Key Points
IL-4 treatment augments sIgM expression and subsequent downstream signalling in a JAK3/STAT6 dependent manner within CLL samples.
IL-4 exposure partially opposes the activity of Bruton tyrosine kinase or PI3K inhibitors on sIgM-mediated signalling.
Abstract
Kinase inhibitors targeting the B-cell receptor (BCR) are now prominent in the treatment of chronic lymphocytic leukemia (CLL). We have focused here on interleukin 4 (IL-4), a cytokine that protects normal and malignant B cells from apoptosis and increases surface immunoglobulin M (sIgM) expression on murine splenic B cells. First, we have demonstrated that IL-4 treatment increased sIgM expression in vitro on peripheral blood B cells obtained from healthy individuals. In CLL, IL-4 target genes are overexpressed in cells purified from the lymph nodes of patients compared with cells derived from matched blood and bone marrow samples. As for normal B cells, IL-4 increased sIgM expression on CLL cells in vitro, especially in samples expressing unmutated V-genes. IL-4–induced sIgM expression was associated with increased receptor signalling activity, measured by anti-IgM–induced calcium mobilization, and with increased expression of CD79B messenger RNA and protein, and the “mature” glycoform of sIgM. Importantly, the ability of the BCR-associated kinase inhibitors idelalisib and ibrutinib, approved for treatment of CLL and other B-cell malignancies, to inhibit anti-IgM–induced signalling was reduced following IL-4 pretreatment in samples from the majority of patients. In contrast to stimulatory effects on sIgM, IL-4 decreased CXCR4 and CXCR5 expression; therefore, CLL cells, particularly within the progressive unmutated V-gene subset, may harness the ability of IL-4 to promote BCR signalling and B-cell retention within lymph nodes. Effects of IL-4 were mediated via JAK3/STAT6 and we propose a potential role for JAK inhibitors in combination with BCR kinase inhibitors for the treatment of CLL.
Introduction
The finding that clinical outcome is linked to the immunoglobulin mutational status in chronic lymphocytic leukemia (CLL) focused attention on the role of the B-cell receptor (BCR) in this disease.1-3 The clinical success of the Bruton tyrosine kinase (BTK) inhibitor ibrutinib and the phosphatidylinositol 3-kinase δ (PI3Kδ) inhibitor idelalisib has further highlighted a key role of the BCR. Interestingly, treatment-naive mutated CLL (M-CLL) cases tend to respond more slowly to ibrutinib and to have fewer complete remissions than unmutated V-gene CLL (U-CLL).4 This difference could reflect that, although surface immunoglobulin M (sIgM) expression and signal capacity is downmodulated in all cases compared with normal B cells, U-CLL samples tend to retain higher levels of sIgM expression and signalling capacity compared with M-CLL.5-8 Although the drivers of BCR signalling in CLL are unknown, in vitro investigations suggest microbial-derived antigens, autoantigens, and autonomous signals could be involved.9-12 Antigen/autoantigen binding to the BCR results in phosphorylation of CD79A and CD79B by Src family kinases,13 leading to assembly and activation of the signalosome which comprises proteins essential for BCR-induced signal transduction including BTK and PI3Kδ.3 sIgM expression appears to be the main determinant of variable sIgM-signalling capacity.5 However, the specific factors that may influence sIgM expression in CLL cells are not known.
Binding of interleukin-4 (IL-4) to the IL-4 receptor (IL-4R) on B cells results in JAK1/3-mediated phosphorylation of STAT6 (pSTAT6).14 pSTAT6 dimerizes and translocates to the nucleus, where it induces expression of multiple target proteins (including Bcl-2 family members, Bcl-XL and Mcl-1,15 MHCII and CD23) and promotes immunoglobulin class switching.16,17 Moreover, IL-4 is also important for B-cell proliferation and differentiation, and the formation of the germinal centers in mice.18-20 In CLL samples, IL-4 suppresses basal and chemotherapy-induced apoptosis,15,21-23 probably via increased expression of anti-apoptotic proteins.15
IL-4 is one of the cytokines classically synthesized by CLL T cells.24,25 Importantly, CLL patients with progressive disease have been reported to have a significantly greater number of T cells that spontaneously produced IL-4 compared with patients with nonprogressive disease or healthy individuals.26 CLL cells themselves express a significantly greater number of IL-4Rs compared with normal B cells,27,28 and this is consistent with increased pSTAT6 expression in CLL samples29 consequent to the augmentation of STAT6-dependent signalling pathways. Furthermore, in an adoptive transfer model of CLL, T cells were essential for tumor engraftment.30 Together, these data suggest a role for T cells and perhaps IL-4 in CLL biology.
IL-4 has previously been shown to induce sIgM expression and signalling capacity in murine splenic B cells.31,32 An early study on the effects of IL-4 on anti-IgM–induced proliferation using 6 CLL samples showed a heterogeneous response that could have reflected changes in levels of sIgM, but this was before the description of the 2 subsets of disease.33 Here, we show that IL-4 also augments sIgM expression and signalling in the majority of CLL samples. Importantly, we show that IL-4 partially overcomes ibrutinib- or idelalisib-mediated inhibition of sIgM signalling. Stimulatory effects of IL-4 were reversed using JAK3 or STAT6 inhibitors. These results suggest that the ability of IL-4 to enhance effects of antigen in tissues may have been captured by CLL cells to promote tumorigenesis.
Methods
Patient and normal donor samples
Diagnosis of CLL was according to the International Workshop on Chronic Lymphocytic Leukemia-National Cancer Institute 2008 criteria.34 Seventy-four CLL cases were studied following informed written consent in accordance with ethics committee approvals under the declaration of Helsinki (supplemental Table 1, available on the Blood Web site). Procedures for the isolation of malignant cells and the determination of their purity have been described previously.5 All isolates contained >90% CD19+CD5+ cells. Normal donor peripheral blood mononuclear cells (PBMCs) were obtained from anonymized leukocyte cones from the National Blood Service (Southampton, UK) following ethical approval. PBMCs were obtained using density gradient centrifugation (Lymphoprep), resuspended in cryopreservation medium (90% fetal calf serum, 10% dimethyl sulfoxide) and frozen in liquid nitrogen before use to replicate our procedure with CLL PBMCs.
Cell culture and protein extraction
Flow cytometry
Cells were labeled for 30 minutes at 4°C with antibodies conjugated to various fluorochromes. Data were acquired on a BD FACSCanto II. All mean fluorescence intensity (MFI) and percent positive staining were measured relative to an isotype control. Detection of intracellular calcium was quantitated following incubation with 4 µM Fluo3-AM (Life Technologies, UK) and 0.02% (vol/vol) Pluronic F-127 (Sigma, UK). Results are presented as: % responding cells = ([maximal peak height following anti-IgM treatment – mean of unstimulated samples]/% CD19+ve cells) × 100. A total of 1 µM Ionomycin (Sigma) was added as a positive control. Analysis was performed using FlowJo, version 10.
Biotinylation of cell-surface proteins
Biotinylation, isolation of cell-surface proteins and digestion using endoglycosidase H (EndoH) was performed as previously described.37
Gel electrophoresis and Immunoblotting
Proteins were separated on 12% polyacrylamide gels (Thermo Fisher, UK), transferred to nitrocellulose membranes (GE Healthcare, Buckinghamshire, UK), and probed with anti-HSC70 (Santa Cruz, CA), anti-actin (Sigma Aldrich, Poole, UK), and anti-Bcl-2 (Dako, Glostrup, Denmark) as loading controls. Anti-CD79B was from Abcam. All other antibodies were from Cell Signalling Technology (Hitchin, UK). Bands were detected by incubation with horseradish peroxidase–linked secondary antibodies (Dako), enhanced chemiluminescence reagents (Thermo Scientific, Rockford, IL), and visualized using the ChemiDoc-It imaging system (UVP, UK). Band intensities were quantified using ImageJ and normalized to HSC70, Actin, or Bcl-2 as indicated.
Results
IL-4 enhances sIgM expression in B cells from healthy human donors
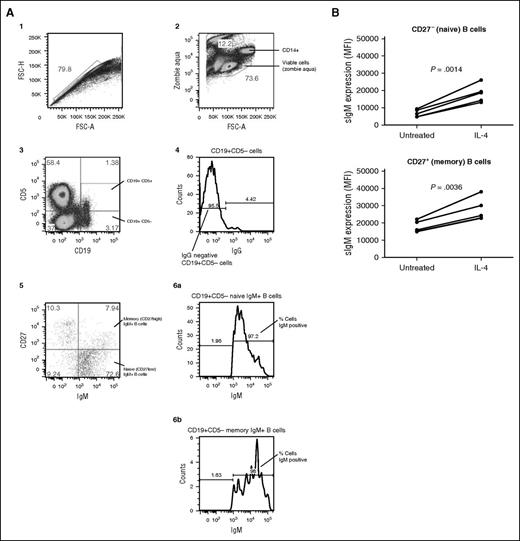
Previous studies have demonstrated that IL-4 enhances sIgM expression in murine B cells.31,32 Before initiating studies in CLL samples, we investigated the effects of IL-4 in normal human B cells. PBMCs were obtained from 5 individual donors and sIgM expression analyzed on the naive (CD19+CD27−) and memory (IgG−CD19+CD27+) B-cell populations using flow cytometry (Figure 1A). PBMCs were treated for 24 hours with IL-4 or the vehicle control (RPMI1640 supplemented with 10% fetal calf serum and antibiotics) before evaluation of sIgM levels. IgM-memory B cells expressed higher levels of sIgM compared with naive B cells, as previously demonstrated38 (Figure 1B). The IL-4–induced fold increase in naive cells (mean 2.7-fold) was significantly greater than that of IgM-memory cells (mean 1.5-fold; P = .0012). Modulation of sIgM was not secondary to effects of IL-4 on cell death because the proportion of dead cells was low (<5%) and not affected by IL-4 in either B-cell subset (data not shown).
IL-4 enhances sIgM expression in B cells from healthy human donors. (A) Depicts the gating strategy for determining the naive and memory B cells from normal donors and described in detail in the supplemental methods. (B) PBMCs were extracted and treated with and without IL-4 (10 ng/mL) for 24 hours. Using the gating strategy in (A), sIgM levels were quantitated by flow cytometry in the naive and memory B cell. Statistical significance was determined by a paired Student t test where indicated (B).
IL-4 enhances sIgM expression in B cells from healthy human donors. (A) Depicts the gating strategy for determining the naive and memory B cells from normal donors and described in detail in the supplemental methods. (B) PBMCs were extracted and treated with and without IL-4 (10 ng/mL) for 24 hours. Using the gating strategy in (A), sIgM levels were quantitated by flow cytometry in the naive and memory B cell. Statistical significance was determined by a paired Student t test where indicated (B).
IL-4 target gene expression in CLL cells in vivo
We performed gene-set enrichment analysis (GSEA)39 to investigate potential effects of IL-4 in CLL patients in vivo. We identified a set of IL-4–regulated genes from a previously published study of CLL cells following treatment in vitro with IL-440 and investigated whether these specific target genes were enriched in the transcriptional profiles of CLL cells purified from lymph node (LN) compared with matched blood and bone marrow.41 GSEA revealed a significant enrichment of expression of IL-4 target genes in LN-derived CLL cells, compared with CLL cells isolated from the other tissues (supplemental Figure 1), consistent with IL-4–dependent transcriptional activity in CLL LN in vivo.
IL-4 enhances expression of sIgM on CLL cells
We investigated the effects of IL-4 on expression of sIgM on CLL cells isolated from the blood of patients. In our initial analysis, CLL samples were treated with or without IL-4 (10 ng/mL), and sIgM expression quantified by flow cytometry at the start of the experiment, and at various times up to 72 hours. A representative sample is shown (Figure 2A) and summarized data are presented (Figure 2B). As previously demonstrated,5 incubation in vitro without IL-4 was associated with a modest increase in sIgM expression, which was most evident at 72 hours. Compared with the vehicle control, IL-4 increased sIgM expression at all time points. Due to potential confounding effects of apoptosis at later time points (48-72 hours), we extended the analysis by investigating effects of IL-4 on sIgM expression at 24 hours in additional samples. In this extended analysis (n = 33 samples), IL-4 increased overall sIgM expression (Figure 2C). However, there was variability in response between individual samples. In contrast to sIgM, IL-4 treatment on the whole did not significantly alter expression of sIgD, although a small subset with sIgD MFI levels >40 (in gray) increased their receptor expression in response to IL-4 (Figure 2D-F). We investigated whether the variation in effects of IL-4 on sIgM expression at 24 hours correlated with CLL prognostic markers. IL-4–increased sIgM expression was significantly greater in U-CLL compared with M-CLL (P = .0016) (Figure 2G) and in ZAP70+ compared with ZAP70− samples (P = .016) (Figure 2H). In contrast, IL-4 effects were not significantly different between CD38+ and CD38− samples (Figure 2I).
IL-4 enhances expression of sIgM, but not sIgD, on CLL cells. Histogram of (A) sIgM and (D) sIgD expression on a representative CLL sample. Graph shows sIg expression at the start of the experiment (black dotted line) and at 24 hours in the presence (solid black line) or absence (dashed line) of IL-4 (10 ng/mL), respectively. Gray histogram, isotype control antibody. (B,E) Results (MFI minus isotype control) for cells treated with IL-4 or left untreated as a control (NA) for up to 72 hours. (C,F) All results expressed as MFI ratios from samples analyzed at 24 hours (C, n = 33; F, n = 29). (G-I) Correlation between the increase in sIgM [(sIgM MFI with IL-4) − (sIgM MFI without IL-4)] and prognostic markers. Samples are categorized according to (G) IGHV mutational status, (H) ZAP70 expression, and (I) CD38 expression. Error bars represent the standard error of the mean (SEM). Statistical significance was determined by either a paired (B,C,E,F) or unpaired (G-I) Student t test or Wilcoxon matched pairs signed rank test.
IL-4 enhances expression of sIgM, but not sIgD, on CLL cells. Histogram of (A) sIgM and (D) sIgD expression on a representative CLL sample. Graph shows sIg expression at the start of the experiment (black dotted line) and at 24 hours in the presence (solid black line) or absence (dashed line) of IL-4 (10 ng/mL), respectively. Gray histogram, isotype control antibody. (B,E) Results (MFI minus isotype control) for cells treated with IL-4 or left untreated as a control (NA) for up to 72 hours. (C,F) All results expressed as MFI ratios from samples analyzed at 24 hours (C, n = 33; F, n = 29). (G-I) Correlation between the increase in sIgM [(sIgM MFI with IL-4) − (sIgM MFI without IL-4)] and prognostic markers. Samples are categorized according to (G) IGHV mutational status, (H) ZAP70 expression, and (I) CD38 expression. Error bars represent the standard error of the mean (SEM). Statistical significance was determined by either a paired (B,C,E,F) or unpaired (G-I) Student t test or Wilcoxon matched pairs signed rank test.
Effects of IL-4 on sIgM expression are mediated via JAK/STAT signalling
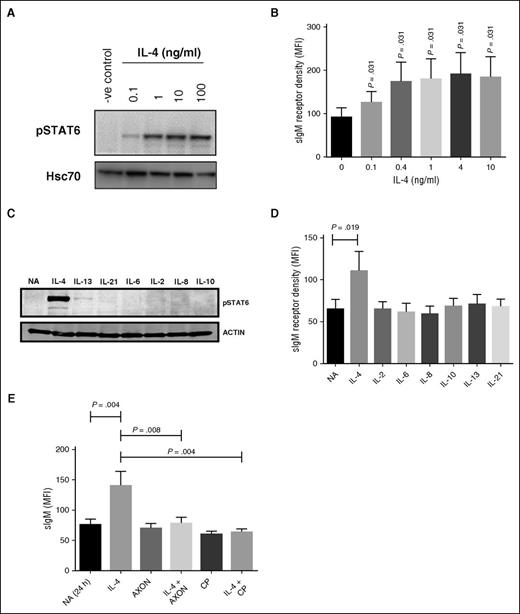
We investigated effects of IL-4 on JAK3-mediated pSTAT6. CLL cells were treated with varying concentrations of IL-4 (0.1–10 ng/mL) for 24 hours, and pSTAT6 quantified by immunoblotting. IL-4 induced a dose-dependent increase in pSTAT6 with a maximal response detected at concentrations >1 ng/mL (Figure 3A) and was paralleled by increased sIgM expression (Figure 3B), but not sIgD (supplemental Figure 2A). We also investigated responses to additional cytokines, including IL-13 (a known inducer of pSTAT6),16 IL-2, IL-6, IL-8, IL-10, and IL-21. Only IL-4 increased pSTAT6 and sIgM expression (Figure 3C-D). The type II IL-4R, which regulates IL-13 signalling, is generally the exclusive receptor expressed by nonhematopoietic cells, and may partially explain the lack of pSTAT6 induced by IL-13.42,43 None of the cytokines altered expression of sIgD (supplemental Figure 2B).
Effects of IL-4 on sIgM expression are mediated via JAK/STAT signaling. CLL samples were treated with IL-4 (0.1-10 ng/mL) for 24 hours and (A) phosphorylated STAT6 (pSTAT6) and (B) sIgM expression analyzed using immunoblotting and flow cytometry, respectively. (A) A representative sample and (B) summarized data (n = 6). (C-D) CLL cells were pretreated with the indicated cytokines and (C) phosphorylated STAT6 (pSTAT6) (n = 9) and (D) sIgM quantified by immunoblotting (n = 9) and flow cytometry (n = 9), respectively. (E) CLL samples (n = 10) were pretreated for 1 hour with the JAK1/3 inhibitor (CP, tofacitinib) or the STAT6 inhibitor (AXON; AS1517499) before IL-4 addition for a further 23 hours. sIgM was quantified by flow cytometry. Graphs show MFI values. Error bars represent the SEM. Statistical significance was determined by a paired Wilcoxon matched pairs signed rank test.
Effects of IL-4 on sIgM expression are mediated via JAK/STAT signaling. CLL samples were treated with IL-4 (0.1-10 ng/mL) for 24 hours and (A) phosphorylated STAT6 (pSTAT6) and (B) sIgM expression analyzed using immunoblotting and flow cytometry, respectively. (A) A representative sample and (B) summarized data (n = 6). (C-D) CLL cells were pretreated with the indicated cytokines and (C) phosphorylated STAT6 (pSTAT6) (n = 9) and (D) sIgM quantified by immunoblotting (n = 9) and flow cytometry (n = 9), respectively. (E) CLL samples (n = 10) were pretreated for 1 hour with the JAK1/3 inhibitor (CP, tofacitinib) or the STAT6 inhibitor (AXON; AS1517499) before IL-4 addition for a further 23 hours. sIgM was quantified by flow cytometry. Graphs show MFI values. Error bars represent the SEM. Statistical significance was determined by a paired Wilcoxon matched pairs signed rank test.
To directly investigate the role of JAK3/STAT6 signalling in IL-4–mediated sIgM induction, CLL cells were pretreated for 1 hour with the JAK3 inhibitor tofacitinib (CP; 10 μM) or the STAT6 inhibitor AS1517499 (AXON; 1 μM) and then incubated for an additional 23 hours in the presence or absence of IL-4 before analysis of sIgM expression. Inhibitor concentrations selected for these experiments were based on initial titration experiments to identify optimal inhibitory concentrations (supplemental Figure 2C-D). AS1517499 and tofacitinib had no effect on sIgM expression when tested alone, but completely blocked the ability of IL-4 to enhance sIgM expression (Figure 3E). The inhibitors had no effects on CLL cell viability at times up to 24 hours (data not shown). Therefore, IL-4-induced increases in sIgM expression appear to be mediated via JAK/STAT signalling.
Increased sIgM expression is associated with higher sIgM signalling capacity in IL-4–treated cells
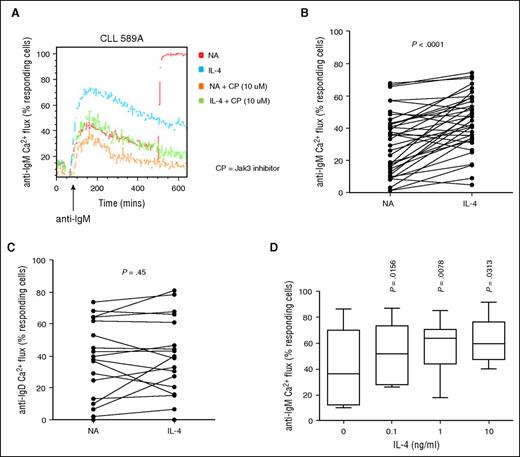
We investigated whether the ability of IL-4 to increase sIgM expression at 24 hours was associated with increased receptor function using intracellular calcium fluxes as a highly quantitative measure of signal responses.5 IL-4 increased anti-IgM–induced calcium mobilization, shown in a representative sample (Figure 4A) and summarized in a larger cohort (Figure 4B), and was accompanied by an increased ability to phosphorylate extracellular signal-regulated kinase (supplemental Figure 2E-F). Sixty-four percent of samples produced >1.2-fold increase in calcium flux, 11% had a 1- to 1.2-fold change. No change or a small reduction <0.3-fold was observed in 24% of samples; however, this was partially confounded due to a small proportion (4/9) of samples that were unable to increase calcium mobilization further because they already produced a substantive signalling response (>50%) before IL-4 addition. Overall, IL-4 treatment did not alter responses to anti-IgD (Figure 4C); however, 4 samples with unmutated IGHV did increase >1.2-fold. Similar to effects on sIgM expression, stimulatory effects of IL-4 on anti-IgM–induced calcium signalling were concentration-dependent (Figure 4D; supplemental Figure 2G), with maximal response at concentrations >1 ng/mL. Moreover, the JAK3 inhibitor tofacitinib also inhibited the stimulatory effects of IL-4 on sIgM signalling (Figure 4A).
Regulation of anti-IgM– and anti-IgD–induced calcium flux by IL-4. CLL samples were treated with IL-4 for 24 hours or left untreated as a control (NA). (A) CLL cells were subsequently treated with the JAK1/3 inhibitor (CP) for 1 hour and then stimulated with soluble anti-IgM and calcium flux assessed by flow cytometry. A representative flow cytometry plot is shown. (B) anti-IgM (n = 38) and (C) anti-IgD (n = 19) signaling responses were quantified using calcium flux analysis. Graphs show fold change in signaling (%responsive cells with IL-4/%responsive cells in the absence of IL-4). Statistical significance of differences are shown. (D) Effect of IL-4 titration on anti-IgM–induced calcium fluxes as previously described. Statistical significance was determined by paired Student t test or Wilcoxon matched pairs signed rank test.
Regulation of anti-IgM– and anti-IgD–induced calcium flux by IL-4. CLL samples were treated with IL-4 for 24 hours or left untreated as a control (NA). (A) CLL cells were subsequently treated with the JAK1/3 inhibitor (CP) for 1 hour and then stimulated with soluble anti-IgM and calcium flux assessed by flow cytometry. A representative flow cytometry plot is shown. (B) anti-IgM (n = 38) and (C) anti-IgD (n = 19) signaling responses were quantified using calcium flux analysis. Graphs show fold change in signaling (%responsive cells with IL-4/%responsive cells in the absence of IL-4). Statistical significance of differences are shown. (D) Effect of IL-4 titration on anti-IgM–induced calcium fluxes as previously described. Statistical significance was determined by paired Student t test or Wilcoxon matched pairs signed rank test.
IL-4 increases CD79B expression
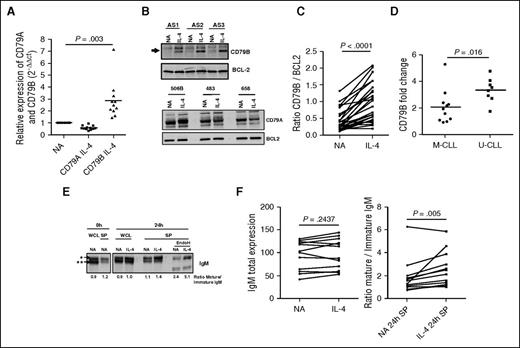
We investigated the effects of IL-4 on expression of CD79A and CD79B, 2 consort molecules that are essential for sIgM expression and signalling.44 β2 microglobulin and Bcl-2 were used to normalize loading for RNA and protein analysis respectively, because their expression did not change upon IL-4 treatment. IL-4 increased expression of CD79B at the RNA (Figure 5A) and total and surface protein (Figure 5B-C; supplemental Figure 3A) at 24 hours. In contrast, IL-4 treatment resulted in a slight reduction in expression of CD79A RNA (Figure 5A) but with no discernible effects on protein expression (Figure 5B). Similar to effects on sIgM expression, effects of IL-4 on CD79B were greater in U-CLL compared with M-CLL samples (Figure 5D). However, no difference in IL-4R expression was observed between U-CLL and M-CLL in a small cohort of CLL (supplemental Figure 3B), indicating IL-4R expression does not explain the differential response between the CLL subsets.
IL-4 increases expression of CD79B and the “mature” glycoform of sIgM. CLL samples were treated with IL-4 for 24 hours or left untreated as a control (NA). (A) CD79A and CD79B mRNA expression was quantified by quantitative polymerase chain reaction (n = 9). β2M was used as the housekeeping gene, and relative expression values were normalized to the untreated cells. (B) Representative sample expressing CD79A and CD79B protein was analyzed by immunoblotting and (C) summarized for CD79B from immunoblotting following Image J quantitation (n = 26). BCL2 expression was analyzed as a loading control. (D) The fold change of CD79B expression between the NA control and IL-4 treated cells was quantified and represented for U-CLL and M-CLL (n = 19). (E) CLL cells were treated with IL-4 and assessed by immunoblotting for total levels of μ-chain (whole cell protein; WCL) or surface protein (SP) μ-chain. EndoH was used to digest the mannosylated µ-chain of IgM (**), whereas the fully glycosylated form (*) was confirmed by resistance to EndoH cleavage. (F) WCL extraction or SP was analyzed by immunoblotting from CLL samples treated in the presence or absence of IL-4. The protein band intensity of mature and immature IgM was quantified using Image J (n = 12). Statistical significance was determined by paired Student t test or Wilcoxon matched pairs signed rank test.
IL-4 increases expression of CD79B and the “mature” glycoform of sIgM. CLL samples were treated with IL-4 for 24 hours or left untreated as a control (NA). (A) CD79A and CD79B mRNA expression was quantified by quantitative polymerase chain reaction (n = 9). β2M was used as the housekeeping gene, and relative expression values were normalized to the untreated cells. (B) Representative sample expressing CD79A and CD79B protein was analyzed by immunoblotting and (C) summarized for CD79B from immunoblotting following Image J quantitation (n = 26). BCL2 expression was analyzed as a loading control. (D) The fold change of CD79B expression between the NA control and IL-4 treated cells was quantified and represented for U-CLL and M-CLL (n = 19). (E) CLL cells were treated with IL-4 and assessed by immunoblotting for total levels of μ-chain (whole cell protein; WCL) or surface protein (SP) μ-chain. EndoH was used to digest the mannosylated µ-chain of IgM (**), whereas the fully glycosylated form (*) was confirmed by resistance to EndoH cleavage. (F) WCL extraction or SP was analyzed by immunoblotting from CLL samples treated in the presence or absence of IL-4. The protein band intensity of mature and immature IgM was quantified using Image J (n = 12). Statistical significance was determined by paired Student t test or Wilcoxon matched pairs signed rank test.
IL-4 increased expression of the “mature” sIgM glycoform
Previous investigations by our group found that the µ-chain of sIgM in CLL cells exists in 2 forms with distinct N-glycosylation patterns: a mature (fully glycosylated) glycoform typical of normal B cells and an immature (high mannose) glycoform more characteristic of IgM in the endoplasmic reticulum (ER), but found on the cell surface after BCR-mediated activation.37 Incubation of CLL cells in vitro led to a restoration in expression of the mature sIgM glycoform.37 To investigate effects of IL-4, we analyzed the expression and glycosylation of µ-chain using whole cell lysates and cell-surface protein-enriched fractions, isolated by biotinylation and streptavidin capture. A representative sample is shown (Figure 5E) and summarized for 12 samples (Figure 5F). Although whole cell expression of µ-chain was not altered, IL-4 significantly increased the relative abundance of the mature µ-chain glycoform on the cell surface as shown by increased expression of a more slowly migrating protein band (Figure 5E). In 1 case with preexisting high levels of the mature glycoform, this was stable (Figure 5F). Incubation with endoglycosidase H to remove terminal mannose selectively removed glycans from the more rapidly migrating protein, confirmed that this glycoform contained high mannose (Figure 5E). Therefore, IL-4 appears to enhance the ability of CLL cells to replace the immature sIgM glycoform with the fully glycosylated form typical of resting normal B cells, but without changes in overall µ-chain expression.
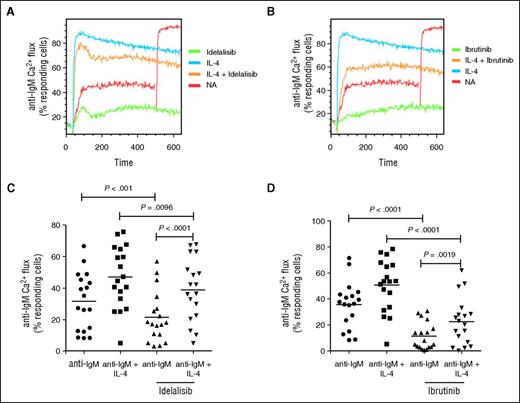
Effect of IL-4 on apoptosis and on inhibition of sIgM signalling by ibrutinib and idelalisib
IL-4 appears to protect against ibrutinib and idelalisib induced apoptosis.45,46 Here we investigated whether IL-4 influenced the response of CLL cells to drug-induced inhibition of the BCR signalling pathway at concentrations in accordance with the literature.45,46 CLL samples were treated with IL-4 for 23 hours and then pretreated with kinase inhibitors for 1 hour before anti-IgM–induced calcium flux. A representative experiment is shown (Figure 6A-B), and the data summarized (Figure 6C-D [idelalisib (5 µM) and ibrutinib (10 µM)]; supplemental Figure 4A-B [1 µM idelalisib or ibrutinib]). The kinase inhibitors significantly reduced CLL viability at 48 hours (supplemental Figure 4C) and anti-IgM–induced signalling in the absence of IL-4 at 1 hour. IL-4 treatment alone increased both CLL viability and anti-IgM–induced calcium responses in the absence and in the presence of inhibitors. Clearly, IL-4 has 2 effects on CLL cells: first in protection against apoptosis per se even in the presence of drugs, and second on the upregulation of sIgM. These may not be tightly linked however, because upregulation of sIgM did not appear to be linked to the level of viability (supplemental Figure 4D). IL-4 treatment had no impact on the expression of BTK or PI3Kδ at the messenger RNA (mRNA) or protein level, indicating that the reduced sensitivity to idelalisib and ibrutinib was not due to reduction in expression of their target protein (data not shown). Therefore, IL-4 may reduce the inhibitory effects of BCR kinase inhibitors.
Effect of IL-4 on inhibition of sIgM signaling by ibrutinib and idelalisib. CLL samples were treated with IL-4 for 23 hours or left untreated as a control (NA) and treated then with ibrutinib or idelalisib for 1 hour. Anti-IgM–induced calcium fluxes were analyzed by flow cytometry. (A-B) Representative samples and (C-D) summarized data (n = 18) are shown. Black icons, U-CLL; gray icons, M-CLL. Graphs show medians and the statistical significance of differences between groups. Statistical significance was determined by paired Student t test or Mann-Whitney U test.
Effect of IL-4 on inhibition of sIgM signaling by ibrutinib and idelalisib. CLL samples were treated with IL-4 for 23 hours or left untreated as a control (NA) and treated then with ibrutinib or idelalisib for 1 hour. Anti-IgM–induced calcium fluxes were analyzed by flow cytometry. (A-B) Representative samples and (C-D) summarized data (n = 18) are shown. Black icons, U-CLL; gray icons, M-CLL. Graphs show medians and the statistical significance of differences between groups. Statistical significance was determined by paired Student t test or Mann-Whitney U test.
The effect of IL-4 on other cell-surface receptors
We investigated whether IL-4 modulated expression of other receptors important for CLL cell behavior. We initially focused on CXCR4, which is thought to play an important role in homing and retention of CLL cells in LNs.47,48 CXCR4 is downmodulated on tissue-localized CLL cells41 (presumably because of ligand-induced receptor endocytosis), and we have shown that culture of CLL cells in vitro is associated with a recovery of CXCR4 expression.49 We therefore investigated whether IL-4 modulated this natural recovery by incubating cells in the presence or absence of IL-4 and quantifying CXCR4 expression by flow cytometry. A representative sample is shown (Figure 7A) and the data summarized (Figures 7B-C; supplemental Figure 5A-C). As previously shown,49 culture in vitro was associated with a substantial increase in CXCR4, which peaked at 24 hours. However, in contrast to effects on sIgM, IL-4 significantly reduced CXCR4 expression. Overall, IL-4 reduced CXCR4 expression by ∼50% at 24 hours (Figure 7C) and by 72 hours CXCR4 expression was almost undetectable (Figure 7B). Effects on CXCR4 were specific for IL-4 among tested cytokines (Figure 7D) and effectively blocked by JAK3 or STAT6 inhibition (Figure 7E). Consistent with reduced CXCR4 expression, IL-4 also reduced CXCL12-dependent CLL cell migration (Figure 7F). In a subset of CLL samples treated with IL-4, we also investigated CXCR4 expression in CD19−CD5+ cells (T cells). In contrast to the CLL cells, IL-4 treatment of CD19−CD5+ cells resulted in an increase in CXCR4 expression (supplemental Figure 5C). This indicated that the effects of IL-4 may be cell specific. The reduction in CXCR4 protein levels in CLL cells was also transcriptionally regulated, because IL-4 also reduced CXCR4 mRNA levels (supplemental Figure 5D). IL-4 also reduced expression of the related chemokine receptor CXCR5 (supplemental Figure 5E-F).
The effect of IL-4 on CXCR4 expression. (A) Histogram showing CXCR4 expression by flow cytometry in a representative CLL sample following treatment in the presence or absence of 10 ng/mL IL-4 for 24 hours. Gray histogram, isotype control; solid black line, 24 hours with IL-4; black dotted line, start of the experiment; dashed line, 24 hours without IL-4. (B,C) Summary of results for all samples analyzed at times up to (B) 72 hours (n = 9) and at (C) 24 hours (n = 21). (D) CLL cells were pretreated with or without the indicated cytokines for 24 hours and CXCR4 expression (n = 9) quantified by flow cytometry. (E) CLL cells were pretreated for 1 hour with the JAK1/3 inhibitor (CP [CP-690550, tofacitinib]) or STAT6 inhibitor (AXON; AS1517499) before IL-4 addition and CXCR4 quantified by flow cytometry at 24 hours. (F) CLL samples (n = 6) were treated with IL-4 for 24 hours and migration to CXCL12 quantified by assessing the number of CD19+CD5+ CLL cells by flow cytometry that passed though the transwell plate. Data are expressed as fold change from NA in the absence of IL-4 (n = 6). Error bars represent the SEM. Statistical significance was determined by paired Student t test or Wilcoxon matched pairs signed rank test.
The effect of IL-4 on CXCR4 expression. (A) Histogram showing CXCR4 expression by flow cytometry in a representative CLL sample following treatment in the presence or absence of 10 ng/mL IL-4 for 24 hours. Gray histogram, isotype control; solid black line, 24 hours with IL-4; black dotted line, start of the experiment; dashed line, 24 hours without IL-4. (B,C) Summary of results for all samples analyzed at times up to (B) 72 hours (n = 9) and at (C) 24 hours (n = 21). (D) CLL cells were pretreated with or without the indicated cytokines for 24 hours and CXCR4 expression (n = 9) quantified by flow cytometry. (E) CLL cells were pretreated for 1 hour with the JAK1/3 inhibitor (CP [CP-690550, tofacitinib]) or STAT6 inhibitor (AXON; AS1517499) before IL-4 addition and CXCR4 quantified by flow cytometry at 24 hours. (F) CLL samples (n = 6) were treated with IL-4 for 24 hours and migration to CXCL12 quantified by assessing the number of CD19+CD5+ CLL cells by flow cytometry that passed though the transwell plate. Data are expressed as fold change from NA in the absence of IL-4 (n = 6). Error bars represent the SEM. Statistical significance was determined by paired Student t test or Wilcoxon matched pairs signed rank test.
The modulatory effects of IL-4 were investigated on additional cell-surface markers including CD19 and the activation markers CD69, CD23, and CD71, which are generally overexpressed on CLL cells compared with normal donor B cells.50 Although CD19 and CD71 expression were unaltered, IL-4 increased expression of CD69 and CD23 (supplemental Figure 6A-D). These data indicate a considerable degree of selectivity among the receptor modulating effects of IL-4.
Discussion
CLL appears to be a BCR-driven malignancy and the expression of sIgM and subsequent downstream signalling are involved in disease behavior. Cases of U-CLL, a subset of poorer prognosis, generally express higher levels of sIgM and subsequently produce a stronger signal compared with M-CLL in response to anti-IgM.3,51 Clearly, there is a potential for microenvironmental factors to influence BCR signalling, and previous studies have shown that BAFF and CD40L promote sIgM-mediated signalling in CLL cells.52 In those studies, regulation appeared to involve an indirect potentiation of intracellular signal transduction via microRNA-155–mediated suppression of the inhibitory phosphatase SHIP1.52 By contrast, IL-4 appears to promote sIgM signalling, at least in part, via direct upregulation of IgM expression at the cell surface.
GSEA revealed a clear signature of IL-4–mediated transcription in the LN of CLL patients, compared with matched blood and bone marrow samples, indicating that IL-4 may be acting on the malignant cells in the LN tissue. The source of IL-4 remains unknown, but CD4+ T cells are detectable, and are likely to be involved.24,53 Follicular helper T (TFH) cells a known source of IL-4 are also increased in the blood of CLL patients and, although unambiguous identification in tissues remains difficult, T cells with features of TFH have been observed in CLL LNs.54 Interestingly, although circulating T cells from CLL patients have increased exhaustion markers and have impaired proliferation, cytotoxicity and synapse formation55,56 they retain their capacity for cytokine production.56
In terms of effects of IL-4 on CLL cells, we found that sIgM, but not sIgD expression, increased following IL-4 treatment in vitro. Because there was no detectable increase in total cellular IgM following IL-4 treatment, the increase in surface expression may be due to “shunting” of IgM from the ER to the cell surface. The relative increase in the fully N-glycosylated glycoform is consistent with this. Our data in CLL cells are similar to those described in mouse splenocytes where IL-4 promoted sIgM expression, leading to enhanced downstream signalling.31,32,57 In normal mouse splenocytes, there was co-amplification of CD79A and CD79B, consort molecules for sIgM expression,31 whereas in CLL, there was a clear accompanying increase in CD79B expression, which is consistent with the reports that CD79B limits BCR assembly in CLL.58 However, CD79A protein expression was more difficult to interpret because of the detection of multiple bands on the immunoblot caused by IL-4 induced glycosylation of this protein. It seems therefore that there may not be a defect in the ability of µ chains to undergo N-glycosylation and assembly with CD79A and CD79B in the ER of CLL cells.58 Instead, there appears to be regulation of the subsequent process of movement through the Golgi complex to the membrane, possibly via IL-4–mediated CD79B upregulation. In contrast, there was little effect of IL-4 on sIgD expression, consistent with the suggested reduced dependence on CD79A and CD79B.59
Expression in CLL cells of several other receptors (CD19, CD71) did not significantly change following IL-4 pretreatment; however, 2 key receptors, CXCR4 and CXCR5, which are involved in cell migration into the LNs were downregulated. In contrast, CXCR4 was upregulated in CD19−CD5+ cells (T cells). These results suggested a possible role by IL-4 in recruiting T cells to and retaining B cells within the LN. This is also consistent with previous reports in which CXCR4 is upregulated by IL-4 in normal peripheral and cord blood T cells.60 Importantly, these data indicated that in CLL the mechanism of IL-4 is both receptor- and cell type–dependent. The IL-4–mediated downregulation of CXCR4 expression and function by CLL cells mirrors the effect of BCR stimulation on this receptor,61,62 suggesting that these influences may be acting together to locate cells to extrafollicular sites63 where enhanced IgM expression then increases antigen engagement and subsequent downstream signalling events.
The effects of IL-4 appear to be common to all B cells, because both naive and memory B cells respond to IL-4 in a similar way to CLL cells; however, naive CD27− B cells responded to a greater extent than CD27+ memory B cells, indicating that the effect of IL-4 may be dependent on the cell of origin. Therefore the higher level of response to IL-4 in U-CLL cases may simply reflect its origin from a naive B cell.49 We propose that CLL cells may highjack the naturally occurring IL-4 signalling pathway to augment BCR signalling and promote B-cell proliferation and survival, especially within the U-CLL subset.
In addition to effects of IL-4 on sIgM expression and function, we have also shown that this cytokine decreases the inhibitory effects of idelalisib or ibrutinib on anti-IgM–induced signalling and protects against BCR kinase inhibitor induced apoptosis. The presence of significant numbers of CD4+ T cells in proliferation centers,53 accompanied by an IL-4 gene signature (shown previously), might lead to an increase in expression of sIgM in nonmobilized tumor cells and therefore could protect a subpopulation from kinase inhibition. The greater effect on U-CLL suggests that CLL cells with more responsive signalling via sIgM engagement6-8 may be more sensitive to IL-4. However, the clinical observation of a reduced effect of inhibitors on disease load in M-CLL might be due to the greater proportion of anergic cells, which respond poorly to BCR signals and to IL-4 and would therefore persist during therapy.51 Because IL-4 effects are mediated by the JAK3/STAT6 pathway, we are currently investigating whether JAK3 inhibition in combination with BCR kinase inhibitors is capable of promoting more durable remissions in vivo in the Eµ-TCL1 mouse model. These data clearly indicate that cytokines operating within the tumor microenvironment enhance BCR signalling, reduce sensitivity to BCR kinase inhibitors, and may have relevance for other B-cell malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients for supplying tissue.
The authors also thank Bloodwise (grants 10034, 10048, and 13036), CLL Global Research Foundation, and Worldwide Cancer Research for supporting this work, and the infrastructure and staff support from a Cancer Research UK center grant (C34999/A18087) and an Experimental Cancer Medicine Centers (ECMC) grant (C24563/A15581). Finally, the authors thank I. Tracy and I. Henderson for the biobanking and extensive characterization of the chronic lymphocytic leukemia samples used in this study (Bloodwise grant 12021, ECMC C24563/A15581).
Authorship
Contributions: M.M.A.-H., M.D.B., J.C.S., N.S., A.D., P.M.W.J., M.S.C., G.P., F.K.S., and A.J.S. designed research and wrote the manuscript. M.M.A.-H., M.D.B., R.D., L.D.S., A.Y., M.L., A.L., S.T., and A.J.S. performed research and analyzed data. F.F. obtained consent from patients, blood samples, and collated and analyzed clinical data. F.K.S., M.S.C., J.C.S., A.D., and G.P. provided critical review, and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew J. Steele, Cancer Sciences Unit, Somers Building (MP824), Southampton General Hospital, Tremona Rd, Southampton SO16 6YD, United Kingdom; e-mail: a.steele@soton.ac.uk.
References
Author notes
M.M.A.-H., M.D.B., and R.D. contributed equally to this study.

![Figure 2. IL-4 enhances expression of sIgM, but not sIgD, on CLL cells. Histogram of (A) sIgM and (D) sIgD expression on a representative CLL sample. Graph shows sIg expression at the start of the experiment (black dotted line) and at 24 hours in the presence (solid black line) or absence (dashed line) of IL-4 (10 ng/mL), respectively. Gray histogram, isotype control antibody. (B,E) Results (MFI minus isotype control) for cells treated with IL-4 or left untreated as a control (NA) for up to 72 hours. (C,F) All results expressed as MFI ratios from samples analyzed at 24 hours (C, n = 33; F, n = 29). (G-I) Correlation between the increase in sIgM [(sIgM MFI with IL-4) − (sIgM MFI without IL-4)] and prognostic markers. Samples are categorized according to (G) IGHV mutational status, (H) ZAP70 expression, and (I) CD38 expression. Error bars represent the standard error of the mean (SEM). Statistical significance was determined by either a paired (B,C,E,F) or unpaired (G-I) Student t test or Wilcoxon matched pairs signed rank test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/24/10.1182_blood-2015-11-682906/4/m_3015f2.jpeg?Expires=1769804308&Signature=oAogtodW~18Ji97AmSTsuxR1vKIwM9k40n3jBU0YEabCERStoj0E5FOCvItUv7-VOcvbAZrxwOcdpXupux1sQ1-9vsQUpmY681ZkRKjV5r4yOGzjraahFgAGzq8wcng9hZvkhsuo~7Ee0YBoKx8HTXmZHBvk6Y6NcrAhXjLLnIjtdgmfjUn0QaKFf4q63bnsyAOedGB68YqNkV3uepMMye4HcO23HdUc-8UpzUYgisOIKQKBIPqfo6evlqz15UB91MGlhqNtpg9buQhUh7wug1~C8mBdjaZ92BNdKL7oT~kEHSID1nL8yvf6GjtG7pybbVTbq-EQkgyVpWH6-kMsQQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. The effect of IL-4 on CXCR4 expression. (A) Histogram showing CXCR4 expression by flow cytometry in a representative CLL sample following treatment in the presence or absence of 10 ng/mL IL-4 for 24 hours. Gray histogram, isotype control; solid black line, 24 hours with IL-4; black dotted line, start of the experiment; dashed line, 24 hours without IL-4. (B,C) Summary of results for all samples analyzed at times up to (B) 72 hours (n = 9) and at (C) 24 hours (n = 21). (D) CLL cells were pretreated with or without the indicated cytokines for 24 hours and CXCR4 expression (n = 9) quantified by flow cytometry. (E) CLL cells were pretreated for 1 hour with the JAK1/3 inhibitor (CP [CP-690550, tofacitinib]) or STAT6 inhibitor (AXON; AS1517499) before IL-4 addition and CXCR4 quantified by flow cytometry at 24 hours. (F) CLL samples (n = 6) were treated with IL-4 for 24 hours and migration to CXCL12 quantified by assessing the number of CD19+CD5+ CLL cells by flow cytometry that passed though the transwell plate. Data are expressed as fold change from NA in the absence of IL-4 (n = 6). Error bars represent the SEM. Statistical significance was determined by paired Student t test or Wilcoxon matched pairs signed rank test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/24/10.1182_blood-2015-11-682906/4/m_3015f7.jpeg?Expires=1769804308&Signature=U~VHIs7kLyrDJbmHh7u-om6XAHuAlqjYgQLTAId6wJRhWxch1Yq8k5Q2RhHwE0fONyvvHHhe974yHIzovpM8ladj84DQl9qzTRCgop0TFrUZ7y4t-Pf7HQtrE1D589gqc1n5iXfmigskUm9jvIW-85yRMA3Ty84E-0T5TWjfD1jxkz9FESho9wsgoCfw1TsCc~eZsK7d700YbH8BcqjFo6X~7frP1NATWrKVuxe5WZTUDKBo2qZTBLe6Hamm~7dlqaJ~LH~5-cArOUlNzNWL2~57fJ65T~y8Q-8dlrg3y8ueMMJrqIir4rZBDap8wHyyKA-ILVQqu-xXis4EgWaarw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal