Key Points
Activation of BCR and canonical NF-κB signaling in the lymph node correlates with survival in MCL.
Mutations and polymorphisms in BCR and NF-κB pathways may confer cell autonomous signaling and affect response to ibrutinib.
Abstract
To interrogate signaling pathways activated in mantle cell lymphoma (MCL) in vivo, we contrasted gene expression profiles of 55 tumor samples isolated from blood and lymph nodes from 43 previously untreated patients with active disease. In addition to lymph nodes, MCL often involves blood, bone marrow, and spleen and is incurable for most patients. Recently, the Bruton tyrosine kinase (BTK) inhibitor ibrutinib demonstrated important clinical activity in MCL. However, the role of specific signaling pathways in the lymphomagenesis of MCL and the biologic basis for ibrutinib sensitivity of these tumors are unknown. Here, we demonstrate activation of B-cell receptor (BCR) and canonical NF-κB signaling specifically in MCL cells in the lymph node. Quantification of BCR signaling strength, reflected in the expression of BCR regulated genes, identified a subset of patients with inferior survival after cytotoxic therapy. Tumor proliferation was highest in the lymph node and correlated with the degree of BCR activation. A subset of leukemic tumors showed active BCR and NF-κB signaling apparently independent of microenvironmental support. In one of these samples, we identified a novel somatic mutation in RELA (E39Q). This sample was resistant to ibrutinib-mediated inhibition of NF-κB and apoptosis. In addition, we identified germ line variants in genes encoding regulators of the BCR and NF-κB pathway previously implicated in lymphomagenesis. In conclusion, BCR signaling, activated in the lymph node microenvironment in vivo, appears to promote tumor proliferation and survival and may explain the sensitivity of this lymphoma to BTK inhibitors.
Introduction
Mantle cell lymphoma (MCL) is an aggressive, largely incurable, subtype of non-Hodgkin lymphoma (NHL).1,2 The genetic hallmark of MCL is the chromosomal translocation t(11;14)(q13;q32), which results in Cyclin-D1 overexpression. The translocation occurs during the pre-B stage of differentiation, and the malignant transformation has been thought to arise within naïve B cells.3 In agreement, in most cases, the tumor cells express an unmutated (germ line configuration) immunoglobulin heavy chain (IGHV) gene. However, recognition of a restricted IGHV repertoire used by some tumors and identification of a less aggressive MCL variant expressing mutated IGHV genes suggest a possible role of antigenic selection in these tumors.4,5
Cyclin-D1 expression alone is not sufficient for malignant transformation, and additional events are required for oncogenesis.3,6 Genomewide studies using next-generation sequencing identified several recurrently mutated genes including ATM and TP53 (DNA damage response); CCND1 and RB1 (cell cycle); MLL2, MLL3, and SMARCA4 (epigenetic modifiers); UBR5 (N-end rule pathway); NOTCH1/2 (Notch signaling pathway); and BIRC3 and TRAF2 (NF-κB pathway).7-9 Notably, most tumors do not contain mutations in signaling pathways amenable to therapeutic targeting.10
Although a few patients present with indolent disease and can be managed with an observant approach,11,12 most have a rapidly progressive disease course requiring urgent intervention.1,2 Despite long-lasting responses achieved with aggressive therapy, late relapses occur.13 Thus, new therapeutic approaches for MCL are urgently needed.1,2 Bortezomib, lenalidomide, and ibrutinib are drugs that the US Food and Drug Administration recently approved for treating MCL.1,2 However, the mechanisms underlying MCL’s sensitivity to these agents are not well understood.10,14 Bortezomib, a proteasome inhibitor, can interfere with NF-κB activation. Although activating mutations in components of the NF-κB pathway have been described in MCL, they preferentially affect the alternative NF-κB pathway and are found in only a small subset of tumors.8,15 Furthermore, bortezomib induces cell death in MCL through oxidative and endoplasmic reticulum stress, resulting in the upregulation of the proapoptotic protein NOXA, a response independent of NF-κB inhibition.16,17 Lenalidomide is an immunomodulatory agent with pleotropic effects that may include immune modulation and disruption of tumor-microenvironment interactions.18,19
Ibrutinib covalently binds Bruton tyrosine kinase (BTK), thereby irreversibly inactivating the kinase.20 BTK is essential for B-cell receptor (BCR) signaling, and loss-of-function mutations lead to the virtual absence of mature B cells. A phase I trial of ibrutinib in patients with relapsed NHL demonstrated a high response rate in MCL, an unexpected finding that was confirmed in a disease-specific phase II trial demonstrating objective responses in 68% of patients, including 21% with complete response, and an overall survival (OS) rate of 58% at 18 months.21,22
Unlike MCL, ibrutinib’s activity in chronic lymphocytic leukemia (CLL) and activated B-cell-like diffuse large B-cell lymphoma (ABC-DLBCL) is consistent with the documented role of BCR signaling in these diseases. In CLL, BCR signaling in the tumor cells is induced within the lymph node (LN) microenvironment.23,24 However, the influence of the microenvironment on the pathogenesis of MCL has not been thoroughly investigated.3 In ABC-DLBCL, chronic active BCR signaling has been linked to somatic mutations in CD79B.25 Moreover, the high efficacy of ibrutinib in Waldenström macroglobulinemia is attributed to activation of BTK through mutant MYD88.26,27 MCL rarely has mutations in CD79B and MYD88,9,28 whereas the reported mutations in TRAF2 and BIRC3 activate the alternative NF-κB pathway, resulting in resistance, rather than sensitivity, to ibrutinib.8 Thus, identification of pathogenic signaling pathways in MCL and the biologic basis for ibrutinib sensitivity require further investigation.
To investigate tumor-microenvironment interactions in vivo and gain further insights into MCL biology, we took advantage of the presence of tumor cells in different anatomic compartments. Specifically, we compared gene expression profiles and the activity of signaling pathways in tumor samples collected from the peripheral blood (PB) and LNs. We found evidence for active BCR and NF-κB signaling that, in most cases, is induced specifically in LN-resident MCL cells and determines OS.
Methods
Study design and patient samples
This translational laboratory study was performed with human LN and PB samples obtained from 43 patients with previously untreated MCL (supplemental Table 1, available on the Blood Web site). Patients were enrolled in a National Cancer Institute Institutional Review Board–approved study (registered with www.clinicaltrials.gov as NCT00114738) and treated with 6 cycles of bortezomib in combination with EPOCH-R (etoposide, prednisone, vincristine, doxorubicin, and rituximab). All samples were obtained with written informed consent. The laboratory study includes all patients with available gene expression data. Confirmatory studies by flow cytometry and in vitro testing were done for patients with sufficient leftover material.
Mononuclear cells from blood and LN single-cell suspension were isolated using density gradient centrifugation with lymphocyte separation medium (ICN Biomedicals). Fresh cells were subjected to CD19 selection using magnetic beads yielding purity >96% (Miltenyi Biotec). Normal (non-MCL) B cells were not detectable in PB samples of 15 of 17 patients analyzed. In 2 patients, normal B cells made up 0.2% and 0.3% of all B cells in the sample. Fixed LN biopsies were stained for Ki67 (DakoCytomation) after deparaffinization and quantified using Aperio Digital Pathology (Leica Microsystems). As previously described, IGHV sequencing was performed on leukemic samples and classified as mutated (<97% homology to germ line) or unmutated (≥97% homology).4
Flow cytometry
Antibodies for cell surface stain were (phycoerythrin [PE]-Cy5 anti-CD19, PE-Cy7 anti-CD5, PE anti-CD69, and PE anti-CD80). After surface staining, cells were washed and suspended in 4% paraformaldehyde (on ice, 1 hour), washed, resuspended in 70% ethanol (at –20°C, 2 hours), washed, incubated with intracellular stain (fluorescein isothiocyanate–anti-Ki67; Alexa Fluor-488–anti-p-SYK, p-PLCγ, p-AKT, p-ERK, and p-p65) for 20 minutes at room temperature, washed, and analyzed on a BD FACSCanto II flow cytometer (BD Biosciences), as described.24 All antibodies were purchased from BD Biosciences, except for Alexa Fluor-488 anti-p-AKT (Life Technologies). Data were analyzed using FlowJo software (Tree Star Inc).
In vitro treatment with ibrutinib, MI-2, and PS1145
Viably frozen PB mononuclear cells and LN single-cell suspensions were exposed to ibrutinib, MI-2, or PS1145 for 48 hours. Triplicates of 500 000 cells per 100 μL were plated in a 96-well plate. We used the CellTiter 96 AQueous Assay dye reduction assay (Promega) to quantify cell viability. Ibrutinib and PS1145, respectively, were purchased from Selleckchem and Sigma-Aldrich, while the MALT1 inhibitor MI-2 was kindly provided by Ari Melnick (Weill Cornell Medical College, New York, New York).
Gene expression profiling
Microarray hybridization was conducted as previously described.24 Briefly, total RNA was extracted using the RNeasy kit (QIAGEN) and profiled on Human Genome U133 Plus 2.0 arrays according to the manufacturer’s instructions (Affymetrix). CEL files were imported into the Affymetrix Expression Console and normalized with the Robust Multiarray Average method. Primary gene expression data of the MCL samples are deposited in Gene Expression Omnibus (GEO) under accession number GSE70910. Consistent with the disease biology, CCND1 mRNA was highly and consistently overexpressed in all the MCL samples compared with CLL samples shown for reference (data available at GEO under accession number GSE21029)24 (supplemental Figure 1).
To derive the NIK gene signature, we extracted the signal intensities of the probe sets from the CEL files of the dataset of GEO accession ID: GSE524358 and normalized them using the Robust Multiarray Average method. Three separate comparisons of the NIK knockdown experiment were analyzed as follows: (1) NIKsh# 100 ng/mL DOX (n = 4) vs NIKsh# 0 ng/mL (n = 4); (2) NIKsh# 100 ng/mL vs NIKsh# 0 ng/mL DOX (n = 4) and NTC (n = 4); and (3) NIKsh# 100 ng/mL DOX (n = 4) vs NTC (n = 4). Using these comparisons, we identified (1) 102 probe sets/77 genes (1.5 fold change [FC], P < .01); (2) 226 probe sets/153 genes (1.5 FC, false discovery rate [FDR] < 0.2); and (3) 105 probe sets/83 genes (1.5 FC, P < .01) with detectable signal intensities in at least half of the arrays, respectively. Forty-five genes were common to all 3 analyses, 28 of which were expressed in both Z-138 and Maver-1 at baseline. These 28 genes were considered the NIK signature genes.
The BCR score in the validation set was computed using gene expression data of MCL LN biopsies reported by Rosenwald et al29 available at http://llmpp.nih.gov/MCL/.
RNA sequencing
Polyadenylated RNA was enriched by 2 rounds of oligo(dT) selection (Invitrogen), followed by PA-seq library construction30 and sequencing on Illumina HiSeq-2000 with paired 50-bp reads. We used MapSplice 231 to map RNA-seq reads to human genome hg19, Samtools package (DHI Group) to remove duplicate reads and create mpileup files, and VarScan package (Genome Institute, Washington University) for INDEL and single-nucleotide variant callings. RNA sequencing data are deposited in GEO under accession number GSE70926. Single-nucleotide variants and INDELs were annotated with Ensembl GRCh37. RNA sequencing data were used only to look for mutations/polymorphisms in candidate genes and were not used toward gene expression, which was based entirely on the Affymetrix microarray data described above.
Statistical analysis
The Student t test (paired or unpaired), Fisher’s exact test, and one-way analysis of variance were used to assess the differences between groups. The probabilities of progression-free survival (PFS) and OS were calculated using the Kaplan-Meier method.32 BCR scores were dichotomized using the optimal cutoff values for predicting progression and overall mortality.33 All P values were 2-sided and P values ≤ .05 were considered statistically significant. Analyses were performed using GraphPad Prism (GraphPad Software Inc), JMP software (SAS Institute), and R statistical software 3.2.2 (Institute for Statistics and Mathematics).
Results
Upregulation of survival and proliferation pathways in MCL cells in vivo is dependent on the LN microenvironment
We analyzed tumor samples from 43 previously untreated patients with MCL that were about to initiate therapy (supplemental Table 1). Consistent with the typical clinical presentation, the majority of patients had LN involvement, 40% had leukemic disease, and over 20% had both nodal and leukemic disease. In 34 patients, LN biopsies were obtained: in 30 by core biopsy and in 4 by surgical excision. In 8 patients, tumor cells were obtained concurrently from both blood and LN. Nine patients provided only blood samples. In summary, 55 samples were analyzed: 17 leukemic samples, 34 LN biopsies, and 4 LN single-cell suspensions.
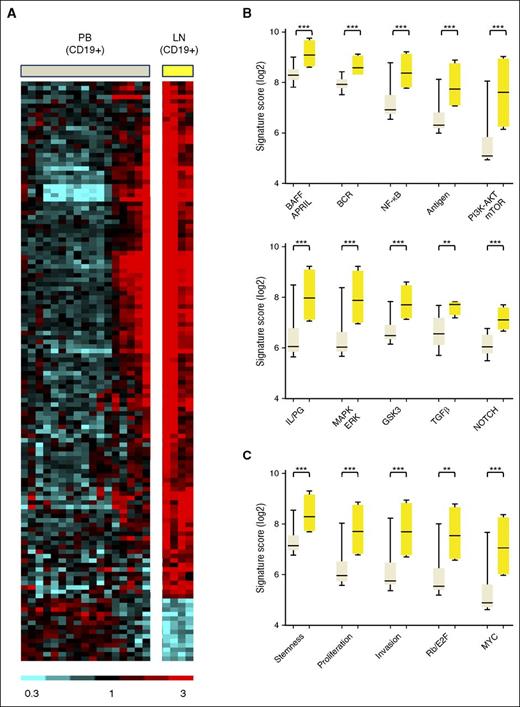
To identify intracellular signaling pathways activated in MCL cells within the lymphoid tissues, we compared gene expression in MCL cells purified by CD19 selection from blood and LN (Figure 1A). By analysis of variance, 130 genes showed at least a 2-fold change in expression with FDR < 0.2. Of these differentially expressed genes, 116 were overexpressed in the LN-derived tumor cells, whereas only 14 were overexpressed in the PB-derived tumor cells (supplemental Table 2). To identify the biologic basis for the differences in gene expression, we chose Gene Set Enrichment Analysis (GSEA) as employed previously.24 GSEA identified 458 curated gene sets that were enriched in tumor cells in the LN compared with their circulating counterparts in the blood at FDR ≤ 0.01 and normalized enrichment score (NES) ≥1.80. Of these, 166 gene sets relevant to lymphoma reflect activation of signaling pathways and tumor proliferation in the LN. Based on their functional similarities, the gene sets were grouped into 4 distinct categories (Figure 1B-C; supplemental Table 3): “Signaling and Interaction with the Microenvironment” comprising 27 gene sets, including BCR, NF-κB, and PI3K-AKT-mTOR (Figure 1B); “Proliferation and Malignancy” comprising 103 gene sets (Figure 1C); “DNA Damage Pathways” (19 gene sets); and “Other” (17 gene sets).
Gene expression differs between MCL cells in PB and LN due to activation of signaling pathways in the LN. (A) Heat map of 130 genes that were differentially expressed in purified MCL cells obtained from PB (PBT, n = 17) and LN (LNT, n = 4) in 18 patients (>2-fold change, FDR < 0.2). Gene expression data were median-centered. Relative expression is indicated by the color scale. (B and C) GSEA identified a large number of upregulated gene sets in LNT compared with PBT. Accordingly, 166 relevant gene sets were selected having FDR ≤ 0.01, NES ≥1.80, and ≥10 “leading edge genes” (the genes of a given gene set most significantly differentially expressed in the experimental data), then grouped into 4 categories based on their functional similarities (supplemental Table 3). Two of these 4 categories are displayed: (B) “Signaling and interaction with the microenvironment” and (C) “Proliferation/malignancy”. Within each subcategory, the “signature score” of the gene set with the highest NES was calculated and displayed. This signature score was computed as the average of the mRNA expression level of the leading edge genes of a given gene set. The signature scores were compared between PBT and LNT samples using unpaired Student t test. AKT, protein kinase B; APRIL, a proliferation inducing ligand; BAFF, B-cell activating factor; E2F, E2F transcription factor; ERK, extracellular signal regulated kinase; GSK3, glycogen synthase kinase 3; IL, interleukin; LNT, purified tumor cells from LN; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin; MYC, v-myc avian myelocytomatosis viral oncogene homolog; PBT, purified tumor cells from PB; PG, prostaglandin; PI3K, phosphatidylinositol 3-kinase; Rb, retinoblastoma; TGFβ, transforming growth factor-β. ***P < .001; **P < .01.
Gene expression differs between MCL cells in PB and LN due to activation of signaling pathways in the LN. (A) Heat map of 130 genes that were differentially expressed in purified MCL cells obtained from PB (PBT, n = 17) and LN (LNT, n = 4) in 18 patients (>2-fold change, FDR < 0.2). Gene expression data were median-centered. Relative expression is indicated by the color scale. (B and C) GSEA identified a large number of upregulated gene sets in LNT compared with PBT. Accordingly, 166 relevant gene sets were selected having FDR ≤ 0.01, NES ≥1.80, and ≥10 “leading edge genes” (the genes of a given gene set most significantly differentially expressed in the experimental data), then grouped into 4 categories based on their functional similarities (supplemental Table 3). Two of these 4 categories are displayed: (B) “Signaling and interaction with the microenvironment” and (C) “Proliferation/malignancy”. Within each subcategory, the “signature score” of the gene set with the highest NES was calculated and displayed. This signature score was computed as the average of the mRNA expression level of the leading edge genes of a given gene set. The signature scores were compared between PBT and LNT samples using unpaired Student t test. AKT, protein kinase B; APRIL, a proliferation inducing ligand; BAFF, B-cell activating factor; E2F, E2F transcription factor; ERK, extracellular signal regulated kinase; GSK3, glycogen synthase kinase 3; IL, interleukin; LNT, purified tumor cells from LN; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin; MYC, v-myc avian myelocytomatosis viral oncogene homolog; PBT, purified tumor cells from PB; PG, prostaglandin; PI3K, phosphatidylinositol 3-kinase; Rb, retinoblastoma; TGFβ, transforming growth factor-β. ***P < .001; **P < .01.
Activation of the BCR and canonical NF-κB pathways in MCL cells
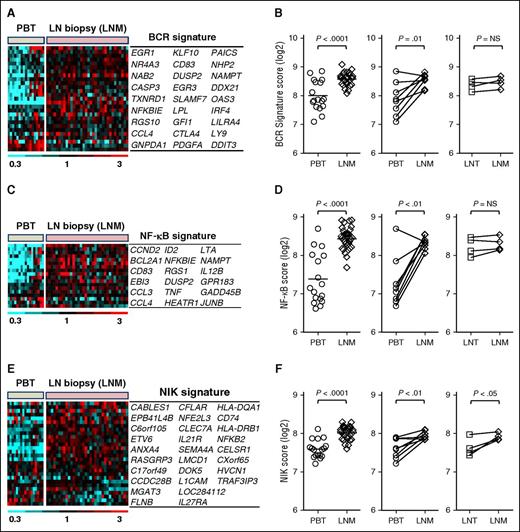
Using experimentally validated BCR and NF-κB gene signatures,24 we investigated the activation of these pathways in a large cohort of patients having contributed LN core biopsies. The BCR gene signature consisting of 27 genes was highly expressed in LN but less so in most leukemic samples (Figure 2A). As a quantitative measure of BCR signaling, we used a gene signature score computed as the averaged expression of all genes in the BCR signature. Across all samples, the median BCR signature score increased in the nodal as compared with the leukemic tumors (P < .0001; Figure 2B, left panel). Ruling out that the observed differences were due to interpatient variability, we specifically confirmed upregulation of BCR signaling in the LN in matched pairs of nodal and leukemic samples obtained concurrently from the same patient (n = 8; Figure 2B, middle panel). Interestingly, in 7 of 17 patients with leukemic MCL, the BCR scores in PB-derived tumor cells were comparable to that in the nodal samples.
Activation of BCR and NF-κB signaling and tumor proliferation in LN-resident cells. (A) Twenty-seven genes comprising a previously validated BCR gene signature are depicted in a heat map. Gene expression data were median-centered and scaled as indicated. Each column represents a patient’s sample; each row represents a gene. Genes are vertically arranged in the corresponding table, following the same order as in the heat map. (B) The BCR signature score was computed by averaging the mRNA expression level of all signature genes. Signature scores between PBT and LNM groups were compared using unpaired Student t test (left panel). BCR scores in matched MCL PBT/LNM (n = 8) and LNT/LNM (n = 4) samples were compared using paired Student t test (right panels). Matched samples were concomitantly collected from the same patient and are connected with a line. (C) Heat map of 18 genes representing the canonical NF-κB pathway (NF-κB signature) generated after a supervised clustering of the samples. Patients’ samples and gene names are arranged in columns and rows, respectively. Genes are vertically arranged in the corresponding table, following the same order as in the heat map. (D) The NF-κB signature scores of MCL PBT, LNM, and LNT were compared as in (B). (E) Heat map of 28 genes representing the alternative NF-κB pathway (NIK signature) generated after a supervised clustering of the samples. Patients’ samples and gene names are arranged in columns and rows, respectively. Genes are vertically arranged in the corresponding table, following the same order as in the heat map. (F) The NIK signature scores of MCL PBT, LNM, and LNT were compared as in (B). LNM, whole lymph node biopsy.
Activation of BCR and NF-κB signaling and tumor proliferation in LN-resident cells. (A) Twenty-seven genes comprising a previously validated BCR gene signature are depicted in a heat map. Gene expression data were median-centered and scaled as indicated. Each column represents a patient’s sample; each row represents a gene. Genes are vertically arranged in the corresponding table, following the same order as in the heat map. (B) The BCR signature score was computed by averaging the mRNA expression level of all signature genes. Signature scores between PBT and LNM groups were compared using unpaired Student t test (left panel). BCR scores in matched MCL PBT/LNM (n = 8) and LNT/LNM (n = 4) samples were compared using paired Student t test (right panels). Matched samples were concomitantly collected from the same patient and are connected with a line. (C) Heat map of 18 genes representing the canonical NF-κB pathway (NF-κB signature) generated after a supervised clustering of the samples. Patients’ samples and gene names are arranged in columns and rows, respectively. Genes are vertically arranged in the corresponding table, following the same order as in the heat map. (D) The NF-κB signature scores of MCL PBT, LNM, and LNT were compared as in (B). (E) Heat map of 28 genes representing the alternative NF-κB pathway (NIK signature) generated after a supervised clustering of the samples. Patients’ samples and gene names are arranged in columns and rows, respectively. Genes are vertically arranged in the corresponding table, following the same order as in the heat map. (F) The NIK signature scores of MCL PBT, LNM, and LNT were compared as in (B). LNM, whole lymph node biopsy.
Three NF-κB gene sets were identified as upregulated in nodal compared with leukemic tumors by GSEA. Next, we used 2 experimentally validated gene signatures to separately probe the activity of the canonical and alternative NF-κB pathways (Figure 2C-F). The canonical NF-κB signature, consisting of 18 genes dependent on IKKβ activation,24,34 was on average 2.1-fold more highly expressed in LN biopsies compared with purified MCL cells (P < .0001). Consistent with BCR-dependent activation of NF-κB signaling, the 2 signature scores were highly correlated (r = 0.88; P < .0001; data not shown).
Akin to the distribution of the BCR score, the NF-κB score clearly dichotomized leukemic samples into 2 subsets: a subset with NF-κB scores in the blood consistently lower than in nodal samples and a second subset with NF-κB scores that were equally high in leukemic and nodal samples (Figure 2D, left panel). Thus, the first set of tumors appears to depend on the LN environment for activation of BCR and NF-κB pathways, whereas in the second set, signaling could be either independent of the LN microenvironment or more sustained once cells exit the tissue.
To probe activation of the alternative NF-κB pathway, we used a gene signature dependent on NIK expression8 (see “Methods” for details). The NIK signature was also more highly expressed in LN biopsies compared with purified MCL cells (average fold change 1.6; P < .0001; Figure 2E-F). Overall, NIK signature genes were less well expressed and showed less variability in expression between patients and anatomic compartments than the canonical NF-κB signature genes. Furthermore, although the BCR and NF-κB signatures were equally expressed in excisional nodal biopsies and tumor cells purified from these biopsies (Figure 2B,D, right panels), the NIK signature was more highly expressed in the LN biopsies compared with matching purified tumor cells (average fold change 1.4; P < .05; Figure 2F, right panel), suggesting that genes regulated through the alternative NF-κB pathway are preferentially expressed by nontumor cells in the LN microenvironment.
Activation and propagation of BCR and NF-κB signaling and tumor proliferation
Antigen-dependent BCR signaling is propagated through phosphorylation of kinases that activate different downstream pathways.20 To query sequential activation steps in the BCR signaling pathway, we used flow cytometry to detect phosphorylation of SYK, PLCγ, ERK, AKT, and the NF-κB transcription factor p65. Two representative patients with available leukemic blood samples and matched LN single-cell suspensions (MCL-21 and MCL-28) were analyzed in detail (Figure 3A-C). In both patients, BCR and NF-κB signature scores in the LN were approximately double the respective scores in the blood (Figure 3A). The LN resident cells also contained more activated signaling proteins at every node of the pathway compared with matched leukemic cells (Figure 3B-C). Levels of p-SYKY348 and the BCR signature score were highly correlated in all 14 patients with available samples (Figure 3D).
Activation of signal transduction components of the BCR and downstream pathways. Phosphorylation of SYK, PLCγ, ERK, AKT, and p65 was assessed by flow cytometry in CD5+/CD19+ MCL cells from the PB and LN. (A) Comparison of the BCR and NF-κB signature scores between PBT and LNT samples concomitantly isolated from 2 patients providing matched cell samples. (B) Flow plots of PB and LN samples from a representative patient are shown. (C) Summary of phosphoprotein analysis (tested in duplicate) for matched PB and LN samples from the 2 patients shown in (A). (D) Pearson correlation r was used to measure the relationship of the percent tumor cells with detectable p-SYK and the corresponding BCR signature score in MCL-PBT (n = 14).
Activation of signal transduction components of the BCR and downstream pathways. Phosphorylation of SYK, PLCγ, ERK, AKT, and p65 was assessed by flow cytometry in CD5+/CD19+ MCL cells from the PB and LN. (A) Comparison of the BCR and NF-κB signature scores between PBT and LNT samples concomitantly isolated from 2 patients providing matched cell samples. (B) Flow plots of PB and LN samples from a representative patient are shown. (C) Summary of phosphoprotein analysis (tested in duplicate) for matched PB and LN samples from the 2 patients shown in (A). (D) Pearson correlation r was used to measure the relationship of the percent tumor cells with detectable p-SYK and the corresponding BCR signature score in MCL-PBT (n = 14).
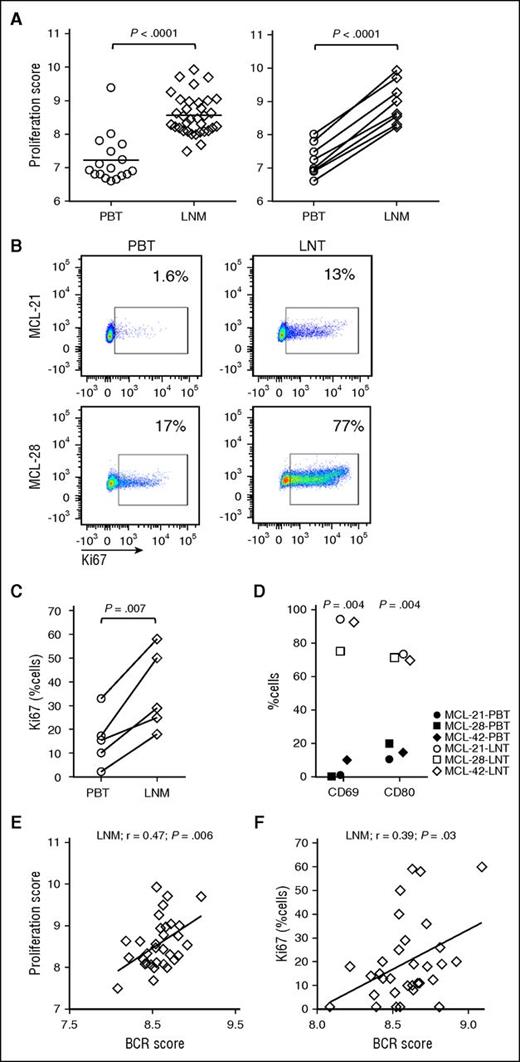
Among the 166 gene sets upregulated in the LN, 52 gene sets were related to tumor proliferation. In addition, a previously reported MCL-derived proliferation signature29 was more highly expressed in nodal compared with leukemic tumors, both among all samples (average fold change 2.5, P < .0001) and among patient-matched samples (n = 8; average fold change 3.2; P < .0001) (Figure 4A). Next, we assessed tumor proliferation by Ki67 expression using immunohistochemistry in LN biopsies and flow cytometry of CD5+/CD19+ gated tumor cells. The proliferation signature score and the number of Ki67-positive tumor cells was highly correlated in both leukemic (r = 0.93; P < .0001) and nodal tumors (r = 0.79; P < .0001) (data not shown). Furthermore, in patients with matched blood and LN samples, Ki67 expression was clearly increased in the nodal compared with the leukemic tumor population (Figure 4B-C). Similarly, expression of activation markers CD69 and CD80 was significantly higher in the nodal tumors (Figure 4D).
The LN microenvironment promotes tumor activation and proliferation. (A) The MCL-proliferation score was compared between PBT and LNM groups using unpaired Student t test (left panel). MCL-proliferation scores in matched MCL PBT/LNM (n = 8) samples were compared using paired Student t test (right panel). (B) Representative PBT and LNT samples from 2 patients with MCL were tested for Ki67 using flow cytometry. (C) Comparison of Ki67-positive cells between PBT and LNM (n = 5) samples concomitantly collected from the same patient. (D) The activation markers CD69 and CD80 were measured using flow cytometry in CD5+/CD19+-gated cells of matched PBT and LNT from 3 patients with MCL. Pearson correlations of the (E) proliferation score in LN biopsies (LNM, n = 34) and (F) the percent Ki67-positive cells in LNM (n = 32) quantified by immunohistochemistry, with the corresponding BCR signature score.
The LN microenvironment promotes tumor activation and proliferation. (A) The MCL-proliferation score was compared between PBT and LNM groups using unpaired Student t test (left panel). MCL-proliferation scores in matched MCL PBT/LNM (n = 8) samples were compared using paired Student t test (right panel). (B) Representative PBT and LNT samples from 2 patients with MCL were tested for Ki67 using flow cytometry. (C) Comparison of Ki67-positive cells between PBT and LNM (n = 5) samples concomitantly collected from the same patient. (D) The activation markers CD69 and CD80 were measured using flow cytometry in CD5+/CD19+-gated cells of matched PBT and LNT from 3 patients with MCL. Pearson correlations of the (E) proliferation score in LN biopsies (LNM, n = 34) and (F) the percent Ki67-positive cells in LNM (n = 32) quantified by immunohistochemistry, with the corresponding BCR signature score.
The BCR score significantly correlated with the proliferation score of nodal samples (Figure 4E;r = 0.47; P = .006), and the percent Ki67-positive cells in the LN (Figure 4F;r = 0.39; P = .03), indicating that tumors with strong BCR signaling are also more proliferative.
Activity of BCR predicts patient survival after cytotoxic therapy
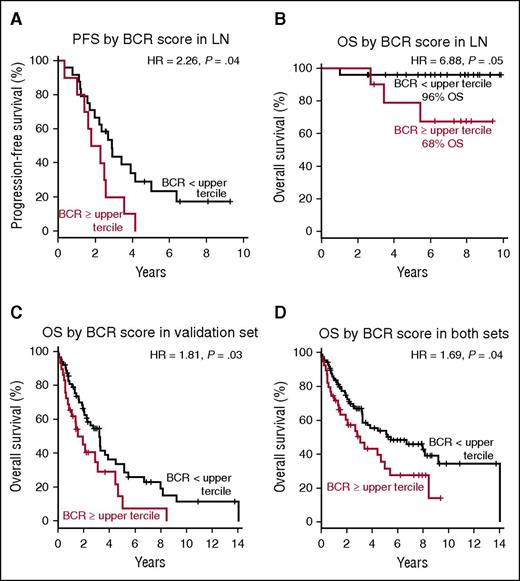
To examine whether the strength of BCR signaling impacts the survival of patients with MCL, we divided patients into 2 groups based on the BCR score in the LN. Patients having a BCR score in the upper tercile had significantly lower PFS (hazard ratio [HR] 2.26; P = .04) and OS (HR = 6.88; P = .05) compared with all other patients (Figure 5A-B). With a median follow-up of 7.5 years, OS for patients with high BCR scores was 68% compared with 96% for patients with lower BCR scores. To validate our finding in an independent set of patients, we computed the BCR score in gene expression data of MCL LN biopsies reported by Rosenwald et al.29 Using the same approach to dichotomize the patients in the validation set, a BCR score in the upper tercile identified patients with inferior survival (HR = 1.81; P = .03; Figure 5C). In the combined patients from both studies (n = 126), the median survival time was 3.1 years for patients with BCR score in the upper tercile compared with 5.5 years for all other patients (HR 1.69; P = .04; Figure 5D). Similarly, the 7 patients whose leukemic samples showed a BCR score above mean had inferior survival compared with the rest of the group, albeit without reaching statistical significance (supplemental Figure 2A-B).
BCR score predicts for PFS and OS in MCL. Probabilities of PFS and OS were compared in subgroups dichotomized by BCR score measured in the LN samples using the log-rank test. The median OS or PFS time was calculated as the smallest survival time when the survival probability is ≤50%. (A) The median PFS time for patients with BCR score within the upper tercile was 2.0 years and was 2.9 years for the remaining patients. (B) At the median follow-up of 7.5 years, the estimated OS for patients with BCR score in the upper tercile was 68% and was 96% for the remaining patients. The median OS time for patients dichotomized by BCR score was (C) in an independent validation set 1.5 years and 3.2 years, respectively, and (D) in both sets combined (n = 126) 3.1 and 5.5 years, respectively.
BCR score predicts for PFS and OS in MCL. Probabilities of PFS and OS were compared in subgroups dichotomized by BCR score measured in the LN samples using the log-rank test. The median OS or PFS time was calculated as the smallest survival time when the survival probability is ≤50%. (A) The median PFS time for patients with BCR score within the upper tercile was 2.0 years and was 2.9 years for the remaining patients. (B) At the median follow-up of 7.5 years, the estimated OS for patients with BCR score in the upper tercile was 68% and was 96% for the remaining patients. The median OS time for patients dichotomized by BCR score was (C) in an independent validation set 1.5 years and 3.2 years, respectively, and (D) in both sets combined (n = 126) 3.1 and 5.5 years, respectively.
Point mutations in signal transduction components of the BCR and NF-κB pathways and response to BTK inhibition
A subset of leukemic samples showed active BCR and NF-κB signaling apparently independent of microenvironmental support. Among 7 samples with BCR scores above mean, the IGHV gene was mutated in 1 (MCL-51), unmutated in 4, and undetermined in 2. All tested samples with BCR scores below mean were IGHV unmutated (supplemental Table S1). We next searched for mutations in genes encoding components of the BCR and NF-κB signaling pathways. Using RNA sequencing, we identified numerous nonsynonymous mutations in 137 genes that encode components of the BCR and NF-κB pathways (supplemental Table 4). Mutations listed in the SNP137 database were deemed polymorphisms and were not considered further. To explore the predicted consequence of mutations on protein structure and function, we used the PolyPhen-2, PROVEAN, Mutation Taster, and PhD-SNP software packages. Mutations that were predicted to have a significant effect on the protein’s structure and/or function by at least 2 programs were considered further and referred to as “relevant mutations” (supplemental Table 5). A total of 12 relevant mutations in 11 genes were identified (Figure 6A). Six of 7 leukemic samples (86%) with a high BCR score carried at least one mutation. By contrast, relevant mutations were identified in only 2 of 9 (22%) samples having a low BCR score (P = .01 by Fisher’s exact test for the overrepresentation of relevant mutations in tumor samples with high BCR score). Only IKBIP V186G and SHARPIN R365H have been previously reported in COSMIC; however, neither has been confirmed as somatic.
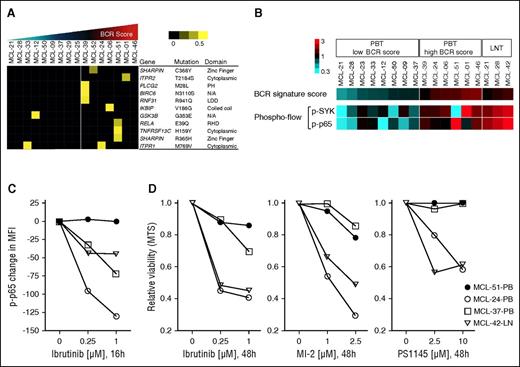
Mutations in signal transduction components, BCR signaling, and sensitivity to BTK inhibition. (A) Nonsynonymous mutations predicted to have a significant effect on the protein by at least 2 software programs were considered “relevant mutations.” Sixteen leukemic MCL samples were subject to RNA sequencing. A set of 137 genes representing the BCR and NF-κB (canonical and alternative) pathways (supplemental Table 4) was tested for point mutations using the following filters: minimum number of reads: 8; minimum percent of mutated reads: 20; duplicate reads excluded; synonymous mutations and mutations listed in the SNP137 database excluded. Samples are displayed according to their ascending BCR signature score and divided in 2 groups: (1) BCR-low, n = 9; and (2) BCR-high, n = 7, based on the BCR score mean. Allele frequency is median-centered and scaled as indicated. Each column represents a patient sample and each row represents a gene. (B) The BCR signature score, and the levels of p-p65 and p-SYK (expressed as log2 of the percent positive cells among CD5+/CD19+-gated cells) were median-centered and displayed according to the color scale. Each column represents a patient’s sample. Leukemic samples are arranged according to their BCR signature score. Suspension cells obtained from LN are shown on the far right for comparison (LNT). (C) Change in p-p65 MFI of 4 samples after exposure to ibrutinib, tested by flow cytometry. Each data point represents the mean of duplicates from 2 separate experiments. (D) MTS assay of samples as in (C) tested in triplicate against increasing concentrations of ibrutinib, the MALT1 inhibitor MI-2, and the IKK inhibitor PS1145 for 48 hours.
Mutations in signal transduction components, BCR signaling, and sensitivity to BTK inhibition. (A) Nonsynonymous mutations predicted to have a significant effect on the protein by at least 2 software programs were considered “relevant mutations.” Sixteen leukemic MCL samples were subject to RNA sequencing. A set of 137 genes representing the BCR and NF-κB (canonical and alternative) pathways (supplemental Table 4) was tested for point mutations using the following filters: minimum number of reads: 8; minimum percent of mutated reads: 20; duplicate reads excluded; synonymous mutations and mutations listed in the SNP137 database excluded. Samples are displayed according to their ascending BCR signature score and divided in 2 groups: (1) BCR-low, n = 9; and (2) BCR-high, n = 7, based on the BCR score mean. Allele frequency is median-centered and scaled as indicated. Each column represents a patient sample and each row represents a gene. (B) The BCR signature score, and the levels of p-p65 and p-SYK (expressed as log2 of the percent positive cells among CD5+/CD19+-gated cells) were median-centered and displayed according to the color scale. Each column represents a patient’s sample. Leukemic samples are arranged according to their BCR signature score. Suspension cells obtained from LN are shown on the far right for comparison (LNT). (C) Change in p-p65 MFI of 4 samples after exposure to ibrutinib, tested by flow cytometry. Each data point represents the mean of duplicates from 2 separate experiments. (D) MTS assay of samples as in (C) tested in triplicate against increasing concentrations of ibrutinib, the MALT1 inhibitor MI-2, and the IKK inhibitor PS1145 for 48 hours.
To investigate which mutations are somatic, we performed Sanger sequencing on tumor and nontumor DNA. Of 8 relevant mutations (RELA E39Q, TNFRSF13C H159Y, IKBIP V186G, PLCG2 M28L, RNF31 R941Q, BIRC6 N3110S, ITPR2 T2184S, and SHARPIN R365H), only the E39Q mutation in RELA in MCL51 was confirmed as somatic (supplemental Figure 3A-H). Previously reported mutations in TRAF2 and BIRC3 were not observed.8
To explore whether mutations in signal transduction components could promote BCR-independent activation of NF-κB, we simultaneously assessed the phosphorylation state of SYK and p65 by flow cytometry (Figure 6B, supplemental Table 6). In nodal tumor cells, both SYK and p65 were highly phosphorylated, consistent with BCR-dependent activation of the canonical NF-κB pathway (Figure 6B). Also, in most leukemic samples, both SYK and p65 were concurrently activated. However, 1 leukemic sample (MCL-51) with 3 relevant mutations had low levels of p-SYK but high levels of p-p65, suggesting that NF-κB activation may occur independently of upstream BCR signaling (Figure 6B).
Next, we tested the effect of BTK inhibition by ibrutinib on p65 activation and viability of primary MCL cells in vitro. In samples having concordant levels of p-SYK and p-p65, ibrutinib reduced p-p65, consistent with BTK-dependent activation of NF-κB downstream of the BCR (Figure 6C) and killed 35% to 50% of the tumor cells within 48 hours (Figure 6D). In contrast, levels of p-p65 and viability of tumor cells from MCL-51 were not substantially affected by ibrutinib (Figure 6C-D). MCL-51 was also resistant to MALT1 and IKK inhibition by MI-2 and PS1145, respectively (Figure 6D).
Discussion
We provide direct in vivo evidence for BCR and canonical NF-κB activation in MCL cells within the LN microenvironment. The activity of BCR signaling directly correlated with increased tumor proliferation and identified a subset of patients with inferior survival, suggesting that activation of BCR signaling drives tumor proliferation and determines clinical outcomes of patients with MCL. Furthermore, we identified mutations in genes encoding components of the BCR and NF-κB pathways that may amplify the cellular response to BCR engagement and promote BTK independent NF-κB activation and ibrutinib resistance.
Ongoing active BCR signaling contrasts with the tenet that MCL is a malignancy of antigen-naïve B cells and provides analogies to CLL and ABC-DLBCL where a role for BCR signaling in lymphomagenesis has been previously recognized.24,25 In contrast to ABC-DLBCL, activating mutations in CD79B are rare in MCL.9,28 Rather, in analogy to CLL, BCR activation in MCL cells appears dependent on tumor-microenvironment interactions in the LN. Although a role of antigen in the ontogeny of MCL has been inferred from the biased IGHV gene repertoire found in a subset of patients,5 this is the first demonstration of ongoing active BCR signaling in tumor cells in vivo. A prior study using gene expression analysis of leukemic samples identified differences between IGHV unmutated and IGHV mutated samples, the latter showing higher expression of genes associated with memory B cells.4 All leukemic cases studied here were of the IGHV unmutated type with the exception of MCL-51, the sample with the RELA mutation. The conclusion that BCR signaling plays a pathogenic role in MCL is consistent with emerging data on the role of SOX11 in this lymphoma.35-37 The transcription factor SOX11 enforces expression of the lineage master regulator PAX5.1,37 PAX5 maintains the B-cell program and prevents terminal plasma-cell differentiation, thereby preserving the cellular responsiveness to antigen induced activation and proliferation.
For the subset of patients in whom expression of BCR and NF-κB target genes appeared to be less dependent on the tissue microenvironment, we considered that intracellular signaling could be due to mutations that activate the BCR in a cell-autonomous fashion or extend the duration of signaling once the cells exit the lymphoid microenvironment. Addressing this possibility, we used RNA sequencing and identified mutations in 11 of 137 genes that encode components of the BCR and NF-κB pathways. Mutations were present in all but 1 of the 7 leukemic samples with high BCR and NF-κB scores. Only the E39Q mutation in RELA was somatic, whereas the others were rare germ line variants. This is the first report of RELA E39Q mutation in cancer. The affected tumor (MCL-51) was unusual for having high levels of p-p65 but low levels of activated SYK and was resistant to inhibition of BTK, MALT1, and NF-κB in vitro. E39 is located in the DNA binding interface of p65 (RELA), and the mutation is predicted to result in a functionally relevant change. Whether this mutation can result in stronger DNA binding or dimerization of the transcription factor remains to be determined. In addition, the RELA mutation in MCL-51 was accompanied by rare germ line variants in TNFRSF13C and SHARPIN, which could alter the cellular signaling capacity. The H159Y mutation in TNFRSF13C, the gene encoding B cell–activating factor receptor, has been associated with the risk of developing B-cell NHL.38 Furthermore, mutations in RNF31 and SHARPIN, both components of the linear ubiquitin chain assembly complex (LUBAC), were identified in 3 leukemic samples. LUBAC aggregates with CARD11-MALT1-BCL10 to activate NF-κB. Recurrent gain-of-function germ line variants in RNF31 were reported in patients with ABC-DLBCL and shown to be required for ABC-DLBCL viability.39 In contrast, we did not identify mutations reported by others in TRAF2 and BIRC38 or any other component of the alternative NF-κB pathway, consistent with the low activity of NIK-dependent signaling in the patients studied here.
All patients who provided samples for this study were subsequently treated, mostly with bortezomib in combination with EPOCH-R. In contrast, the validation set consisted of a group of patients treated in the pre-rituximab era with a variety of different regimens.29 This difference in treatment between the 2 cohorts likely explains why the OS of patients in the validation cohort was shorter than for patients in our study. Notwithstanding, in both cohorts highly active BCR signaling identified a subset of patients with inferior survival. Although we have not shown a direct link between BCR activation in the LN and disease progression, this can be reasonably inferred given the biologic role of BCR signaling in promoting expansion and maturation of antigen-responsive B cells. Furthermore, antigen-dependent BCR activation has been shown to accelerate disease progression in mouse lymphoma models and can drive early stages of mucosa-associated lymphoid tissue lymphomas.40-42
Our analysis of primary tumor samples revealed a previously unappreciated importance of BCR and canonical NF-κB signaling in the pathogenesis of MCL, underscoring the value of specifically targeting these pathways. However, depth and durability of response to BTK inhibition differ considerably between lymphoma subtypes.21,22,43,44 The response rate of 68% to single-agent ibrutinib in relapsed/refractory MCL compares favorably with previously available treatment options for these patients. However, roughly one-third of patients have intrinsically resistant disease whose mechanisms are not well understood. The importance of the RELA E39Q mutation and the germ line polymorphisms in components of the BCR and canonical NF-κB pathways, identified here, await confirmation in patients treated with ibrutinib. To overcome ibrutinib resistance, targeting signal transduction downstream of BTK with inhibitors of LUBAC and/or the MALT1 protease should be explored.39,45
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession numbers GSE70910 and GSE70926).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank their patients for participating and donating the blood and tissue samples to make this research possible. The authors thank Therese White and Margaret Shovlin for assistance in the clinic, Theresa Davis-Hill for preparing LN single-cell suspensions, Jun Zhu and the members of the DNA sequencing and genomics core for assistance with gene expression profiling and RNA sequencing, and Keyvan Keyvanfar for assistance with flow cytometry.
This work was funded by the Intramural Research Programs of the National Heart, Lung, and Blood Institute and the National Cancer Institute, National Institutes of Health.
Authorship
Contribution: N.S.S., S.E.M.H., C.U., and D.B. performed experiments. N.S.S. and A.W. designed the research, analyzed the results, made the figures, and wrote the manuscript. S.P. performed and analyzed the immunohistochemistry experiments. D.L. performed the biostatistical analysis and contributed to microarray and sequencing data analysis. A.M. and L.F. provided MI-2. S.B. and A.N. performed IGHV sequencing. N.S.S., A.W., M.A.W., M.S., J.G., C.G., M.R., K.D., and W.H.W. contributed to the study design and to manuscript writing. All authors read and approved the final manuscript.
Conflict-of-interest disclosure: A.W. received institutional research funding from Pharmacyclics. W.H.W. is an inventor on a patent application regarding the use of ibrutinib in ABC-DLBCL. N.S.S. has served on advisory boards and on the speaker's bureau for Pharmacyclics.
Correspondence: Adrian Wiestner, Hematology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bld 10, CRC 3-5140, 10 Center Dr, Bethesda, MD 20892-1202; e-mail: wiestnera@mail.nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal