Key Points
C1q can form a multimolecular signaling complex with HMGB1, RAGE, and LAIR-1 in lipid rafts.
C1q and HMGB1 together promote monocytes to differentiate to an anti-inflammatory phenotype.
Abstract
A healthy immune system results from a balance of stimulatory and inhibitory pathways that allow effective responses to acute insults, without descending into chronic inflammation. Failed homeostasis is characteristic of autoimmune diseases such as systemic lupus erythematosus. Although HMGB1 induces proinflammatory M1-like macrophage differentiation, we describe a mechanism by which C1q modulates this activity and collaborates with HMGB1 to induce the differentiation of monocytes to anti-inflammatory M2-like macrophages. These anti-inflammatory macrophages are unresponsive to dendritic cell induction factors, effectively removing them from participation in an adaptive immune response. This pathway is mediated through a complex with RAGE and LAIR-1 and depends on relative levels of C1q and HMGB1. Importantly, these data provide insight into a homeostatic mechanism in which C1q and HMGB1 can cooperate to terminate inflammation, and which may be impaired in C1q-deficient patients with autoimmune disease.
Introduction
The ability of the mammalian immune system to avoid self-reactivity relies on a fine balance of multiple interrelated signaling pathways. When the inhibitory pathways are inadequate, autoreactivity and inflammation result in disease. In particular, systemic lupus erythematosus (SLE) is characterized by activation of endosomal Toll-like receptors (TLRs), leading to an immunogenic and inflammatory milieu1 in which monocytes/macrophages and dendritic cells (DCs) contribute to systemic inflammation and tissue injury.2-5 To date, therapeutic strategies for SLE have been largely palliative or rely on nonspecific immunosuppressive drugs with serious toxicities. Developing a targeted therapeutic requires a better understanding of the molecular mechanisms through which the body achieves a natural program of quiescence and of how this process is disrupted in SLE.
Molecules that may contribute to immune dysregulation in SLE include High Mobility Group Box 1 (HMGB1) and the first component of the complement system, C1q. HMGB1 is an evolutionarily ancient DNA-binding nucleoprotein.6,7 It can be released passively from dying cells or actively secreted by monocytes, macrophages, and myeloid DCs and function as a damage-associated molecular pattern (DAMP).8,9 HMGB1 functions as a critical cofactor for the activation of endosomal TLRs in SLE.10,11 When bound to the Receptor for Advanced Glycation Endproducts (RAGE), it facilitates the transport of RNA and DNA to endosomal TLRs, leading to the production of type 1 interferon (IFN), IFN-inducible genes, and proinflammatory cytokines, which skew the differentiation of monocytes toward proinflammatory macrophages.9,12-15 Significantly elevated serum levels of HMGB1 have been demonstrated in SLE patients,16,17 and administration of antibodies against HMGB1 confer protection against tissue injury in some experimental models of autoimmune disease and inflammation.18 HMGB1 has also been shown to suppress inflammation.19,20 It is unclear how it can elicit both pro- and anti-inflammation responses.
C1q is a similarly evolutionarily conserved molecule well known to possess immunosuppressive properties distinct from its role in initiating the complement cascade.21,22 It is a 460-kDa protein formed by 6 heterotrimeric subunits containing an N-terminal collagen-like sequence and a C-terminal globular region.23 It binds to pathogen-associated molecular patterns and endogenous DAMPs, including antibody-antigen complexes, myelin, and β-amyloid (Aβ)24 and can decrease myeloid cell activation and monocyte to DC differentiation. Patients with active SLE have lower levels of C1q both because the immune complexes in SLE consume complement components and because SLE patients produce antibodies that target C1q.25 Although rare, C1q deficiency is the strongest genetic risk factor for SLE,26,27 and a polymorphism that associates with decreased production confers risk for SLE.28,29 Recently, it was demonstrated that apoptotic cells bound by C1q suppress human macrophage and DC-mediated T helper 17 (Th17) and Th1 cell activation.30
Leukocyte-Associated Ig-like Receptor-1 (LAIR-1; CD305), a transmembrane protein of the immunoglobulin superfamily, is a high-affinity receptor for C1q.31 Several functions of C1q are mediated by its binding to LAIR-1, such as inhibition of monocyte-to-DC differentiation and plasmacytoid DC activation, functions that help explain the contribution of C1q deficiency to SLE pathogenesis.
Here, we investigated the possibility of a homeostatic relationship between C1q and HMGB1 and demonstrate a specific C1q-HMGB1 interaction in which C1q binds to HMGB1 and catalyzes formation of a multimeric protein complex comprising HMGB1, C1q, LAIR-1, and RAGE. This complex triggers monocytes to acquire an anti-inflammatory (M2-like) phenotype, upregulating the expression of CD163 and several anti-inflammatory molecules, including Programmed Death-Ligand 1 (PD-L1), Mer tyrosine-kinase (Mer), and Interleukin-10 (IL-10). These anti-inflammatory macrophages fail to differentiate to DCs, blocking the downstream adaptive immune response. Thus, we have identified a mechanism by which C1q levels modulate the inflammatory activity of HMGB1, a mechanism that is impaired in SLE due to genetic or acquired C1q deficiency.
Methods
Additional methods are presented in the supplemental Methods, available on the Blood Web site.
Reagents
Human C1q was obtained from Complement Technology (#A099). C1q tail was purified from whole C1q as previously described.31 Recombinant HMGB1 (Calmodulin Binding Protein Epitope, Cbp tagged) and reduced or oxidized forms of HMGB1 were generated as previously described.32,33 Human recombinant (r) RAGE was purchased from PROSPEC. rLAIR-2 and rLAIR-1 were purchased from R&D Systems. Complement C1 was purchased from EMD Millipore. Anti-C1q and anti-C1r antibodies were purchased from Abcam. Alexa Fluor 555 and the EZ-Link Sulfo-NHS-LC Biotinylation kit were purchased from Thermo Fisher Scientific. Purified proteins and culture reagents were endotoxin tested (<0.1 EU/mL) using a Limulus Amebocyte Lysate assay kit performed following the manufacturer’s instructions (Lonza).
Lipid raft fractionation
Lipid rafts from HMGB1-, C1q-, or HMGB1- and C1q-treated monocytes (5 × 106) were prepared as described.34
Phosphoimmunoreceptor array
Tyrosine phosphorylation of LAIR-1 was determined by human phosphoimmunoreceptor array (Proteome Profiler Array; R&D Systems) according to the manufacturer’s protocol and as described.31
Statistical analysis
Numerical values represent the mean of at least 3 independent experiments in which each condition was tested in triplicate. Variance of mean values between 2 groups was analyzed by the Student t test for unpaired observations. Group differences were tested with one-way analysis of variance (ANOVA) followed by Bonferroni’s correction for multiple comparisons, *P < .05; **P < .01; ***P < .001. Statistical analyses were performed using Prism 6 (Graphpad software).
Results
C1q inhibits HMGB1-induced monocyte activation
We have previously demonstrated that C1q inhibits the differentiation of monocytes to DCs and the activation of plasmacytoid DCs by engaging LAIR-1.31 To test whether C1q can inhibit the proinflammatory effects of HMGB1 on monocytes, we incubated cultures of human monocytes isolated from peripheral blood of healthy volunteers in the presence or absence of HMGB1, with and without C1q, and assessed downstream cytokine production. Cultures were performed in serum-free medium to avoid contamination by C1q in serum and to permit accurate control of the concentration of C1q. Reported levels for C1q in SLE are often <50 μg/mL (50-80 nM) and are between 50 and 150 μg/mL (100-300 nM) in control serum.35 The amount of HMGB1 in SLE serum is increased compared with healthy controls36 ; however, the HMGB1 enzyme-linked immunosorbent assay is an inadequate instrument for measuring HMGB1 levels in active SLE patients.17 Therefore, we chose to use a saturating concentration of HMGB1 (3 μg/mL) and C1q levels up to 50 ug/mL. As anticipated, the addition of HMGB1 dramatically increased the transcription and secretion of type 1 IFN, IFN-inducible genes, and NFκB-dependent proinflammatory cytokines, as previously reported7 (Figure 1A-B). Although C1q did not alter transcription of these genes in the absence of HMGB1, the addition of human C1q to HMGB1 counteracted the HMGB1-mediated cytokine transcription and protein secretion in a dose-dependent manner (0-50 μg/mL of C1q) (Figure 1A-B). Increasing the levels of HMGB1 abrogated the inhibitory effect of C1q (supplemental Figure 1A), suggesting that the ratio of C1q to HMGB1 is critical.
C1q inhibits expression of HMGB1-induced IFN-α, MX1, and inflammatory cytokines by human monocytes. (A) Messenger RNA (mRNA) levels for IFN-α, MX1, IL-6, and TNF-α in monocytes stimulated with or without HMGB1 (3 μg/mL) and various concentrations of C1q (μg/mL) for 6 hours in serum-free medium (mean ± standard deviation [SD] of triplicates, N = 3). (B) HMGB1 (3 μg/mL) -induced IL-6 and TNF-α secretion were reduced in the presence of various concentrations of C1q, assessed at 24 hours (mean ± SD of duplicates, N = 3). (C) IFN-α, MX1, IL-6, and TNF-α mRNA expression by monocytes transfected with control siRNA or LAIR siRNA and treated with C1q (25 μg/mL), HMGB1 (3 μg/mL) for 6 hours (mean ± SD of triplicates, N = 3). Data are expressed as fold induction relative to controls. R.E., relative expression. ns, not significant. *P < .05; **P < .01; ***P < .001 (one-way ANOVA with Bonferroni’s correction for multiple comparisons).
C1q inhibits expression of HMGB1-induced IFN-α, MX1, and inflammatory cytokines by human monocytes. (A) Messenger RNA (mRNA) levels for IFN-α, MX1, IL-6, and TNF-α in monocytes stimulated with or without HMGB1 (3 μg/mL) and various concentrations of C1q (μg/mL) for 6 hours in serum-free medium (mean ± standard deviation [SD] of triplicates, N = 3). (B) HMGB1 (3 μg/mL) -induced IL-6 and TNF-α secretion were reduced in the presence of various concentrations of C1q, assessed at 24 hours (mean ± SD of duplicates, N = 3). (C) IFN-α, MX1, IL-6, and TNF-α mRNA expression by monocytes transfected with control siRNA or LAIR siRNA and treated with C1q (25 μg/mL), HMGB1 (3 μg/mL) for 6 hours (mean ± SD of triplicates, N = 3). Data are expressed as fold induction relative to controls. R.E., relative expression. ns, not significant. *P < .05; **P < .01; ***P < .001 (one-way ANOVA with Bonferroni’s correction for multiple comparisons).
We next assessed the involvement of LAIR-1 in this C1q-mediated inhibition of HMGB1-induced monocyte activation. We observed a lack of effect of C1q on monocyte activation when monocytes were treated with LAIR-1-specific small interfering RNA (siRNA) (Figure 1C). C1q also failed to inhibit monocyte activation in the presence of the soluble decoy receptor LAIR-2 (supplemental Figure 1B). To study monocytes in mixed cell populations, we incubated peripheral blood mononuclear cells with HMGB1, C1q, or both, and analyzed the adherent cell population for cytokine induction. HMGB1 induced IFN, IL-6, and tumor necrosis factor-α (TNF-α); C1q inhibited this HMGB1-mediated activation (supplemental Figure 1C). These results indicate that C1q can modulate immune homeostasis by inhibiting HMGB1-induced monocyte activation through engaging LAIR-1.
C1q inhibits HMGB1 internalization
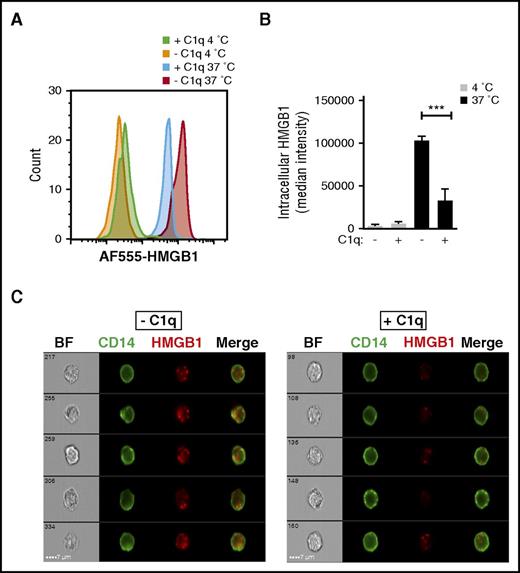
HMGB1 has an important role in potentiating the innate immune response to foreign (and endogenous) nucleic acids by transporting them into the cytoplasm of immune cells, such as monocytes and DCs, where they bind to endosomal TLRs.11,17 Because C1q inhibited HMGB1-induced cytokine secretion, we asked whether this might result from a C1q-mediated inhibition of the internalization of HMGB1. We incubated AF555-HMGB1-labeled HMGB1 with freshly isolated monocytes in the presence or absence of C1q and performed imaging flow cytometry. At 4°C, there was essentially no internalization of HMGB1 and little binding. Monocytes incubated with HMGB1 at 37°C accumulated significantly less cytosolic HMGB1 in the presence of C1q than in its absence (Figure 2).
C1q inhibits HMGB1 internalization by human monocytes. (A) Human monocytes were incubated with AF555-labeled HMGB1 with (+C1q) or without C1q (−C1q) for 30 minutes at 4°C or 37°C and stained with fluorescein isothiocyanate–anti-CD14 antibody. Imaging flow cytometry was performed on an ImageStreamX Mark II. Intensity of AF555-HMGB1 of a CD14+ population in each condition is shown. (B) The Internalization Wizard following membrane masking was used to quantify intracellular HMGB1. The median fluorescent intensity from 3 independent experiments is shown. ***P < .001. (C) Representative images of monocytes that internalized HMGB1 (red) without C1q (left) and in the presence of C1q (right).
C1q inhibits HMGB1 internalization by human monocytes. (A) Human monocytes were incubated with AF555-labeled HMGB1 with (+C1q) or without C1q (−C1q) for 30 minutes at 4°C or 37°C and stained with fluorescein isothiocyanate–anti-CD14 antibody. Imaging flow cytometry was performed on an ImageStreamX Mark II. Intensity of AF555-HMGB1 of a CD14+ population in each condition is shown. (B) The Internalization Wizard following membrane masking was used to quantify intracellular HMGB1. The median fluorescent intensity from 3 independent experiments is shown. ***P < .001. (C) Representative images of monocytes that internalized HMGB1 (red) without C1q (left) and in the presence of C1q (right).
C1q’s immunoregulatory function requires RAGE
It was previously reported that the internalization of HMGB1 is RAGE dependent.37 To determine whether C1q inhibits RAGE-dependent HMGB1-mediated activation, we incubated monocytes from the spleens of both wild-type and RAGE-deficient mice33 with HMGB1 in the presence or absence of C1q. HMGB1 induced cytokine expression in both wild-type and RAGE-deficient monocytes (supplemental Figure 2A) in agreement with previous reports that disulfide HMGB1 signals through TLR4 and MD2 as well as through RAGE.33,38 Coincubation with C1q led to a reduction in HMGB1-induced proinflammatory cytokine gene transcription in wild-type cells, but not in RAGE-deficient cells, demonstrating that C1q inhibits an HMGB1:RAGE pathway, but not an HMGB1:TLR4 pathway. To confirm this observation, we incubated monocytes with lipopolysaccharide, which activates TLR4, and showed that C1q does not block this pathway of activation (supplemental Figure 2B).
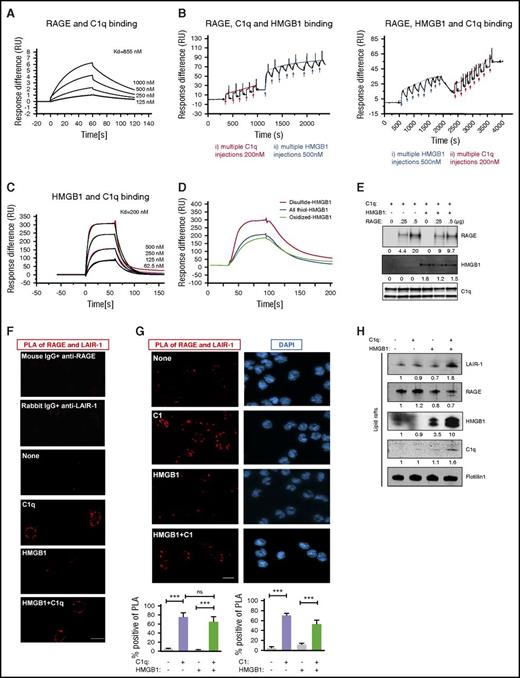
Because C1q inhibited the activation of RAGE by HMGB1 and because both C1q and HMGB1 have been shown to bind RAGE directly,39,40 we asked whether C1q prevented the interaction of HMGB1 with RAGE. For these experiments, we employed a surface plasmon resonance (SPR) assay. C1q bound RAGE in a dose-dependent manner (the equilibrium dissociation constant [KD] = 855 nM; Figure 3A). We next asked whether C1q prevented an interaction of HMGB1 with RAGE, or whether a trimolecular complex of C1q, HMGB1, and RAGE could form. RAGE was immobilized on the sensor chip, and C1q was introduced until the chip was saturated; then HMGB1 was added to form a RAGE-C1q-HMGB1 complex (Figure 3B left). Because HMGB1 is significantly smaller than C1q, displacement of C1q by HMGB1 would have produced a lower signal after addition of HMGB1; this did not occur. Qualitatively similar results were observed when HMGB1 was added to immobilize RAGE followed by C1q (Figure 3B right). In both cases, C1q, HMGB1, and RAGE formed a trimolecular complex. Interestingly, C1q also bound directly to HMGB1 in a concentration-dependent manner (KD = 200 nM; Figure 3C), preferentially binding to the disulfide form of HMGB1, the form that functions as a cytokine (Figure 3D; supplemental Figure 2B). We confirmed the existence of a trimolecular complex by immobilizing C1q on beads, incubating with saturating amounts of HMGB1, followed by increasing concentrations of RAGE (Figure 3E). RAGE bound to C1q beads even in the presence of saturating amounts of HMGB1. We do not know from these studies whether C1q binds directly to both RAGE and HMGB1 at distinct binding sites or whether HMGB1 forms a bridge between C1q and RAGE; nonetheless, the data provide evidence that C1q, RAGE, and HMGB1 all interact.
C1q, HMGB1, RAGE, and LAIR-1 form a multimolecular complex in lipid rafts. (A) SPR assay of C1q and RAGE binding; KD = 855 nM. Experiments were repeated 3 times. (B) SPR assay of RAGE-C1q-HMGB1 trimolecular complex. RAGE (500 nM) was immobilized onto a CM5 chip and the first analyte (C1q, 200 nM) was added to saturation. HMGB1 (500 nM) was added to the RAGE-C1q complex in multiple pulses (left). Alternatively, HMGB1 was added to immobilized RAGE until the chip was saturated, followed by C1q in multiple pulses (right). N = 3. (C) SPR assay for HMGB1 and C1q binding; KD = 200 nM. N = 4. (D) SPR assay of HMGB1 in different redox states and C1q binding. C1q (200 nM) was immobilized onto a CM5 chip, and disulfide-, all thiol-, or oxidized-HMGB1 (500 nM) was added. N = 3. (E) C1q-coated beads were incubated with saturating amounts of HMGB1, followed by 250 ng or 500 ng of RAGE. Complexes were analyzed by western blot using anti-RAGE, anti-Cbp tag antibody for HMGB1 or infrared-labeled streptavidin for biotinylated-C1q. N = 3. ***P < .001 (one-way ANOVA). (F) Human monocytes were treated with HMGB1 (3 μg/mL) in the absence or presence of C1q (25 μg/mL) for 15 minutes at 37°C. Colocalization of LAIR-1 and RAGE on the plasma membrane was assessed by PLA. Red dots (PLA positive), representing colocalization of RAGE and LAIR-1, are only seen in the presence of C1q, with or without HMGB1. Percentage of PLA-positive cells in total cells was counted in several random fields in 3 independent experiments. (G) PLA assay using C1 (25 μg/mL) complex instead of C1q. N = 3. (H) Lipid raft fractions from monocytes treated with HMGB1 (3 μg/mL), in the absence or presence of C1q (25 μg/mL), for 15 minutes at 37°C were concentrated and analyzed by western blot for LAIR-1, RAGE, HMGB1, C1q, or Flotillin1 as a lipid raft marker. N = 3. DAPI, 4′,6-diamidino-2-phenylindole.
C1q, HMGB1, RAGE, and LAIR-1 form a multimolecular complex in lipid rafts. (A) SPR assay of C1q and RAGE binding; KD = 855 nM. Experiments were repeated 3 times. (B) SPR assay of RAGE-C1q-HMGB1 trimolecular complex. RAGE (500 nM) was immobilized onto a CM5 chip and the first analyte (C1q, 200 nM) was added to saturation. HMGB1 (500 nM) was added to the RAGE-C1q complex in multiple pulses (left). Alternatively, HMGB1 was added to immobilized RAGE until the chip was saturated, followed by C1q in multiple pulses (right). N = 3. (C) SPR assay for HMGB1 and C1q binding; KD = 200 nM. N = 4. (D) SPR assay of HMGB1 in different redox states and C1q binding. C1q (200 nM) was immobilized onto a CM5 chip, and disulfide-, all thiol-, or oxidized-HMGB1 (500 nM) was added. N = 3. (E) C1q-coated beads were incubated with saturating amounts of HMGB1, followed by 250 ng or 500 ng of RAGE. Complexes were analyzed by western blot using anti-RAGE, anti-Cbp tag antibody for HMGB1 or infrared-labeled streptavidin for biotinylated-C1q. N = 3. ***P < .001 (one-way ANOVA). (F) Human monocytes were treated with HMGB1 (3 μg/mL) in the absence or presence of C1q (25 μg/mL) for 15 minutes at 37°C. Colocalization of LAIR-1 and RAGE on the plasma membrane was assessed by PLA. Red dots (PLA positive), representing colocalization of RAGE and LAIR-1, are only seen in the presence of C1q, with or without HMGB1. Percentage of PLA-positive cells in total cells was counted in several random fields in 3 independent experiments. (G) PLA assay using C1 (25 μg/mL) complex instead of C1q. N = 3. (H) Lipid raft fractions from monocytes treated with HMGB1 (3 μg/mL), in the absence or presence of C1q (25 μg/mL), for 15 minutes at 37°C were concentrated and analyzed by western blot for LAIR-1, RAGE, HMGB1, C1q, or Flotillin1 as a lipid raft marker. N = 3. DAPI, 4′,6-diamidino-2-phenylindole.
C1q bridges RAGE and LAIR-1
Because the C1q binds RAGE through its globular head39 while the C1q collagen tail binds LAIR-1,29 the next question was whether C1q might crosslink LAIR-1 to RAGE on the surface of monocytes. We used a proximity ligation assay (PLA) to investigate the localization of RAGE and LAIR-1 in the absence and presence of C1q. Polymerase-amplified fluorescence, indicative of RAGE-LAIR-1 proximity, was only detected in the presence of C1q, with or without HMGB1 (Figure 3F). Because C1q binds both RAGE and HMGB1 through its globular head, the fact that HMGB1 does not alter the C1q-mediated crosslinking of LAIR-1 and RAGE suggests that RAGE and HMGB1 bind C1q on different regions of the globular head. This interpretation agrees well with the SPR results, which are also consistent with distinct binding sites for HMGB1 and C1q on RAGE.
Because ∼80% of C1q forms a complex with C1r and C1s in blood in the presence of calcium,41,42 we performed a dot blot in the presence of calcium and demonstrated that C1 detected with an anti-C1r antibody bound to LAIR-1 (supplemental Figure 1D). Moreover, C1 complex bridged RAGE and LAIR-1 (Figure 3H). To ascertain the function of colocalization of RAGE and LAIR-1, we tested their localization in lipid rafts, which are membrane compartments that regulate signal transduction.43 We fractionated cell lysates by sucrose gradient ultracentrifugation and analyzed fractions by slot blot using anti-LAIR-1 or anti-RAGE antibodies (supplemental Figure 3A). Lipid rafts were identified by staining with cholera toxin subunit (supplemental Figure 3A). RAGE is present in both lipid raft and non–lipid raft fractions. When we focused on lipid raft fractions using western blot, we observed more LAIR-1 in lipid rafts in the presence of both HMGB1 and C1q (Figure 3H). In addition, membrane-bound C1q and HMGB1 were detected in the lipid rafts, confirming that complexes were formed on the membrane (Figure 3H). C1q tail, which lacks a globular head and cannot bridge RAGE and LAIR-1, did not recruit LAIR-1 into lipid rafts even in the presence of HMGB1 (supplemental Figure 3B). These results suggest that C1q can create a tetramolecular complex with LAIR-1, HMGB1, and RAGE, bringing these molecules together in lipid rafts.
Phosphorylation of RAGE and LAIR-1 differs in the presence of HMGB1 and C1q
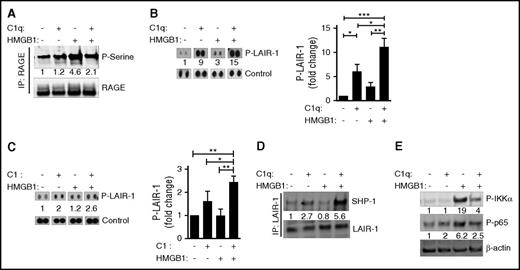
To understand how the colocalization of RAGE and LAIR-1 might affect downstream signaling pathways, we examined the phosphorylation of several molecules on primary human monocytes in the presence of HMGB1, C1q, or both. We confirmed a previous report that HMGB1 enhances phosphorylation of RAGE, which others have shown is mediated by protein kinase Cζ,44 and demonstrated that the HMGB1-induced phosphorylation is diminished in the presence of C1q (Figure 4A). We observed that both C1q and C1 alone, or in the presence of HMGB1, induced the phosphorylation of LAIR-131 (Figure 4B-C). Because phosphorylation on both immunoreceptor tyrosine-based inhibitory motifs (ITIMs) recruits SHP-1 to LAIR-1,45,46 the increased recruitment of SHP-1 to LAIR-1 suggests that C1q plus HMGB1 leads to more LAIR-1 molecules that are phosphorylated on both ITIMs (Figure 4D). Finally, C1q inhibited the downstream activation and nuclear translocation of NFκB that is induced by HMGB1. We observed diminished HMGB1-induced phosphorylation of IKKα and diminished nuclear localization of p65 in the presence of C1q (Figure 4E; supplemental Figure 4). Thus, in the presence of both C1q and HMGB1, RAGE is dephosphorylated; SHP-1 is recruited to LAIR-1, and NFκB activity is inhibited.
C1q dephosphorylates RAGE and recruits SHP-1 to LAIR-1. Human monocytes were treated with C1q (25 μg/mL) and/or HMGB1 (3 μg/mL) for 15 minutes at 37°C. (A) Total cell lysates were subjected to immunoprecipitation with antibodies to RAGE followed by immunoblotting with antibodies specific for phosphoserine (top) or RAGE (bottom). Numbers indicate the relative signal intensity. N = 3. (B) Total cell lysates were subjected to immunophosphorylation array to observe the phosphorylation of LAIR-1 ITIM motifs. Relative quantification of phosphorylation of LAIR-1 was normalized to control spots (relative fold induction) from 4 independent experiments. (C) Human monocytes were treated with C1 complex (25 μg/mL) and/or HMGB1 (3 μg/mL) for 15 minutes at 37°C. Immunophosphorylation array was performed to observe the phosphorylation of LAIR-1 ITIM motifs. N = 3. *P < .05; **P < .01; ***P < .001 (one-way ANOVA). (D) Total cell lysates were immunoprecipitated with antibodies to LAIR-1 followed by immunoblotting with antibodies for SHP-1 (top) or LAIR-1 (bottom). N = 4. (E) Human monocytes were treated with HMGB1 and/or C1q for 3 hours, and cell lysates were subjected to immunoblotting with antibodies for activated IKKα (P-IKKα, top), p65 (P-p65, middle), or β-actin. N = 3.
C1q dephosphorylates RAGE and recruits SHP-1 to LAIR-1. Human monocytes were treated with C1q (25 μg/mL) and/or HMGB1 (3 μg/mL) for 15 minutes at 37°C. (A) Total cell lysates were subjected to immunoprecipitation with antibodies to RAGE followed by immunoblotting with antibodies specific for phosphoserine (top) or RAGE (bottom). Numbers indicate the relative signal intensity. N = 3. (B) Total cell lysates were subjected to immunophosphorylation array to observe the phosphorylation of LAIR-1 ITIM motifs. Relative quantification of phosphorylation of LAIR-1 was normalized to control spots (relative fold induction) from 4 independent experiments. (C) Human monocytes were treated with C1 complex (25 μg/mL) and/or HMGB1 (3 μg/mL) for 15 minutes at 37°C. Immunophosphorylation array was performed to observe the phosphorylation of LAIR-1 ITIM motifs. N = 3. *P < .05; **P < .01; ***P < .001 (one-way ANOVA). (D) Total cell lysates were immunoprecipitated with antibodies to LAIR-1 followed by immunoblotting with antibodies for SHP-1 (top) or LAIR-1 (bottom). N = 4. (E) Human monocytes were treated with HMGB1 and/or C1q for 3 hours, and cell lysates were subjected to immunoblotting with antibodies for activated IKKα (P-IKKα, top), p65 (P-p65, middle), or β-actin. N = 3.
Monocytes exposed to both HMGB1 and C1q express a novel set of genes
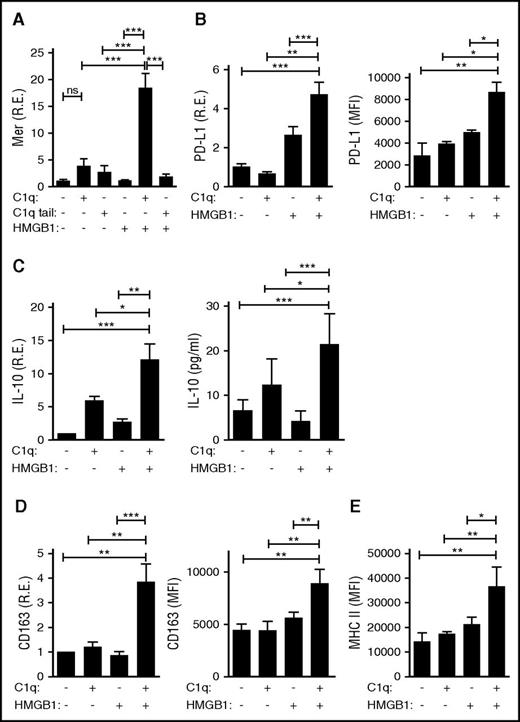
To assess whether HMGB1 and C1q might further alter the activity of monocytes, primary human monocytes were assessed for expression of immunomodulatory proteins after incubation for 24 hours alone or in the presence of HMGB1, C1q, or both. Exposure to both HMGB1 and C1q induced the transcription and protein expression of a variety of anti-inflammatory molecules, including Mer, a receptor tyrosine kinase important in the clearance of apoptotic debris (Figure 5A); PD-L1, a molecule that suppresses T-cell activation (Figure 5B); IL-10, an anti-inflammatory cytokine (Figure 5C); and CD163 and major histocompatibility complex class II (MHCII), markers of anti-inflammatory macrophages (Figure 5D-E). These findings demonstrate that when C1q crosslinks LAIR-1 with RAGE and HMGB1, it engages a program of monocyte differentiation into anti-inflammatory macrophages. Importantly, exposure to HMGB1 and C1q tail did not mediate similar effects (Figure 5A), confirming that C1q must interact with RAGE as well as LAIR-1 to induce anti-inflammatory gene expression and that coincident but uncoordinated signaling through RAGE and LAIR-1 is not sufficient. Moreover, C1 complex, like C1q, suppressed the induction of proinflammatory cytokines by HMGB1 and induced Mer expression, a marker of anti-inflammatory macrophages (supplemental Figure 1E). Thus, C1 was functionally active.
HMGB1 and C1q induce anti-inflammatory molecules and promote an anti-inflammatory macrophage phenotype. Human monocytes, treated with C1q (25 μg/mL) or C1q tail (53 μg/mL) and/or HMGB1 (3 μg/mL) for 24 hours, were processed for mRNA and protein. (A) Mer tyrosine kinase as assessed by quantitative polymerase chain reaction (qPCR) (left). R.E., relative expression. N = 3. (B) PD-L1 was assessed by qPCR. N = 4. (C) IL-10 was assessed by qPCR (left) and enzyme-linked immunosorbent assay (right). N = 4. (D) CD163 was assessed by qPCR (left) and flow cytometry (right). N = 3 for left; N = 3 for right. (E) HLA-DR (MHCII) was determined by flow cytometry. N = 3. All data represent the mean ± standard error of the mean (SEM) of independent experiments in which each condition was tested in triplicate. Statistical analysis was performed by one-way ANOVA. ns, not significant; *P < .05; **P < .01; ***P < .001.
HMGB1 and C1q induce anti-inflammatory molecules and promote an anti-inflammatory macrophage phenotype. Human monocytes, treated with C1q (25 μg/mL) or C1q tail (53 μg/mL) and/or HMGB1 (3 μg/mL) for 24 hours, were processed for mRNA and protein. (A) Mer tyrosine kinase as assessed by quantitative polymerase chain reaction (qPCR) (left). R.E., relative expression. N = 3. (B) PD-L1 was assessed by qPCR. N = 4. (C) IL-10 was assessed by qPCR (left) and enzyme-linked immunosorbent assay (right). N = 4. (D) CD163 was assessed by qPCR (left) and flow cytometry (right). N = 3 for left; N = 3 for right. (E) HLA-DR (MHCII) was determined by flow cytometry. N = 3. All data represent the mean ± standard error of the mean (SEM) of independent experiments in which each condition was tested in triplicate. Statistical analysis was performed by one-way ANOVA. ns, not significant; *P < .05; **P < .01; ***P < .001.
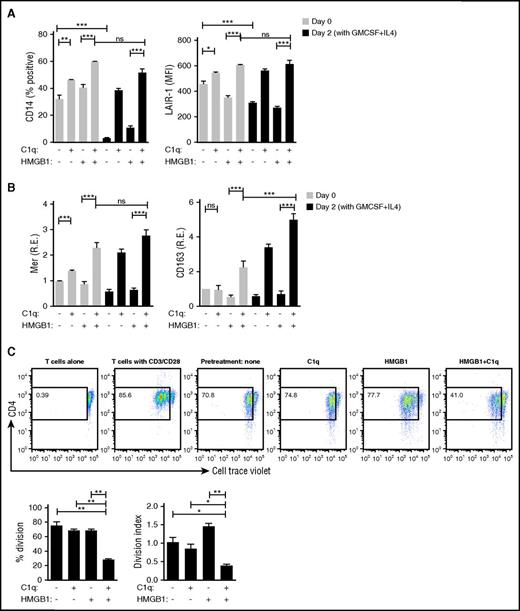
Monocyte differentiation to DCs is prevented by HMGB1 and C1q
As an early step toward an adaptive immune response, monocytes can differentiate into DCs and function as antigen-presenting cells.47 In view of the anti-inflammatory state that C1q and HMGB1 elicited in monocytes, we wondered whether monocytes previously exposed to HMGB1 and C1q would lose their ability to differentiate into DCs. Monocytes previously exposed for 24 hours to either C1q alone or C1q and HMGB1 retained expression of CD14 and LAIR-1, but expression was significantly higher in monocytes exposed to both HMGB1 and C1q (Figure 6A). In contrast, CD14 and LAIR-1 downregulation, indicating monocyte to DC differentiation, occurred in previously untreated monocytes or monocytes previously treated with HMGB1 (Figure 6A). Markers for anti-inflammatory macrophages, Mer and CD163, were consistently enhanced by HMGB1 and C1q and were further enhanced 2 days after incubation with cytokines that induce monocyte to DC differentiation (granulocyte-macrophage colony-stimulating factor [GM-CSF] and IL-4) (Figure 6B), demonstrating that exposure to HMGB1 and C1q drives monocytes to become anti-inflammatory macrophages that do not differentiate to antigen-presenting cells in the presence of GM-CSF and IL-4.
HMGB1 and C1q inhibit plasticity of macrophages. DC differentiation induced by GM-CSF and IL-4 was assessed by flow cytometry. Monocytes were treated with C1q and/or HMGB1 for 24 hours (day 0) and then further cultured with GM-CSF and IL-4 for 2 days (day 2). (A) High levels of CD14 and LAIR-1 denote suppression of DC differentiation. Data represent the mean ± SEM of 3 independent experiments. Statistical analysis was performed by one-way ANOVA. ns, not significant. *P < .05; **P < .01; ***P < .001. (B) Transcription of Mer and CD163 was measured by qPCR. Data represent the mean ± SEM of 3 independent experiments in which each condition was tested in triplicate. Statistical analysis was performed by one-way ANOVA. ns, not significant. *P < .05; **P < .01; ***P < .001. (C) Mixed lymphocyte reaction. Monocytes were exposed to HMGB1 (3 μg/mL), C1q (25 μg/mL), or both for 24 hours in X-Vivo 15 serum-free medium, washed, and further incubated for 2 days in X-Vivo 15 medium. Cell Trace Violet–stained allogeneic primary human CD4 T cells and anti-CD3 (1 μg/mL) were added (2:1). T cells alone in the presence of anti-CD3 antibody or T cells with antibodies to CD3/CD28 were negative and positive controls, respectively. After 4 days, the nonadherent cells, which stained with anti-CD4 antibodies, were assessed by flow cytometry. Live CD4+ T cells were analyzed. Bar graphs represent the mean ± SEM of percentage having undergone division (left) and division index (right). N = 3. *P < .05; **P < .01; ***P < .001 (one-way ANOVA).
HMGB1 and C1q inhibit plasticity of macrophages. DC differentiation induced by GM-CSF and IL-4 was assessed by flow cytometry. Monocytes were treated with C1q and/or HMGB1 for 24 hours (day 0) and then further cultured with GM-CSF and IL-4 for 2 days (day 2). (A) High levels of CD14 and LAIR-1 denote suppression of DC differentiation. Data represent the mean ± SEM of 3 independent experiments. Statistical analysis was performed by one-way ANOVA. ns, not significant. *P < .05; **P < .01; ***P < .001. (B) Transcription of Mer and CD163 was measured by qPCR. Data represent the mean ± SEM of 3 independent experiments in which each condition was tested in triplicate. Statistical analysis was performed by one-way ANOVA. ns, not significant. *P < .05; **P < .01; ***P < .001. (C) Mixed lymphocyte reaction. Monocytes were exposed to HMGB1 (3 μg/mL), C1q (25 μg/mL), or both for 24 hours in X-Vivo 15 serum-free medium, washed, and further incubated for 2 days in X-Vivo 15 medium. Cell Trace Violet–stained allogeneic primary human CD4 T cells and anti-CD3 (1 μg/mL) were added (2:1). T cells alone in the presence of anti-CD3 antibody or T cells with antibodies to CD3/CD28 were negative and positive controls, respectively. After 4 days, the nonadherent cells, which stained with anti-CD4 antibodies, were assessed by flow cytometry. Live CD4+ T cells were analyzed. Bar graphs represent the mean ± SEM of percentage having undergone division (left) and division index (right). N = 3. *P < .05; **P < .01; ***P < .001 (one-way ANOVA).
We further tested this by using monocytes exposed to HMGB1, C1q, or both as antigen-presenting cells in an allogeneic mixed lymphocyte reaction. Proinflammatory macrophages, previously exposed to HMGB1, induced the strongest mixed lymphocyte reaction and the highest division index (the average number of divisions that a cell has undergone assessed with a standard algorithm48 ); anti-inflammatory macrophages, previously exposed to both HMGB1 and C1q, were poor stimulators of T cells (Figure 6C), presumably due to a high level of PD-L1 and IL-10 secretion (Figure 5B-C) despite high levels of MHCII (Figure 5E).
Discussion
It is well established that the healthy mammalian immune system is in a state of dynamic equilibrium, where activating stimuli are constantly balanced by negative feedback loops and inhibitory molecules to maintain a healthy homeostasis. Because autoreactive lymphoid cells have been shown to persist in healthy adults, and myeloid cells respond to DAMPs as well as pathogen-associated molecular patterns, these regulatory mechanisms are of paramount importance in keeping a state of immune quiescence and avoiding unwanted autoimmunity.
This study reveals previously unknown effects of HMGB1 and C1q on human monocyte activation and differentiation in inflammatory settings and in SLE. HMGB1 and C1q have opposing effects on human monocytes with HMGB1 inducing a proinflammatory phenotype. More surprisingly, their combined function results in the differentiation to a cell with the characteristics of an anti-inflammatory macrophage, and which cannot differentiate into a DC. Thus, HMGB1 plus C1q–exposed monocytes are effectively removed from forming the bridge to an adaptive immune response. These observations help reconcile the apparently contradictory studies showing that HMGB1 can be both pro- and anti-inflammatory. HMGB1 may trigger proinflammatory macrophages during the onset of inflammation, but monocytes may be skewed to anti-inflammatory macrophages as C1q levels rise.
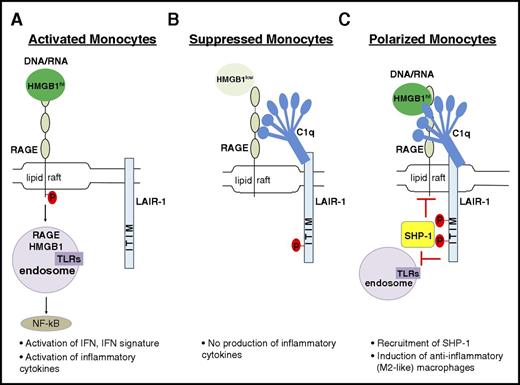
Although we are aware there are many receptors for both HMGB1 and C1q,18,23 we propose a model in which they can interact in a tetramolecular complex with RAGE and LAIR-1 (Figure 7). C1q crosslinks LAIR-1 with RAGE and induces the colocalization of these receptors in lipid rafts. RAGE is the major transporter of HMGB1 and its cargo into the cytosol; internalization of HMGB1 is significantly inhibited in the presence of C1q (Figure 2). Although C1q alone can bind LAIR-1 on monocytes and lead to its phosphorylation, presumably by Hck,45 a more suppressive role of C1q depends on costimulation of monocytes with HMGB1 leading to enhanced recruitment of SHP-1. In our experimental condition, C1q did not suppress HMGB1 activation of RAGE−/− monocytes nor did it suppress lipopolysaccharide-mediated activation of human monocytes, suggesting that C1q specifically suppresses RAGE-mediated activation. We believe that HMGB1-mediated activation of TLR4 is not seen in monocytes incubated with HMGB1 and C1q because HMGB1 may preferentially bind RAGE, perhaps because that interaction is avidity enhanced by the presence of C1q. Alternatively, the recruitment of SHP-1 to LAIR-1 may block the activation of NFκB by TLR4 as well as by RAGE. This issue requires further exploration (Figure 7C).
Model showing how C1q utilizes a natural pathway to dampen inflammation. (A) DNA/RNA-associated HMGB1 is internalized and activates endosomal TLRs, leading to downstream activation of NFκB to induce proinflammatory macrophages. (B) In the presence of C1q without inflammation (basal levels of HMGB1), C1q and LAIR-1 signaling prevents proinflammatory cytokine production. (C) In inflammation with high levels of HMGB1, C1q mediates differentiation of anti-inflammatory macrophages by crosslinking RAGE and LAIR-1 in lipid rafts to facilitate SHP-1 binding to LAIR-1 via phosphorylated ITIMs. Activated SHP-1 may suppress directly or indirectly the pathways downstream of RAGE activation.
Model showing how C1q utilizes a natural pathway to dampen inflammation. (A) DNA/RNA-associated HMGB1 is internalized and activates endosomal TLRs, leading to downstream activation of NFκB to induce proinflammatory macrophages. (B) In the presence of C1q without inflammation (basal levels of HMGB1), C1q and LAIR-1 signaling prevents proinflammatory cytokine production. (C) In inflammation with high levels of HMGB1, C1q mediates differentiation of anti-inflammatory macrophages by crosslinking RAGE and LAIR-1 in lipid rafts to facilitate SHP-1 binding to LAIR-1 via phosphorylated ITIMs. Activated SHP-1 may suppress directly or indirectly the pathways downstream of RAGE activation.
Our model suggests a multimodal function of C1q. In the absence of inflammatory stimuli, C1q represses the proinflammatory properties of the low levels of HMGB1 that are normally present in human serum (Figure 7B). However, in the presence of higher levels of HMGB1, C1q acts as a molecular switch that drives monocyte differentiation to an anti-inflammatory cell type (Figure 7C). When HMGB1 levels are very high, C1q fails to dampen HMGB1-mediated inflammation (Figure 7A).49 In summary, we postulate that HMGB1 and C1q cooperate in an inflammatory setting to terminate inflammation by recruiting SHP-1 to prevent NFκBp65 activation, by inducing anti-inflammatory macrophage differentiation with expression of suppressive molecules such as Mer, PD-L1, and IL-1050 and by inhibiting the differentiation of monocytes into DCs and preventing their participation in an adaptive immune response. It is also reported that C1q stimulation leads to the activation of predominantly p50p50 homodimers, thereby competing with transcriptionally active p65p50 and resulting in reduced proinflammatory cytokine production.51 In our studies, we did not observe that C1q enhanced transforming growth factor-β transcription as might have been expected. This observation is consistent with a previous study that showed that HMGB1 alone can increase transforming growth factor-β expression in a RAGE-dependent pathway in renal tubular epithelial cells.52 Finally, it is important to note that we do not know the relative amount of C1q to C1 in inflamed tissues and whether there may be functional differences between the 2 that we did not detect in our studies. Thus, whether this pathway of inflammation resolution functions differently depending on the ratio of C1q to C1 requires further study.
These findings emphasize the importance of generating therapeutic approaches to selectively engage RAGE and LAIR-1 to target DAMP-mediated inflammation while preserving other protective immune responses.
It is well established that SLE pathology begins after class switching of autoantibodies from immunoglobulin M (IgM) to IgG, and that IgM autoantibodies can protect against disease onset.53 Our data explain how IgM immune complexes engaging C1q and LAIR-1 would suppress innate inflammation, whereas IgG immune complexes, directly engaging Fc receptors, would provoke an inflammatory response. Interestingly, it has been reported54 that immunoglobulin class switching to IgG can be facilitated through engagement of HMGB1 to TLR2. Whether C1q can alter this activity of HMGB1 is not known.
In blood, circulating C1q engages LAIR-1 and maintains monocyte quiescence.31 When increased levels of HMGB1 are present as a consequence of tissue damage or infection, these cells may differentiate toward macrophages or DCs and migrate to where C1q can be actively secreted by myeloid cells to dampen immune activation. Indeed, we hypothesize that infiltrating monocytes/anti-inflammatory macrophages engage in the resolution of inflammation, whereas tissue-resident myeloid cells may not. Because tissue resident cells will experience LAIR-1 activation through extracellular matrix collagen, which fails to crosslink LAIR-1 to RAGE, they may not differentiate into anti-inflammatory macrophages. Finally, it is interesting to consider whether the same paradigm may exist for other C1q binding partners such as Aβ and S100, which also bind RAGE.55-57 It has been shown, for example, that C1q binds Aβ and that C1q promotes neuroprotection in Alzheimer disease.58
Considering that the overwhelming proportion of C1q-deficient patients manifest an autoimmune disease, this immune-regulatory mechanism of C1q is evidently of great importance in safeguarding an appropriately regulated immune response. Moreover, the fact that motifs within C1q and HMGB1 can activate an unappreciated natural program of immune suppression raises the exciting possibility of harnessing this pathway to develop novel mechanism-based lupus therapeutics.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Huan Yang for discussions, Susan Malkiel for proofreading the manuscript, and Erin Emmons for technical assistance.
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (K01AR065506 [M.S.]; R01AR057084 [B.D.]) and by a Shared Instrumentation Grant program of the National Institutes of Health (S10 RR033072 [Y.A.-A.]). M.S. was a recipient of an Arthritis Foundation Fellowship.
Authorship
Contribution: M.S., A.P., and B.D. planned the research, analyzed data, and wrote the manuscript; M.S., A.P., and M.H. performed the experiments; J.S., F.S.-S., and B.T.V. analyzed data; T.R.C., Y.A.-A., U.A., and K.J.T. analyzed data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Betty Diamond, The Feinstein Institute for Medical Research, Center for Autoimmune and Musculoskeletal Diseases, 350 Community Dr, Manhasset, NY 11030; e-mail: bdiamond@northwell.edu.
References
Author notes
M.S. and A.P. contributed equally to this study.

![Figure 1. C1q inhibits expression of HMGB1-induced IFN-α, MX1, and inflammatory cytokines by human monocytes. (A) Messenger RNA (mRNA) levels for IFN-α, MX1, IL-6, and TNF-α in monocytes stimulated with or without HMGB1 (3 μg/mL) and various concentrations of C1q (μg/mL) for 6 hours in serum-free medium (mean ± standard deviation [SD] of triplicates, N = 3). (B) HMGB1 (3 μg/mL) -induced IL-6 and TNF-α secretion were reduced in the presence of various concentrations of C1q, assessed at 24 hours (mean ± SD of duplicates, N = 3). (C) IFN-α, MX1, IL-6, and TNF-α mRNA expression by monocytes transfected with control siRNA or LAIR siRNA and treated with C1q (25 μg/mL), HMGB1 (3 μg/mL) for 6 hours (mean ± SD of triplicates, N = 3). Data are expressed as fold induction relative to controls. R.E., relative expression. ns, not significant. *P < .05; **P < .01; ***P < .001 (one-way ANOVA with Bonferroni’s correction for multiple comparisons).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/18/10.1182_blood-2016-05-719757/4/m_2218f1.jpeg?Expires=1769128974&Signature=ZkRWVNQZJDBRtJoHisLHT-aVTLVOWHGbMCA8AsE-0UXwOK0i8xyjvOmpP17Tu5bKmek9AFmE9MufolrlJN6ZdBuiZRJidc1wHkoswKFVKN891GcE3R4mipNHLOhwKtrZjFuXsYpCvFUaywHpmzyN5e~EEIOOEugIKZRey~CSuJod3z8icZYIF1LmhbLJeKc8WUvlhM1DReA7DsFhd~-Jgp1OOKuZNyFQ1sh-MpzTx5NcQ9CET3qywsmsWb9kG~Uv3gW74-7oP3DvmNJdoSDDNBFowxB4s-k-5j7HUoC5g65Qqnf4k20lr-6Xg7I6qibTmFvswO8P4Vck-azIDv8cHA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)