Key Points
We develop and validate Karyogene, a comprehensive one-stop diagnostic platform for the genomic analysis of myeloid malignancies.
Karyogene simultaneously detects substitutions, insertions/deletions, translocations, copy number and zygosity changes in a single assay.
Abstract
The diagnosis of hematologic malignancies relies on multidisciplinary workflows involving morphology, flow cytometry, cytogenetic, and molecular genetic analyses. Advances in cancer genomics have identified numerous recurrent mutations with clear prognostic and/or therapeutic significance to different cancers. In myeloid malignancies, there is a clinical imperative to test for such mutations in mainstream diagnosis; however, progress toward this has been slow and piecemeal. Here we describe Karyogene, an integrated targeted resequencing/analytical platform that detects nucleotide substitutions, insertions/deletions, chromosomal translocations, copy number abnormalities, and zygosity changes in a single assay. We validate the approach against 62 acute myeloid leukemia, 50 myelodysplastic syndrome, and 40 blood DNA samples from individuals without evidence of clonal blood disorders. We demonstrate robust detection of sequence changes in 49 genes, including difficult-to-detect mutations such as FLT3 internal-tandem and mixed-lineage leukemia (MLL) partial-tandem duplications, and clinically significant chromosomal rearrangements including MLL translocations to known and unknown partners, identifying the novel fusion gene MLL-DIAPH2 in the process. Additionally, we identify most significant chromosomal gains and losses, and several copy neutral loss-of-heterozygosity mutations at a genome-wide level, including previously unreported changes such as homozygosity for DNMT3A R882 mutations. Karyogene represents a dependable genomic diagnosis platform for translational research and for the clinical management of myeloid malignancies, which can be readily adapted for use in other cancers.
Introduction
Advances in genomics have defined many of the clinically significant gene mutations in human cancers. In the myeloid malignancies acute myeloid leukemia (AML) and the related myelodysplastic syndromes (MDS), individual cancers harbor a small number of driver mutations, however more than 50 genes are recurrently mutated across cases. Additionally, as in other cancers, the nature of mutations is diverse and ranges from nucleotide (nt) substitutions and insertions/deletions (indels), to large-scale changes such as chromosomal deletions, duplications, and translocations. Because many of these changes influence patient prognosis and/or predict response to therapy, their detection at the time of diagnosis represents an important clinical need.
To address this need, a number of methodologies for the simultaneous analysis of multiple target genes have been developed.1-4 However, traditional diagnostic approaches such as karyotyping and fluorescence in situ hybridization (FISH), also remain critical to the complete characterization of AML and a number of important mutations such as internal tandem duplications (ITD) of Fms-like tyrosine kinase 3 (FLT3) (FLT3-ITD) and partial tandem duplications (PTD) of mixed-lineage leukemia (MLL) (MLL-PTD) genes are difficult to detect using conventional next-generation sequencing (NGS)-based approaches.1,5 Furthermore, copy neutral loss-of-heterozygosity (CN-LOH) events, a frequent and prognostically significant class of mutations in AML,6-8 are not detectable by mainstream diagnostic platforms. Although whole genome and exome sequencing can capture many of the target mutations, they both remain costly, analytically intensive, and unable to reliably detect translocations and zygosity changes in their standard formats. Furthermore, they can both fail to detect low-burden subclonal mutations with clinical significance, such as those affecting TP53.9 Therefore, there is a pressing need for a robust and accessible platform that can comprehensively characterize the diverse types of mutations in myeloid malignancies to guide clinical decision-making.
In order to address this unmet clinical need, we have developed Karyogene, a one-stop diagnostic method employing targeted capture followed by NGS coupled with a bespoke suite of novel and recently developed bio-informatic tools for the simultaneous detection of substitutions, indels, chromosomal translocations, and genome-wide copy number and zygosity changes. We describe and validate this diagnostic platform using 62 AML and 50 MDS diagnostic samples previously characterized using conventional diagnostic approaches. Our results show that Karyogene performs remarkably well in detecting these diverse mutation classes and can also identify novel mutations, including an MLL–diaphanous-related formin 2 (DIAPH2) fusion and CN-LOH of mutations involving DNMT3A R882. The approach represents a significant advance toward bringing genomics to the diagnosis of myeloid malignancies and can easily be adapted for use in other cancers.
Methods
DNA samples
Diagnostic bone marrow (BM) DNA samples from 62 unselected AML patients were obtained from 2 centers: Hospital de la Santa Creu I Sant Pau, Barcelona, Spain and Addenbrooke’s Hospital, Cambridge, United Kingdom. Of these, 24 patients also had paired remission samples. Diagnostic BM DNA samples from 50 MDS patients, enriched for cases with cytogenetic abnormalities, were extracted from cytogenetic pellets stored at −20°C in methanol/acetic acid at the Haemato-oncology Diagnostic Service, Addenbrooke’s Hospital and genomic DNA was extracted using a Qiagen DNeasy Kit as per the manufacturer’s instructions. Included in the study were also cord blood samples (n = 7), and blood granulocyte and mononuclear cell DNA from unselected adults without evidence of hematologic abnormalities (n = 33). (See supplemental Table 1, available on the Blood Web site, for characteristics of the 181 samples used in the study). Samples were obtained with written informed consent and appropriate ethics committee approval (approval reference numbers: 07/MRE05/44 or CEIC-11/2012, and EC/15/092/4214).
Bait design for targeted DNA capture
A custom library of 53 613 oligonucleotide baits was designed using SureDesign software (ELID reference: 0479081; SureSelect, Agilent Technologies) to capture the following: (1) all exons of 49 genes known to be recurrently mutated in myeloid malignancies (Table 1). The exon co-ordinates were downloaded from BioMart release 68 (http://www.ensembl.org/biomart/martview/), RefSeq release 54 (http://www.ncbi.nlm.nih.gov/refseq/), CCDS release 9 (http://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi), Gencode release 12 (http://www.gencodegenes.org/releases/12.html), and Vega release 48 (http://vega.sanger.ac.uk/index.html), and overlapping coordinates were merged into the longest possible consensus sequence for which overlapping 120-nt baits were created, starting every 30 bp (supplemental Figure 1A). Baits were designed using SureDesign to include 10 bp flanking regions at the 5′ and 3′ ends of each exon and bait overlap with repetitive regions was limited to a maximum of 20 bp. (2) Previously identified intronic breakpoints at both partner genes for detection of PML-RARA t(15;17), CBFB-MYH11 (inv[16]), RUNX1-RUNXT1 t(8;21), and at the MLL gene for detection of MLL translocations with any partner (Table 1; supplemental Table 2). These regions were covered with overlapping 120 bp baits starting every 40 bp (supplemental Figure 1B); and (3) 9111 single nucleotide polymorphisms (SNPs) with minor allele frequency (MAF) of 0.40 to 0.45 across diverse human population cohorts, spaced on average every 300 kb on all autosomes and on the X chromosome. Of these, 135 were discarded because they gave less than 10 reads in two or more normal samples leaving 8976 for analysis, of which 8673 gave consistent results in normal samples and were used for copy number calls (see supplemental Methods for details). Each SNP location was covered by 3 overlapping 120 nt baits (supplemental Figure 1C). The size of total target region was 2.3 Mbp. The least stringent repeat masking option was selected in SureDesign to avoid placing baits on highly repetitive or low complexity regions. The replication of individual baits was adjusted depending on the guanine-cytosine content of the target regions using the SureDesign maximize performance bait boosting option for all targets, with the exception for breakpoint probes where balanced boosting was selected (Agilent Technologies). For access to the bait design, see https://github.com/karyogene/diagnostic-tool.
Genomic loci captured and analyzed by the Karyogene platform
| I. Coding mutations in 49 genes . | |||||
|---|---|---|---|---|---|
| Gene ID . | Chromosome . | Position (Mb)* . | Gene ID . | Chromosome . | Position (Mb)* . |
| MSTP9 | 1 | 17.1 | WT1 | 11 | 32.4 |
| NRAS | 1 | 115.2 | MLL | 11 | 118.3 |
| DNMT3A | 2 | 25.5 | CBL | 11 | 119.1 |
| DDX18 | 2 | 118.5 | ETV6 | 12 | 11.8 |
| SF3B1 | 2 | 198.2 | KRAS | 12 | 25.4 |
| IDH1 | 2 | 209.1 | PTPN11 | 12 | 112.9 |
| CBLB | 3 | 105.3 | FLT3 | 13 | 28.6 |
| GATA2 | 3 | 128.1 | DIS3 | 13 | 73.3 |
| MUC4 | 3 | 195.4 | IDH2 | 15 | 90.6 |
| KIT | 4 | 55.5 | CREBBP | 16 | 3.7 |
| TET2 | 4 | 106.1 | TP53 | 17 | 7.6 |
| DDX4 | 5 | 55 | NF1 | 17 | 29.4 |
| CSF1R | 5 | 149.4 | SRSF2 | 17 | 74.7 |
| NPM1 | 5 | 170.8 | MUC16 | 19 | 8.9 |
| DAXX | 6 | 33.2 | CEBPA | 19 | 33.8 |
| ZAN | 7 | 100.3 | ASXL1 | 20 | 30.9 |
| EZH2 | 7 | 148.5 | PTPRT | 20 | 40.7 |
| CSMD1 | 8 | 2.8 | RUNX1 | 21 | 36.1 |
| RAD21 | 8 | 117.8 | U2AF1 | 21 | 44.5 |
| JAK2 | 9 | 49.8 | BCOR | X | 39.9 |
| PRUNE2 | 9 | 79.2 | KDM6A | X | 44.7 |
| WAC | 10 | 28.8 | SMC1A | X | 53.4 |
| PTEN | 10 | 89.6 | STAG2 | X | 123.1 |
| SMC3 | 10 | 112.3 | PHF6 | X | 133.5 |
| NUP98 | 11 | 3.7 | |||
| I. Coding mutations in 49 genes . | |||||
|---|---|---|---|---|---|
| Gene ID . | Chromosome . | Position (Mb)* . | Gene ID . | Chromosome . | Position (Mb)* . |
| MSTP9 | 1 | 17.1 | WT1 | 11 | 32.4 |
| NRAS | 1 | 115.2 | MLL | 11 | 118.3 |
| DNMT3A | 2 | 25.5 | CBL | 11 | 119.1 |
| DDX18 | 2 | 118.5 | ETV6 | 12 | 11.8 |
| SF3B1 | 2 | 198.2 | KRAS | 12 | 25.4 |
| IDH1 | 2 | 209.1 | PTPN11 | 12 | 112.9 |
| CBLB | 3 | 105.3 | FLT3 | 13 | 28.6 |
| GATA2 | 3 | 128.1 | DIS3 | 13 | 73.3 |
| MUC4 | 3 | 195.4 | IDH2 | 15 | 90.6 |
| KIT | 4 | 55.5 | CREBBP | 16 | 3.7 |
| TET2 | 4 | 106.1 | TP53 | 17 | 7.6 |
| DDX4 | 5 | 55 | NF1 | 17 | 29.4 |
| CSF1R | 5 | 149.4 | SRSF2 | 17 | 74.7 |
| NPM1 | 5 | 170.8 | MUC16 | 19 | 8.9 |
| DAXX | 6 | 33.2 | CEBPA | 19 | 33.8 |
| ZAN | 7 | 100.3 | ASXL1 | 20 | 30.9 |
| EZH2 | 7 | 148.5 | PTPRT | 20 | 40.7 |
| CSMD1 | 8 | 2.8 | RUNX1 | 21 | 36.1 |
| RAD21 | 8 | 117.8 | U2AF1 | 21 | 44.5 |
| JAK2 | 9 | 49.8 | BCOR | X | 39.9 |
| PRUNE2 | 9 | 79.2 | KDM6A | X | 44.7 |
| WAC | 10 | 28.8 | SMC1A | X | 53.4 |
| PTEN | 10 | 89.6 | STAG2 | X | 123.1 |
| SMC3 | 10 | 112.3 | PHF6 | X | 133.5 |
| NUP98 | 11 | 3.7 | |||
| II. Chromosomal rearrangements | III. Copy number and zygosity changes | ||||
| PML-RARA | t(15:17) | Genome-wide polymorphic SNPs (n = 9111) with | |||
| CBFB-MYH11 | inv(16) | MAF 0.40 to 0.45 in 3 continental cohorts | |||
| RUNX1-RUNXT1 | t(8;21) | ||||
| MLL fusions | 11q23 rearrangements | ||||
| II. Chromosomal rearrangements | III. Copy number and zygosity changes | ||||
| PML-RARA | t(15:17) | Genome-wide polymorphic SNPs (n = 9111) with | |||
| CBFB-MYH11 | inv(16) | MAF 0.40 to 0.45 in 3 continental cohorts | |||
| RUNX1-RUNXT1 | t(8;21) | ||||
| MLL fusions | 11q23 rearrangements | ||||
CRCh37/hg19.
DNA target enrichment and sequencing
DNA fragmentation, library preparation, indexing, and solution phase hybrid capture were performed according to the manufacturer’s instructions (Agilent Technologies). The 181 indexed samples were sequenced across 9 lanes of Illumina HiSeq 2000 (75 bp paired-end) and FASTQ files aligned to GRCh37/hg19 human reference sequence (2009) using Burrows–Wheeler Alignment (http://bio-bwa.sourceforge.net/bwa.shtml). All samples were also aligned using version 0.7.6.2 of the Sequence Mapping and Alignment Tool (SMALT) aligner (http://smalt.sourceforge.net) for the purposes of translocation detection.
Translocation detection using SMALT-finder of inversion and translocations (FIT)
In order to detect translocation breakpoints, paired-end reads were aligned to the human reference genome (Hg19) using SMALT version 0.7.6.2, which reports paired read alignments by individual alignment scores and has a mode with enhanced sensitivity for “split” read alignments. The exact breakpoints were identified from chimeric reads using the in-house written software FIT (https://github.com/gt1/FIT). A minimum of 3 independent supporting chimeric reads was required to call a translocation (see supplemental Methods for a more detailed description).
Detection of nt substitutions and indels
Substitutions and indels (small indels) involving exons of the 49 genes studied here were detected using Mutation Identification and Analysis Software (MIDAS), an in-house perl script previously designed and validated to detect such mutations without the need for matched normal comparisons.1 Briefly, MIDAS was adjusted to report positions covered by at least 2 independent high-quality reads (sequencing and mapping quality >20 and with no additional mismatches or indels in the same read) reporting a different base to the reference genome. Mutations near polynucleotide tracks or with a clear read position or read orientation bias were removed. In the case of indels, at least 5 independent reads reporting the indel were required in the tumor sample, as well as the absence of any evidence of the indel in the normal sample. CORDG1 DNA (normal DNA from a cord blood sample) was used as a normal sample in all the comparisons. All variants present in the 1000 Genomes database were removed. NPM1 exon 12 4-nt insertions/duplications were also searched for using a highly sensitive and specific tool we described recently.10 Variant calls supported by a variant allele frequency (VAF) of ≥0.05 (5%) were cross-referenced against the Catalogue of Somatic Mutations in Cancer (COSMIC) database (http://cancer.sanger.ac.uk/cosmic/). Missense, frameshift, or nonsense mutations at VAF >0.1 and not present in COSMIC or within ± 10 bases of a COSMIC mutation were reported only if they affected genes known to be targeted by somatic mutations at multiple sites throughout their length (ie, CEBPA, TET2, DNMT3A BCOR, TP53, PHF6, STAG2, RAD21, and SMC1A). To minimize the likelihood of reporting inherited variants, non-hotspot mutations were also manually checked to confirm they were previously reported as somatic and if they were not, we only reported them if their VAF was <0.47 or >0.53 (but <0.98). Mutations were annotated against the transcript in which the mutation is predicted to have the most deleterious effect. Annotations for DNMT3A and KIT mutations were manually changed after mutation calling to match their commonly used reference transcripts. Mutation calls were compared with the known molecular information derived by the diagnostic laboratories using conventional molecular methods including melt curve analysis, real-time polymerase chain reaction (PCR), and gel electrophoresis and capillary sequencing (supplemental Table 3). The MIDAS software can be downloaded from https://github.com/karyogene/diagnostic-tool.
MLL-PTD and FLT3-ITD detection using novel “tandem finder” algorithms
MLL can be mutated via intragenic, PTDs involving exons 3-9, 3-10, or 3-11.11 Because MLL exon 3 is always involved, we designed the MLL Tandem Finder (M-TAFI), a tool comparing the relative coverage for MLL exons 3 to 27 in each sample (supplemental Figure 2). The 2 exons were chosen because of their large size (exon 3: 2654 bp and exon 27: 4249 bp) and very uniform coverage ratio in samples lacking MLL-PTD, including those with MLL fusions or other cytogenetic abnormalities. M-TAFI, a tool based on SAMtools12 (http://samtools.sourceforge.net and http://www.htslib.org/) is available from https://github.com/karyogene/diagnostic-tool.
FLT3-ITDs are in-frame duplications of varying length (3 to >200 nt), within exon 14 or 15 of the gene. They are difficult to detect through analysis of short-read NGS with conventional bio-informatic tools, mainly because of misalignment and/or binning of mutant reads. In order to optimize FLT3-ITD detection, we developed the FLT3 Tandem Finder (F-TAFI), a new bio-informatic tool that extracts sequences with at least partial mapping to FLT3 exons 14 and 15, and generates an overlap graph equivalent to de novo regional assembly. This identified instances when overlapping FLT3 sequences formed “bubbles” or “loops,” indicating the presence of an ITD (supplemental Methods). The F-TAFI software is available from https://github.com/gt1/alternatives.
Copy number and LOH analysis using cloneHD
For genome-wide copy number and zygosity analysis, we analyzed 8673 highly polymorphic SNPs, with an MAF of 0.40 to 0.45 across diverse human populations (9 ethnic populations over 3 continents; supplemental Table 4) to maximize the number of informative (heterozygous) individuals across ethnic groups. Sequencing data from the targeted SNPs were used to derive copy number and identify areas of LOH using cloneHD, a probabilistic algorithm designed for subclone reconstruction from data generated by high-throughput DNA sequencing, that can be used for analysis of copy number, B-allele status, and single nucleotide variant (SNV) genotype.13 A panel of 40 normal samples sequenced using the same bait set were used as a control set to standardize for coverage bias during sequencing and pull-down (supplemental Methods). Copy number outputs were compared with the results of diagnostic cytogenetic and FISH data for each patient (supplemental Table 3).
Validation of somatic mutations: SNVs, indels, duplications, and translocations
Mutations affecting NPM1, FLT3, CEBPA, IDH1, and WT1 were validated by comparison with pre-derived diagnostic data. Additionally, a subgroup of mutations affecting different genes was validated using PCR and MiSeq as described before10 (supplemental Figure 3). Validation of PML-RARA, RUNX1-RUNXT1, CBFB-MYH11, and MLL rearrangement calls was by comparison of translocation breakpoints with pre-derived cytogenetic and FISH diagnostic data (supplemental Table 5).
Validation of the MLL-DIAPH2 fusion gene
PCR and real-time PCR with AML DNA and complementary DNA (cDNA), respectively, were used. DNA primers were: P1: (TAAAATTACAAATGGAAAGGACA) and P2: (TGTCATTTCACATTCCTCCCA); and cDNA primers were P3: (GGAAGTCAAGCAAGCAGGTC) and P4: (CCTTCATGGCCAAAGTTGTT). PCR products were sequenced using Sanger sequencing.
Results
Sequencing data were aligned using Burrows–Wheeler Alignment, and separately by SMALT and analyzed as described in Figure 1. Average coverage was ≥30× for 94% of target exons and 98% of target SNPs, with 75% of exons/SNPs covered at ≥70×. Coverage statistics per exon for each of the 49 genes captured is given in supplemental Appendix 1.
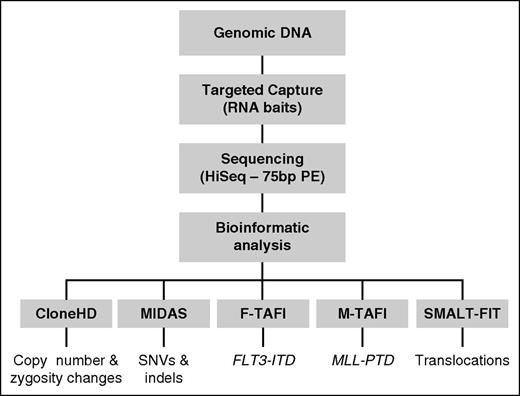
Outline of the Karyogene workflow. Genomic DNA was processed to capture target loci using RNA baits and sequenced on a HiSeq 2000 sequencer as described in “Methods.” Sequencing data were mapped to the genome and analyzed through the indicated software to detect the corresponding types of mutations. The bait design underpinning these is described in supplemental Figure 1. HD, high definition; PE, paired-end.
Outline of the Karyogene workflow. Genomic DNA was processed to capture target loci using RNA baits and sequenced on a HiSeq 2000 sequencer as described in “Methods.” Sequencing data were mapped to the genome and analyzed through the indicated software to detect the corresponding types of mutations. The bait design underpinning these is described in supplemental Figure 1. HD, high definition; PE, paired-end.
Substitutions and indels
Using the approach described in “Methods,” we identified 2185 on-target variants, of which 792 had a VAF ≥0.05. After excluding silent mutations and probable inherited variants, we were left with 218 substitutions/indels of which 155 had been previously reported in myeloid malignancies. Among 62 AML samples, the 4 most common coding mutations identified affected FLT3 (n = 18), NPM1 (n = 13), DNMT3A (n = 14), CEBPA (n = 10), IDH1 (n = 7), and NRAS or KRAS (n = 11) (Figure 2; supplemental Table 6). By comparison with conventional diagnostics performed a priori, we detected 13/13 NPM1, 12/12 FLT3-ITD (size range, 18 to 106 bp), 5/5 IDH1R132, 4/4 CEBPA, 3/3 MLL-PTD, 2/2 IDH2 R140Q, and 1/1 IDH2R172K mutations. A number of variants were called that were not reported in COSMIC and involved genes known to be affected by mutations at multiple positions (eg, DNMT3A, TET2, CEBPA, RUNX1, NF1, STAG2, PHF6, and ZAN). An unselected set of variants were also validated using PCR followed by MiSeq sequencing (supplemental Figure 3), as was 1 AML sample (AML_125_a) with co-existent mutations in IDH1 R132H (VAF 0.23) and IDH2 R140Q (VAF 0.22), given the reported mutual exclusivity of IDH1/2 mutations in AML.14 Patients with translocations generally had fewer coding mutations, although these could be of prognostic significance (eg, KIT exon 8 mutations in patients with CBFB-MYH11 fusions).15,16 Among 24 AML patients with paired diagnostic-remission samples, we identified 5 patients with detectable driver mutations present in the remission sample, involving DNMT3A (×2), ASXL1, IDH2, or RUNX1 (supplemental Table 7). In 4/5 instances, there was a significant reduction in the VAF at remission, but this was not the case for the AML74a/AML74b pair in which the VAF of an ASXL1 mutation was not reduced by chemotherapy. Interestingly, the VAFs of this mutation suggested that it was not part of the leukemic clone or this may represent an artifact.
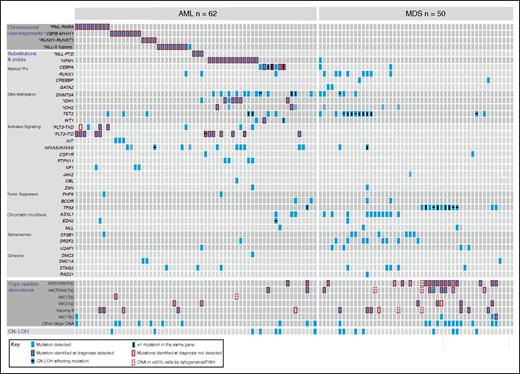
Genomic characterization of myeloid malignancies using Karyogene. Individual AML (n = 62) and MDS (n = 50) samples are represented in columns and genetic mutations in rows. AML samples were unselected whereas MDS samples were pre-selected to harbor chromosomal copy number changes. Mutations are grouped into chromosomal translocations (top), substitutions and indels (middle), CNAs (bottom), and CN-LOH events (bottom row). Clinically relevant CNAs are depicted in separate rows and “other large CNAs” refers to changes affecting regions larger than 3 mbp (described in detail in supplemental Figure 3). The presence of mutations in different contexts is indicated according to the key (bottom left). TF, transcription factor.
Genomic characterization of myeloid malignancies using Karyogene. Individual AML (n = 62) and MDS (n = 50) samples are represented in columns and genetic mutations in rows. AML samples were unselected whereas MDS samples were pre-selected to harbor chromosomal copy number changes. Mutations are grouped into chromosomal translocations (top), substitutions and indels (middle), CNAs (bottom), and CN-LOH events (bottom row). Clinically relevant CNAs are depicted in separate rows and “other large CNAs” refers to changes affecting regions larger than 3 mbp (described in detail in supplemental Figure 3). The presence of mutations in different contexts is indicated according to the key (bottom left). TF, transcription factor.
Among 50 MDS samples, selected to be enriched for cases with abnormal karyotypes, the most common mutations affected TP53 (n = 16), TET2 (n = 18), SRSF2 (n = 8), and ASXL1 (n = 11). Of note, 6 of the 11 ASXL1 mutations identified in our MDS samples were frameshift mutations at c.1927 due to an insertion of a guanine (G) leading to p.G643fs*15 (equivalent to c.1934dupG; p.G646GWfs*12). Although we did not identify this mutation in any of the 40 normals sequenced, it is possible this result may be artifactual as reported for G insertions in this G-rich region.17 Notably, all cases with co-existing deletions of chromosome 5 and of 1 other chromosome (eg, chromosome 7) also harbored mutations in TP53. Specific patterns of mutational co-occurrence such as SRSF2 and TET2, and of mutual exclusivity among mutations affecting spliceosome genes were observed as previously described18,19 (Figure 2).
Detection of FLT3-ITD and MLL-PTD
FLT3-ITD mutations are prognostically important,20-22 but difficult to identify reliably using conventional short-read NGS data.1,2,5 To address this, we developed F-TAFI, a novel bio-informatic tool that uses a de novo graph-based assembly-like approach to identify sequence “loops” within FLT3 exons 14 and 15, and this detected all 12 cases of FLT3-ITDs in our samples without false positives among 171 FLT3-ITD–negative samples (Figure 2; supplemental Methods). MLL-PTD is also associated with an adverse prognosis23-25 and cannot be detected by standard mutational callers, because it does not change the exonic nt sequence. To detect these mutations, we developed M-TAFI, a distinct bio-informatic approach used to derive an MLL exon3/exon27 coverage ratio from our sequencing data. In our analysis, M-TAFI detected all 3 known cases of MLL-PTD in our 181 samples, without false-positive results (supplemental Figure 4).
Copy number and LOH analysis using cloneHD
To detect copy number and LOH changes, we captured and analyzed sequencing data from highly polymorphic SNPs distributed across all chromosomes except Y using cloneHD.13 The depth at each SNP locus (supplemental Table 8) was calculated as the average depth over the segment targeted in the pull-down minus 10 bp at either end. We further selected for each sample a subset of SNPs that were germ line heterozygous. These read depth, and SNP data were used to generate genome-wide copy number and zygosity values for each sample (supplemental Methods), which were compared with the results of diagnostic cytogenetic and FISH data. This identified 44/47 clinically significant copy number changes that were present in ≥20% of cells at diagnosis, namely del(5)/del(5q) (18/18), del(7)/del(7q) (8/8), del(20q) (6/7), trisomy 8 (10/12), del(13q) (1/1), and del(17p) (1/1) (Figure 2; supplemental Figure 5). Additionally, we identified 3 further cases of 17p deletion, which were not detected cytogenetically (2 of which also harbored TP53 mutations) as well as many smaller genomic deletions and amplifications (Figure 2; supplemental Figure 5).
Furthermore, we identified 18 CN-LOH events in 15 samples, including 11 cases involving known somatic driver mutations (3 TP53, 3 TET2, 2 DNMT3A R882, 1 FLT3-ITD, 1 NRAS, and 1 EZH2) (Figure 2; supplemental Figure 5). In 9 of these 11 cases, the VAF of these mutations was >70% indicating duplication of the mutated allele. The 2 cases of chromosome 2p CN-LOH were seen in association with DNMT3A R882C (VAF 0.97) (Figure 3), and R882H (VAF 0.72) mutations were of particular interest because we could not identify previous published reports of CN-LOH affecting DNMT3A R882 mutations. Additional examples of cloneHD outputs are shown in supplemental Figure 6.
Example of cloneHD output for an MDS sample. (A) Read depth of genome-wide SNP loci (top) and the posterior probability of copy number state of the inferred clone (bottom) in sample MDS108; with karyotype 47, XY, +8, add (13q)[12]. Chromosomes 1 to 22 and chromosome X (23) are depicted. For chromosome 8 and for 13q, copy number gains reflect the karyotype as does the reduced coverage for X. (B) Genome-wide BAF for MDS108 (top) and posterior probability of the B-allele state of the inferred clone (bottom). The B-allele states of 0/2 at 2p and 11q indicate a loss of heterozygosity in these regions, thus in keeping with CN-LOH in these regions. This region includes the DNMT3A gene and CN-LOH explains the high VAF (0.97) for the R882C mutation that was also detected in MDS108. BAF, B-allele fraction.
Example of cloneHD output for an MDS sample. (A) Read depth of genome-wide SNP loci (top) and the posterior probability of copy number state of the inferred clone (bottom) in sample MDS108; with karyotype 47, XY, +8, add (13q)[12]. Chromosomes 1 to 22 and chromosome X (23) are depicted. For chromosome 8 and for 13q, copy number gains reflect the karyotype as does the reduced coverage for X. (B) Genome-wide BAF for MDS108 (top) and posterior probability of the B-allele state of the inferred clone (bottom). The B-allele states of 0/2 at 2p and 11q indicate a loss of heterozygosity in these regions, thus in keeping with CN-LOH in these regions. This region includes the DNMT3A gene and CN-LOH explains the high VAF (0.97) for the R882C mutation that was also detected in MDS108. BAF, B-allele fraction.
Identification of AML-associated chromosomal translocations and identification of the novel fusion gene MLL-DIAPH2
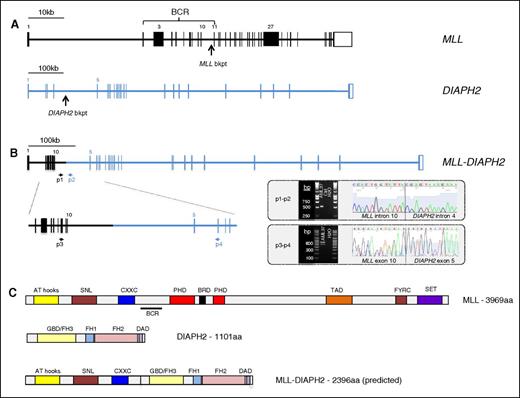
We used targeted pull-down to capture previously identified recurrent breakpoint regions and analyzed sequence reads mapping to these regions using the SMALT-FIT platform. This detected all instances of one of the four common AML-associated translocations, namely t(15;17)/PML-RARA (9/9), inv(16)/CBFB-MYH11 (8/8), t(8;21)/RUNX1-RUNXT1 (4/4), and MLL fusions (8/8); as well as 1 patient with an MLL translocation not identified at diagnosis (see supplemental Table 5 for coordinates of all 30 breakpoints identified in this study). The partner gene was identified in all 9 cases with MLL fusions, with 7/9 involving well-known partners. In 1 case the partner, FLNA, has been described only in 2 cases of infant AML, but never in adult,26,27 and in another, the partner DIAPH2, was novel (Figure 4). There were no false-positive results among the 181 samples analyzed.
Identification of the novel fusion gene MLL-DIAPH2 in an AML sample with t(X;11)(q13;q23). (A) Structure of the MLL (KMT2A) and DIAPH2 genes indicating the DNA breakpoint regions in MLL intron 10 and DIAPH2 intron 4 in this patient with a t(X;11)(q13;q23). (B) Structure of the MLL-DIAPH2 fusion gene verified using PCR amplification and Sanger sequencing of leukemic DNA using primers 1 and 2 (p1 and p2) and cDNA using primers 3 and 4 (p3 and p4). Gel electrophoresis and Sanger sequencing of the PCR product from each experiment are shown delineating translocation breakpoint in DNA sequence (intron 10 of MLL and intron 4 of DIAPH2), and in cDNA (exon 10 of MLL and exon 5 of DIAPH2). (C) Protein structure of MLL, DIAPH2, and (predicted) MLL-DIAPH2 fusion. AT, adenine-thymine hook DNA-binding; BCR, breakpoint cluster region; bkpt, breakpoint; BRD, bromodomain; CXXC, cysteine-X-X-cysteine; DAD, diaphanous autoregulatory domain; FH1-3, formin homology 1-3; FYRC, phenylalanine (F)/tyrosine (Y)-rich C-terminal; GBD, rho GTPase-binding; PHD, plant homeodomain; SET, Su(var)3-9, Enhancer-of-zeste and Trithorax.
Identification of the novel fusion gene MLL-DIAPH2 in an AML sample with t(X;11)(q13;q23). (A) Structure of the MLL (KMT2A) and DIAPH2 genes indicating the DNA breakpoint regions in MLL intron 10 and DIAPH2 intron 4 in this patient with a t(X;11)(q13;q23). (B) Structure of the MLL-DIAPH2 fusion gene verified using PCR amplification and Sanger sequencing of leukemic DNA using primers 1 and 2 (p1 and p2) and cDNA using primers 3 and 4 (p3 and p4). Gel electrophoresis and Sanger sequencing of the PCR product from each experiment are shown delineating translocation breakpoint in DNA sequence (intron 10 of MLL and intron 4 of DIAPH2), and in cDNA (exon 10 of MLL and exon 5 of DIAPH2). (C) Protein structure of MLL, DIAPH2, and (predicted) MLL-DIAPH2 fusion. AT, adenine-thymine hook DNA-binding; BCR, breakpoint cluster region; bkpt, breakpoint; BRD, bromodomain; CXXC, cysteine-X-X-cysteine; DAD, diaphanous autoregulatory domain; FH1-3, formin homology 1-3; FYRC, phenylalanine (F)/tyrosine (Y)-rich C-terminal; GBD, rho GTPase-binding; PHD, plant homeodomain; SET, Su(var)3-9, Enhancer-of-zeste and Trithorax.
Discussion
We describe Karyogene, a genomic analysis platform based on targeted DNA capture followed by sequencing and bespoke bio-informatic analysis based on open-source software tools. We show that the platform efficiently identifies all major categories of somatic mutations found in AML and MDS without the requirement for a matched normal sample as a comparator.
With regards to substitutions and indels, we accurately detected all pre-detected instances of NPM1, FLT3, IDH1, IDH2, and CEBPA mutations, as well as other nt substitutions and indels with established prognostic significance including those affecting TP53, DNMT3A, ASXL1, KIT, SRSF2, and SF3B1. Notably, this was done by comparison with the same unmatched normal comparator for all samples, as would be practical in a diagnostic context. Our approach to the filtration of SNVs and indels reduced the likelihood of misreporting inherited variants as somatic as much as possible; although such an event could not be completely ruled out without the use of paired germ line DNA as a matched comparator. Additionally, we show in a subset analysis of 24 samples with a matched “normal” comparator (remission BM), that such a paired comparison risks filtering out key leukemic mutations from the diagnostic sample. In fact, we found that the comparison between the 24 matched diagnosis-remission pairs in our study, missed clinically important mutations in 5/24 cases affecting DNMT3A (×2), IDH2, RUNX1, and ASXL1, as these mutations persisted in the remission sample. Additionally, using our novel bio-informatic approaches for detecting tandem duplications, we correctly identified all instances of FLT3-ITD and MLL-PTD in our samples; mutations that have previously proven difficult to detect using conventional NGS bio-informatic approaches.1,2,5
A number of different approaches have been described for the detection of chromosomal translocations in NGS sequencing data28,29 by searching for discordant paired-end reads and in some cases, also for split reads. Many of these algorithms display very good sensitivity in detecting translocations and inversions in mappable parts of the genome, but perform less well when repetitive regions are involved and often have a low specificity.29 In order to maximize the accuracy of calls and reach the level required for clinical diagnosis, we focused on detecting the 4 most common translocations in AML/MDS (Table 1), which represent >80% of AML-associated translocations and >90% of those with clinical significance.30 This enabled us to limit the size of our bait-set and to develop a targeted algorithm (SMALT-FIT), which achieved 100% specificity and sensitivity for their detection. Furthermore, we identified 9/9 MLL fusion partners, including the novel partner DIAPH2. DIAPH2, located on Xq21, encodes a member of the diaphanous subfamily of the formin homology family of proteins, which are key regulators of fundamental actin-driven cellular processes conserved from yeast to humans.31,32 Formins have been linked to the progression of cancer,33,34 including hematologic cancer35,36 and even myeloid malignancy37 ; however, DIAPH2 itself has not previously been specifically linked to oncogenesis.
Copy number abnormalities/aberrations (CNAs) and zygosity changes are key determinants of prognosis in many cancers, including AML and MDS. In current diagnostic practice, large-scale genomic gains and losses are detected using karyotyping or FISH,38 but more subtle changes go undetected, as does CN-LOH. To enable the detection of these mutations as part of a single diagnostic tool, we selected 9111 SNPs for targeted capture. These were chosen to have high MAFs (0.40 to 0.45) in multiple human populations, to increase the likelihood of heterozygosity across ethnic groups. Reads mapping to these SNPs were analyzed using cloneHD to derive genome-wide copy number estimates without the need for a matched normal/remission sample. To test the effectiveness of our approach, we deliberately studied several MDS cases with chromosomal abnormalities (Figure 2) and successfully identified 93% (44/47) of clinically relevant large chromosomal abnormalities involving >20% of cells, namely all such cases of del(5)/del(5q) (18/18) and del(7)/del(7q) (8/8), and the majority of del(20q) (6/7), trisomy 8 (10/12), del(13q) (1 of 1), and del(17p) (1 of 1). The 3 missed CNAs (2 cases of trisomy 8 and 1 case of del20q) affected ≤35% of cells and 2/3 were detected using FISH probes rather than karyotyping, leaving some uncertainly about the extent of the genomic gain/loss. In addition, we identified several smaller areas of deletions or amplifications including 3 cases of del(17p), which were not detected cytogenetically (Figure 2; supplemental Figure 5).
Furthermore, we detected several CN-LOH mutations, often involving (duplicating) mutations in genes such as TP53 (17p), TET2 (4q), and FLT3-ITD (13q). Among these, we identified 2 cases of CN-LOH at 2p, 1 involving DNMT3A R882C in a chronic myelomonocytic leukemia (VAF 0.97) (Figure 3) and the other, a DNMT3A R882H in an AML (VAF 0.72). Homozygosity for somatic DNMT3A non-R882 mutations has been reported in AML in association with chromosome 2p CN-LOH.14,39 However, DNMT3A R882 mutations are thought to have dominant negative effects on wild-type expression40 and therefore are not normally found in a homozygous or compound heterozygous state, despite representing 60% of all DNMT3A mutations in AML.14,41,42 In a recent paper, only 1 of 172 cases of DNMT3A R882H or R882C had a VAF >0.6.42 The finding that CN-LOH can sometimes duplicate R882 mutations indicates that homozygosity at this codon is not detrimental to leukemic cells as has been hypothesized. In fact, another possible case of R882 homozygosity was reported recently.42
In conclusion, we report a methodology for the integrated diagnostic work-up of myeloid malignancies, capable of capturing the majority of clinically significant somatic mutations in a single assay and without the need for a matched normal sample, while also enabling the identification of previously undescribed mutations such as novel MLL gene fusions. Importantly, although here we sequenced 181 samples across 9 lanes of a high-throughput platform (HiSeq 2000), smaller numbers of samples can be processed in an identical way and sequenced by lower-throughput sequencers (eg, MiSeq, NextSeq, or other). This would allow a diagnostic laboratory to study 5 to 20 samples once or twice weekly and reduce “sample to report” turnaround time to less than 14 days (less than 10 days for twice weekly runs), thus integrating comfortably into a clinical service. Also, the approach can be easily adapted for use in other malignancies by changing the gene targets and, if relevant, the chromosomal breakpoints for capture. The set of polymorphic SNPs validated here can be used unaltered for the detection of copy number and LOH mutations in other cancers and even for the detection of LOH in constitutional disorders, although bespoke selection of SNPs or an increase in sequencing depth could improve detection of smaller areas of copy number change within selected regions or of smaller subclones. The ability of Karyogene to detect copy number changes with a sensitivity that is at least equivalent to conventional karyotyping, which is expensive and labor-intensive, is an important advantage that is likely to make cost calculations favorable for most integrated diagnostic laboratories. Karyogene represents an important advance that can accelerate the introduction of genomics to clinical diagnosis.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Servicio Santander Supercomputación for their support, the Cambridge Blood and Stem Cell Biobank, the Cambridge Cancer Molecular Diagnosis Laboratory, and the Cambridge Biomedical Research Centre (National Institute for Health Research, United Kingdom) for help with sample collection and processing.
This study was supported by a Wellcome Trust Clinician Scientist Fellowship (100678/Z/12/Z) (T. McKerrell), the Wellcome Trust Sanger Institute (WT098051), and an educational grant from Celgene (ref: 51261). G.S.V. is funded by a Wellcome Trust Senior Fellowship in Clinical Science (WT095663MA), and work in his laboratory is also funded by Bloodwise and the Kay Kendall Leukaemia Fund. A.J.W. is supported by a Specialist Programme from Bloodwise (12048) and by the Medical Research Council (MC_U105161083). I.V. is funded by the Spanish Ministerio de Economía y Competitividad subprograma Ramón y Cajal.
Authorship
Contribution: G.S.V. conceived and designed the study; G.S.V. and T. McKerrell supervised the study, analyzed the data, and wrote the manuscript; T. McKerrell performed experimental procedures; I.V., T. Moreno, H.P., J.M.L.D., G.T., Z.N., and V.M. wrote scripts and performed bio-informatic analysis; J.S., J.N., J.C., B. Huntly, T.F., M.S., A.J.W., P.C., J.B., C.H., B.M., D.B., A.B., and T. McKerrell contributed to sample acquisition and subject recruitment; B. Herman and D.F. contributed to bio-informatic analysis and bait design; V.C. and C.T.-S. identified polymorphic SNPs used in Karyogene; and N.B., N.M., R.R., C.G., N.P., and M.A.Q. contributed to study strategy, and to technical and analytical aspects.
Conflict-of-interest disclosure: G.S.V. is a consultant for and holds stock in Kymab Ltd, and receives an educational grant from Celgene. The remaining authors declare no competing financial interests.
Correspondence: George S. Vassiliou, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SA, United Kingdom; e-mail: gsv20@sanger.ac.uk.

![Figure 3. Example of cloneHD output for an MDS sample. (A) Read depth of genome-wide SNP loci (top) and the posterior probability of copy number state of the inferred clone (bottom) in sample MDS108; with karyotype 47, XY, +8, add (13q)[12]. Chromosomes 1 to 22 and chromosome X (23) are depicted. For chromosome 8 and for 13q, copy number gains reflect the karyotype as does the reduced coverage for X. (B) Genome-wide BAF for MDS108 (top) and posterior probability of the B-allele state of the inferred clone (bottom). The B-allele states of 0/2 at 2p and 11q indicate a loss of heterozygosity in these regions, thus in keeping with CN-LOH in these regions. This region includes the DNMT3A gene and CN-LOH explains the high VAF (0.97) for the R882C mutation that was also detected in MDS108. BAF, B-allele fraction.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/1/10.1182_blood-2015-11-683334/4/m_e1f3.jpeg?Expires=1769110392&Signature=LWo8fD6akGF-zBFoE45Vb4hqo9SetRrOBkWN0gKF-r~KbJaJheRu6hMw6eIrxC6Y4rXpBWKAxHlHBS3778cA8JELWfFv6PdLX-IN7J2DMi8UgEwbCzTVWW-YtqZBVeNI2y374a3KLZEa0QwI-Sr7IL6CVvYgWhZqhpFILvUS-CM04AaeArlYwmgvraCMm4lbXeQtw1YPKLEL2AlpEhcufwaP3HopKgCSUEsyZVz2FZWioE2E8~9PuI3QTHOxlAoLFi~wt8DQr6dNe9K9jwS~CLbYKboZM0Gd6JB5j-DO8demF~lwWKKvbksEQJ75Gnk7Le9DwykKivh5pGgQno2lNg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal