (Figure 3D and not shown). These data suggest that Notch inhibition targets LICs and may explain the strong selection for suppressing DNMAML-GFP and activating Notch during T-ALL development.
Myc, but not Akt, can relieve the selective pressure for Notch activation during leukemogenesis
We next sought to determine the basis of the strong selection for Notch1 activation. Specifically, we tested the ability of 2 downstream events, Myc upregulation and Akt activation, to substitute for Notch signaling. We chose Myc and Akt as both can induce T-ALL in mouse models55,56 and multiple studies link these targets to Notch activation in T-ALL.23,28-31
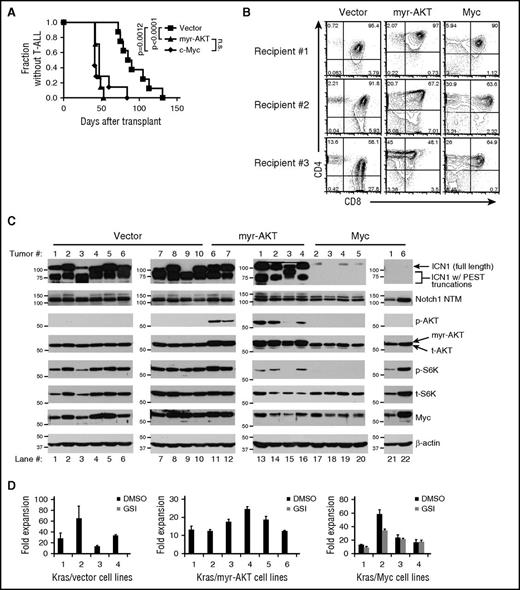
BM progenitors from LCR mice were transduced with retroviruses expressing control vector, Myc, or myristoylated AKT (myr-AKT) and then transferred to lethally irradiated LC littermates. Myc and myr-AKT shortened the median time to leukemic presentation from ∼87.5 days to ∼41 days and ∼47 days, respectively, as compared with control virus (Figure 5A). CD4 and CD8 expression of the various Myc, myr-AKT, and empty vector T-ALLs were variable and consistent with developmental arrest at the ISP to DP stages (Figure 5B). These data show that both Myc and myr-AKT accelerate KrasG12D-initiated T-ALL development.
Ectopic expression of Myc, but not myr-AKT, bypasses the need for Notch activation and generates lymphomas resistant to Notch inhibition. Lethally irradiated LC recipient mice were reconstituted with 5FU-treated donor LCR BM cells after transduction with retroviral supernatants expressing empty vector (N = 8), myr-AKT (N = 7), or Myc (N = 7). (A) Kaplan-Meier curve showing fraction of mice without T-ALL after BMT. P values (log-rank test) were <.0001 (vector vs myr-AKT), .0012 (vector vs Myc), and not significant (n.s.) (myr-AKT vs Myc). (B) Flow cytometric analysis of CD4 and CD8 expression of vector, myr-AKT, and Myc-transduced thymic lymphomas. (C) Western blot analysis of vector, myr-AKT, and Myc-transduced T-ALLs (for antibodies and blotting procedures, see “Methods”). ICN1, staining observed with an antibody against the V1744 epitope, which only recognizes the γ-secretase cleaved ICN1; Notch1 NTM, staining observed with an antibody against the C-terminal region of Notch1, which recognizes the furin-processed NTM; p-AKT, staining observed with an antibody specific for AKT phosphorylated on S473; p-S6K, staining observed with an antibody specific for S6K phosphorylated on Thr389; t-AKT, total AKT; t-S6K, total S6K. The arrow indicates ICN1 likely resulting from 5′ rearrangements or other mutations as described in “Results.” The multiple bands within the brackets likely result from combined PEST mutations and 5′ rearrangements or other mutations as described in “Results.” (D) Primary T-ALL cell lines derived from vector, myr-AKT, and Myc T-ALLs were cultured ex vivo for 3 weeks. These cell lines were then treated with GSIs (1 μM JC-19) for 6 days and counted on day 6. Fold expansion was calculated based on the initial day 0 cell counts. DMSO, dimethyl sulfoxide.
Ectopic expression of Myc, but not myr-AKT, bypasses the need for Notch activation and generates lymphomas resistant to Notch inhibition. Lethally irradiated LC recipient mice were reconstituted with 5FU-treated donor LCR BM cells after transduction with retroviral supernatants expressing empty vector (N = 8), myr-AKT (N = 7), or Myc (N = 7). (A) Kaplan-Meier curve showing fraction of mice without T-ALL after BMT. P values (log-rank test) were <.0001 (vector vs myr-AKT), .0012 (vector vs Myc), and not significant (n.s.) (myr-AKT vs Myc). (B) Flow cytometric analysis of CD4 and CD8 expression of vector, myr-AKT, and Myc-transduced thymic lymphomas. (C) Western blot analysis of vector, myr-AKT, and Myc-transduced T-ALLs (for antibodies and blotting procedures, see “Methods”). ICN1, staining observed with an antibody against the V1744 epitope, which only recognizes the γ-secretase cleaved ICN1; Notch1 NTM, staining observed with an antibody against the C-terminal region of Notch1, which recognizes the furin-processed NTM; p-AKT, staining observed with an antibody specific for AKT phosphorylated on S473; p-S6K, staining observed with an antibody specific for S6K phosphorylated on Thr389; t-AKT, total AKT; t-S6K, total S6K. The arrow indicates ICN1 likely resulting from 5′ rearrangements or other mutations as described in “Results.” The multiple bands within the brackets likely result from combined PEST mutations and 5′ rearrangements or other mutations as described in “Results.” (D) Primary T-ALL cell lines derived from vector, myr-AKT, and Myc T-ALLs were cultured ex vivo for 3 weeks. These cell lines were then treated with GSIs (1 μM JC-19) for 6 days and counted on day 6. Fold expansion was calculated based on the initial day 0 cell counts. DMSO, dimethyl sulfoxide.
We next assessed Notch1 activation in empty vector, Myc, and myr-AKT T-ALLs by performing western blot analysis (Figure 5C). All of the control and myr-AKT tumors had high levels of 1 or more ICN1 polypeptides. In murine T-ALL, high levels of ICN1 strongly correlate with the presence of 5′ Notch1 deletions and other mutations that target the NRR16 whereas the presence of small ICN1 species, alone or in combination with normal-sized ICN1, is consistent with the presence of truncating PEST domain mutations.9,50,54 By inference, the existence of several different ICN1 polypeptides in most empty vector and myr-AKT tumors means that several independent tumors or tumor subclones with different combinations of Notch1 mutations are present in these mice. By contrast, all Myc tumors had very low or undetectable levels of ICN1, despite containing similar levels of Notch1 (Notch1 transmembrane subunit [NTM]; Figure 5C), the transmembrane subunit of mature, resting Notch1 receptors. Importantly, the amount of Myc in the Myc-transduced tumors was similar to the levels in the control and myr-AKT tumors; hence, retroviral transduction did not drive Myc expression to levels higher than those found in Notch1-mutated tumors. Confirming that myr-AKT was functional, 5 of the 6 myr-AKT tumors had levels of phosphorylated AKT and S6 kinase that greatly exceeded those seen in vector or Myc-transduced KrasG12D tumors (Figure 5C). Further confirmation of the Notch independence of Myc tumors, but not AKT tumors, was provided by DNA sequencing, which identified activating PEST mutations in ∼89% of control tumors, ∼71% of myr-AKT tumors, and none of the Myc tumors (Table 2). Together, these data show that enforced expression of Myc but not myr-AKT circumvents the need for Notch activation during Kras-mediated leukemogenesis.
Frequency of PEST mutations in KrasG12D-initiated T-ALL tumors complemented with MigR1 (control), myr-AKT, or Myc
| Tumor . | PEST mutation* . | Mutational consequence† . |
|---|---|---|
| Control-1 | DelTACACA, InsAGGG @7282 | Δ2340 |
| Control-3 | None‡ | WT |
| Control-4 | Deletion TGGTCCCACCCATGACC 7604 | Δ2447 |
| Control-5 | Deletion CGGCTGGCAACACA 7345 | Δ2361 |
| Control-6 | Insertion AGTACCCC 7513 | Δ2416 |
| Control-7 | Insertion C 7524 | Δ2420 |
| Control-8 | Insertion AA/Deletion G 7346 | Δ2361 |
| Control-9 | Substitution C to G 7326 | Δ2354 |
| Control-10 | Insertion GGGG 7460 | Δ2399 |
| myr-AKT-1 | InsAGCC@7345 | Δ2361 |
| myr-AKT-2 | DelG, InsCC@7346 | Δ2361 |
| myr-AKT-3 | DelCA, InsGGGACCAA@7519 | Δ2419 |
| myr-AKT-4 | InsCC@7346 | Δ2361 |
| myr-AKT-5 | None | WT |
| myr-AKT-6 | None‡ | WT |
| myr-AKT-7 | DelG, InsCA@7317 | Δ2351 |
| Myc-1 | None | WT |
| Myc-2 | None | WT |
| Myc-3 | None | WT |
| Myc-4 | None | WT |
| Myc-5 | None | WT |
| Myc-6 | None | WT |
| Myc-7 | None | WT |
| Tumor . | PEST mutation* . | Mutational consequence† . |
|---|---|---|
| Control-1 | DelTACACA, InsAGGG @7282 | Δ2340 |
| Control-3 | None‡ | WT |
| Control-4 | Deletion TGGTCCCACCCATGACC 7604 | Δ2447 |
| Control-5 | Deletion CGGCTGGCAACACA 7345 | Δ2361 |
| Control-6 | Insertion AGTACCCC 7513 | Δ2416 |
| Control-7 | Insertion C 7524 | Δ2420 |
| Control-8 | Insertion AA/Deletion G 7346 | Δ2361 |
| Control-9 | Substitution C to G 7326 | Δ2354 |
| Control-10 | Insertion GGGG 7460 | Δ2399 |
| myr-AKT-1 | InsAGCC@7345 | Δ2361 |
| myr-AKT-2 | DelG, InsCC@7346 | Δ2361 |
| myr-AKT-3 | DelCA, InsGGGACCAA@7519 | Δ2419 |
| myr-AKT-4 | InsCC@7346 | Δ2361 |
| myr-AKT-5 | None | WT |
| myr-AKT-6 | None‡ | WT |
| myr-AKT-7 | DelG, InsCA@7317 | Δ2351 |
| Myc-1 | None | WT |
| Myc-2 | None | WT |
| Myc-3 | None | WT |
| Myc-4 | None | WT |
| Myc-5 | None | WT |
| Myc-6 | None | WT |
| Myc-7 | None | WT |
Numbers correspond to nucleotide positions in Notch1 complementary DNA.
Numbers indicate amino acid residues in Notch1 at which mutations occur.
Truncated Notch1 protein was detected by western blot (Figure 5C).
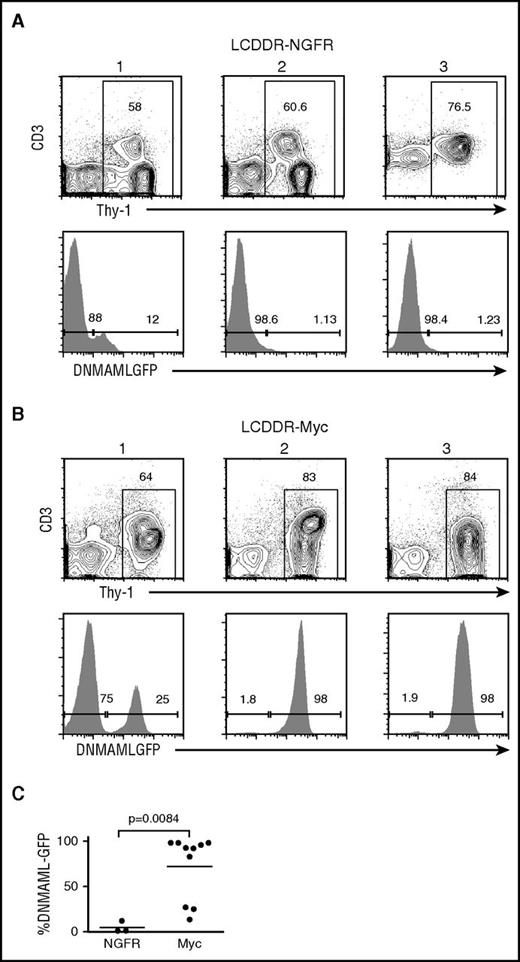
Because Myc substitutes for Notch activation, we predicted that the Myc-transduced tumors would be Notch-independent. To test this possibility, we cultured primary T-ALL cells from control, myr-AKT, and Myc tumors in the presence of vehicle or GSI. As expected, GSI markedly inhibited growth of control and myr-AKT cell lines, but had only minimal effects on the growth of Myc cell lines (Figure 5D). Furthermore, Myc-transduced BM progenitors from LCDDR mice had significantly higher DNMAML-GFP expression (Figure 6), emphasizing that Myc expression in these tumors relieves the selection pressure for Notch1 activation.
Ectopic Myc expression counters the loss of DNMAML-GFP expression in LCDDR tumors. Lethally irradiated LC recipient mice were reconstituted with 5FU-treated donor LCDDR BM cells after transduction with empty NFGR vector (negative control) or Myc-IRES-NGFR vector expressing retrovirus. The resulting splenic lymphoma cells in NGFR-transduced (A) and Myc-transduced (B) mice were analyzed for Thy-1 and CD3ε expression by flow cytometry. Three representative tumors are shown. DNMAML-GFP expression within the Thy-1+ gate is also shown and displayed as a scatterplot (C). P value was .0084 for the NGFR vs Myc comparison (Student t test).
Ectopic Myc expression counters the loss of DNMAML-GFP expression in LCDDR tumors. Lethally irradiated LC recipient mice were reconstituted with 5FU-treated donor LCDDR BM cells after transduction with empty NFGR vector (negative control) or Myc-IRES-NGFR vector expressing retrovirus. The resulting splenic lymphoma cells in NGFR-transduced (A) and Myc-transduced (B) mice were analyzed for Thy-1 and CD3ε expression by flow cytometry. Three representative tumors are shown. DNMAML-GFP expression within the Thy-1+ gate is also shown and displayed as a scatterplot (C). P value was .0084 for the NGFR vs Myc comparison (Student t test).
Discussion
Although activating NOTCH1 mutations occur frequently in both human T-ALL and murine models, the requirement for Notch signals in T-ALL models induced by other oncogenes is less clear.17,57 This is an important therapeutic issue, as it may shed light on whether resistance to Notch inhibitors will depend on reactivation of Notch or other components of the Notch signaling axis or de novo activation of other pathways that circumvent Notch. To address this question, we investigated T-ALL initiation in mice expressing a conditional KrasG12D transgene in early thymocytes under control of Lck-cre. These mice develop highly penetrant T-ALL that acquire activating Notch1 mutations at high frequency.44 This mouse model is relevant to human T-ALL, as the Ras pathway is activated in ∼50% of human T-ALLs.58-61 To determine the importance of Notch activation in the pathogenesis of these tumors, we expressed the pan-Notch inhibitor, DNMAML-GFP, in these mice. Surprisingly, instead of utilizing alternative signaling pathways, the T-ALL cells acquired activating Notch1 mutations and suppressed the DNMAML-GFP allele (even when present at homozygous dosage). These data show that there is high selective pressure for Notch1 activation, suggesting that Notch1 signals are not easily duplicated or replaced through other mechanisms.
Notably, however, although NOTCH1 mutations are found in >50% of human T-ALL, Notch mutation/activation is not ubiquitous. Based on our studies, it seems possible that Notch-independent human tumors may arise due to alternative events that upregulate MYC, an idea that will require precise quantitation of MYC expression in human T-ALLs and correlation with driver mutations in NOTCH1 and other genes. Indeed, given that Myc can be dysregulated through many mechanisms, including Myc gene rearrangements,62 it is not clear why Notch activation is the predominant mechanism in T-ALL, particularly because it appears that T-ALL cells can access alternative epigenetic states that relieve the dependence on Notch for Myc expression.35,63 Such alternative mechanisms may also contribute to the ultimate failure of Notch inhibitors in most mouse models and in patients, an idea that remains to be tested. Because our model relies on an oncogenic Ras allele, it also suggests that Myc (either by itself or through Notch) complements Ras to drive T-ALL development. Consistent with this idea, murine retroviral mutagenesis experiments revealed a propensity for Rasgrp1 (Ras guanyl releasing protein 1) and Myc insertions within the same T-ALL tumor.64
Our results should not be taken as evidence that other Notch targets (such as PI3K/AKT/mTOR) are dispensable for T-ALL development; indeed, multiple studies show that mTOR activation is essential for T-ALL cell survival.23,27,36 Accordingly, treatment of KrasG12D-activated T-ALL cell lines with PI3K/mTOR inhibitors significantly inhibits cell growth (Chiang et al44 and Kindler et al,65 and data not shown). Rather, our results indicate that Notch targets apart from Myc cannot fully explain the selective pressure to activate Notch during leukemogenesis. T-ALL cells apparently can find alternative mechanisms to turn on these targets (or possibly complementary ones) in the absence of Notch activation. For example, at least some Myc-transduced LCDDR tumors activate mTOR through Notch-independent mechanisms (Figure 5C). The mechanism is not known, but there are multiple possibilities, as mTOR is downstream of many physiological growth factor signaling pathways (eg, T-cell receptor, platelet-derived growth factor receptor, Kit) and can be activated through aberrant regulation (eg, phosphatase and tensin homolog inactivation). Furthermore, our results show that Notch inhibition decreases, but does not eliminate, LIC activity, suggesting that effective targeting of LICs will likely require additional agents besides Notch inhibitors.
In summary, our results show that there is a strong selective pressure to activate Notch in the KrasG12D-induced T-ALL mouse model. Activating Notch improves cell survival and increases both proliferation and leukemia-initiating cell activity. We also show that Myc induction is a critical Notch function in T-ALL pathogenesis. Although Ras signaling is activated in ∼50% of patient samples, it remains to be seen whether Notch and Myc play similar roles in other T-ALL–prone genetic backgrounds and other Notch-dependent cancers. Our study adds to the accumulating evidence that support targeting the Notch/Myc axis in human T-ALL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Eden Childs and Candice Romany for excellent technical assistance and Lili Tu, David Tuveson, Gary Koretsky, Yueming Li, and Michael Tomasson for invaluable reagents. The authors are grateful to the following University of Pennsylvania cores: the Mouse Husbandry Core, the Abramson Cancer Center Flow Cytometry Core (P30CA016520), and the Abramson Family Cancer Research Institute Core.
This work was supported by the National Institutes of Health (NIH) National Cancer Institute (NCI) T32CA009140 and the American Cancer Society (ACS) PF-15-065-01-TBG (S.J.S.); NIH NCI R01CA196604, the ACS, the Rally Foundation for Childhood Cancer Research, Alex’s Lemonade Stand Foundation, and the Concern Foundation (M.Y.C.); and NIH National Institute of Allergy and Infectious Diseases R01AI047833, NIH NCI P01CA119070 (W.S.P. and J.C.A.).
Authorship
Contribution: M.Y.C. designed the study, performed most of the experiments, and wrote the manuscript; Q.W., A.C.G., S.J.S., L.X., and O.S. designed and performed experiments; J.C.A. and W.S.P. designed the study, supervised research, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark Y. Chiang, University of Michigan School of Medicine, 2043 Taubman BSRB, 109 Zina Pitcher Pl, Ann Arbor, MI 48109; e-mail: markchia@umich.edu; or Warren S. Pear, University of Pennsylvania School of Medicine, 556 BRB II/III, 421 Curie Blvd, Philadelphia, PA 19104; e-mail: wpear@mail.med.upenn.edu.