Key Points
This study is the first to show that genome-editing approaches can modify multilineage, long-term repopulating cells in a large animal model.
We demonstrate that the persistence of genome-edited hematopoietic stem cells can be tracked in vivo in a mutation-specific manner.
Abstract
Genome editing in hematopoietic stem and progenitor cells (HSPCs) is a promising novel technology for the treatment of many human diseases. Here, we evaluated whether the disruption of the C-C chemokine receptor 5 (CCR5) locus in pigtailed macaque HSPCs by zinc finger nucleases (ZFNs) was feasible. We show that macaque-specific CCR5 ZFNs efficiently induce CCR5 disruption at levels of up to 64% ex vivo, 40% in vivo early posttransplant, and 3% to 5% in long-term repopulating cells over 6 months following HSPC transplant. These genome-edited HSPCs support multilineage engraftment and generate progeny capable of trafficking to secondary tissues including the gut. Using deep sequencing technology, we show that these ZFNs are highly specific for the CCR5 locus in primary cells. Further, we have adapted our clonal tracking methodology to follow individual CCR5 mutant cells over time in vivo, reinforcing that CCR5 gene-edited HSPCs are capable of long-term engraftment. Together, these data demonstrate that genome-edited HSPCs engraft, and contribute to multilineage repopulation after autologous transplantation in a clinically relevant large animal model, an important step toward the development of stem cell-based genome-editing therapies for HIV and potentially other diseases as well.
Introduction
Genome editing represents the next generation of potential gene therapy-mediated treatments for human disease.1 The current paradigm centers on the use of integrating or non-integrating viral vectors to deliver a transgene of interest into a selected cell type; however, these methods must overcome potential concerns associated with insertional oncogenesis, and immune reactions that may impact safety and efficacy.2,3 In contrast, genome-editing strategies use transient expression of an engineered, site-specific endonuclease capable of inducing a DNA double-strand break. Resolution of the targeted DNA double-stand break via the nonhomologous end joining pathway is error-prone and can be used to generate targeted mutations, leading to a loss-of-function, or in some cases gain-of-function mutations in the disease-relevant targeted gene.4 Homologous recombination can also be exploited as a strategy to dictate the precise repair outcome at the nuclease-targeted locus.5 In both contexts, nuclease-based gene editing is advantageous over traditional viral vector-based methods because the genetic intervention targets the specific locus of interest.
A potential drawback of site-specific genome-editing techniques is that stable expression of a therapeutic transgene may have unintended proximal or distal effects on host gene expression. This limitation has been overcome by the identification and characterization of “Safe Harbor Loci,” where targeted gene insertion is least likely to impinge on endogenous transcriptional activity. The adeno-associated virus integration site 1 (AAVS1) locus is the best characterized of these loci.6-8 Recent studies suggest that the C-C chemokine receptor 5 (CCR5) locus, which has been extensively evaluated as a gene-editing target in HIV infection, may also function as a safe harbor locus, expanding its utility beyond the spectrum of infectious disease.9,10
Nonobese diabetic/severe combined immunodeficiency gamma (NSG)/interleukin-2rγnull and related immunodeficient mouse models can be used to model a broad array of human pathologies, including infectious diseases such as HIV-1.11 The mouse model, however, has limitations for the study of human stem cell transplants, with respect to CD34+ cell engraftment (and differentiation into all possible blood lineages), modeling the immunologic impacts of HIV infection, and in targeting of clinically relevant HIV reservoirs. In contrast, nonhuman primates (NHPs) allow rigorous evaluation of long-term engraftment of all blood lineages similar to the human setting, and have been used extensively in highly clinically relevant HIV/AIDS modeling.12 Indeed, we focused on the use of NHP models in gene therapy-based strategies to combat diverse human diseases (including HIV/AIDS), predominantly through the use of viral vector-mediated strategies.13-15 Here, we extend these studies to the evaluation of the safety and feasibility of genome-edited hematopoietic stem cells (HSCs) in NHPs. Specifically, we investigate the ability of zinc finger nucleases (ZFNs) to target and edit the CCR5 locus in CD34+ hematopoietic stem and progenitor cells (HSPCs) isolated from pigtailed macaques (Macaca nemestrina), and the ability of these cells to engraft and differentiate long-term following autologous HSC transplant.
Materials and methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (”The Guide”). The protocol was approved by the Institutional Animal Care and Use Committees of the Fred Hutchinson Cancer Research Center and University of Washington (Protocol #3235-03). For details of animal husbandry and care, see supplemental Methods, available on the Blood Web site.
Ex vivo HSPC engineering and autologous transplantation
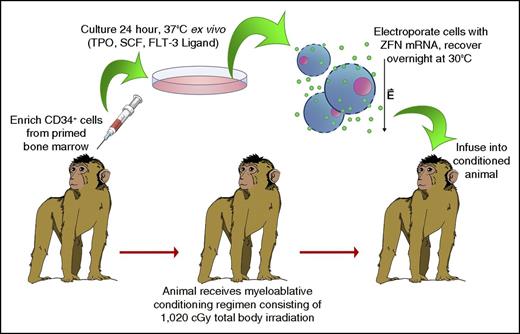
Autologous HSC transplants were conducted consistent with our previously published protocols and as described in Figure 1.16 CD34+ HSPCs were electroporated following enrichment from BM and overnight culture in Iscove's Modified Dulbecco's medium + 10% fetal bovine serum, 1% penicillin/streptomycin, and 100 ng/μL each of recombinant human stem cell factor, thrombopoietin, and FMS-like tyrosine kinase 3 ligand. ZFN messenger RNA (mRNA) (TriLink BioTechnologies) was added to cells resuspended to 1 × 107 cells/mL in Cytoporation Media T (Harvard Apparatus, Holliston, MA) at a final concentration of 125 μg/mL for each ZFN mRNA. Electroporation was conducted using an AgilePulse MAX machine and 5 mL, 6-mm gap width electroporation chambers (Harvard Apparatus), using a single 750V pulse, 0.8 milliseconds in duration. Following electroporation, cells were rested for 10 minutes, extracted from electroporation chambers, plated into fresh media, and recovered overnight in a 30°C, 3% CO2 incubator. The next day, cells were harvested, counted, resuspended to 5 × 106 cells/mL, and pulsed for 2 hours in 10 μM prostaglandin E2 on ice. Cells were then resuspended in Hanks balanced salt solution containing 2% autologous serum, and infused into the animal. During the 48-hour ex vivo HSPC culture period and prior to cell infusion, animals received a fractionated dose of 1020 cGy total body irradiation (TBI). See supplemental Methods for further details on sample collections and analyses following cell infusion.
Schematic of electroporation-based autologous transplant protocol. Primed bone marrow (BM) aspirates were collected from 4 pigtailed macaques, and CD34+ cells were isolated as described previously. Following overnight ex vivo culture, cells were electroporated in the presence of mRNA encoding CCR5-targeting ZFNs, and incubated overnight at 30°C. The next day, cells were collected, pulsed with prostaglandin E2, and infused into the autologous animal. During the 48-hour ex vivo culture period, the animal received a fractionated dose of 1020 cGy TBI. FLT-3, FMS-like tyrosine kinase 3; SCF, stem cell factor; TPO, thrombopoietin.
Schematic of electroporation-based autologous transplant protocol. Primed bone marrow (BM) aspirates were collected from 4 pigtailed macaques, and CD34+ cells were isolated as described previously. Following overnight ex vivo culture, cells were electroporated in the presence of mRNA encoding CCR5-targeting ZFNs, and incubated overnight at 30°C. The next day, cells were collected, pulsed with prostaglandin E2, and infused into the autologous animal. During the 48-hour ex vivo culture period, the animal received a fractionated dose of 1020 cGy TBI. FLT-3, FMS-like tyrosine kinase 3; SCF, stem cell factor; TPO, thrombopoietin.
ZFN assays
Cells were collected at 1, 2, and 5 days post-electroporation for analysis of ZFN protein expression and gene disruption. For western blots, 5 × 105 cells were washed once in phosphate-buffered saline, and were snap frozen as a dry pellet. Total cell lysates were made using RIPA Lysis Buffer with protease and phosphatase inhibitors (Santa Cruz Biotechnology, Santa Cruz, CA). Western blotting was performed essentially as described.17 Primary antibodies were against the ZFN FokI domain18 or Actin (Pierce, Rockford, IL), followed by secondary IRDye-conjugated antibodies, and imaging on an Odyssey Fluorescence Imager (LI-COR, Lincoln, NE). CCR5 mutation measurements were made using the Surveyor Nuclease Detection Kit (Transgenomic Inc, Omaha, NE) as described by the manufacturer. Polymerase chain reaction (PCR) was performed with Phusion Polymerase (Thermo Scientific, Waltham, MA). Macaque CCR5 primer sequences were rmC5_Cel_916F: 5′-ACTGTTTGCATTCATGGTGG-3′ and rmC5_Cel_1537R: 5′-GCTGCAGGTGTAATGAAGACC-3′.
Deep sequencing
To evaluate levels of both on- and off-target genome modifications, loci of interest were first recovered via PCR using the primers shown in supplemental Table 1. For off-target analysis of M nemestrina samples, genomic DNA (gDNA) was first subjected to a whole genome amplification using a REPLI-g Kit (Qiagen), then the amplified gDNA was PCR amplified using the adaptor primers (one-stage amplification) except for off-target sites #11 and #16, which were first amplified using the out-out primers as listed (two-stage amplification). The two-stage amplification protocol was also used for CCR5 amplification in all other experiments. These procedures yielded amplicons sufficiently small (<180 bp target-specific sequences) for complete sequencing using a barcoded, paired-end MiSeq protocol, essentially as described by the manufacturer (Illumina, San Diego, CA).
Off-target analyses
LLC-MK2 cells were electroporated with ZFN mRNA and recovered at 30°C or 37°C for 24 hours. Cells were then cultured at 37°C for 2 to 3 days and collected for gDNA purification, and subsequent MiSeq analysis to evaluate levels of genome modification at each potential off-target site (see supplemental Methods). M nemestrina CD34+ cells were similarly electroporated and recovered at 30°C, followed by transplantation into the autologous recipient or culture at 37°C. Cells were collected at indicated time points, and subjected to gDNA purification and MiSeq analysis to evaluate levels of genome modification. For each potential off-target site, the statistical test described by Guilinger et al19 was applied to the number of sequences scored as genetic insertions or deletions (indels) and the total number of sequences for both the ZFN-treated sample and the cognate mock-treated control. A Bonferroni-corrected P value of ≤.05 was considered as significant.
Results
ZFNs disrupt macaque CCR5 in CD34+ cells ex vivo
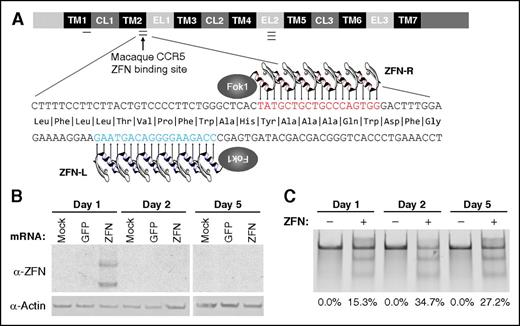
The experimental workflow for these autologous transplant experiments is shown in Figure 1. Our first goal was to demonstrate efficient targeting of macaque CCR5 by ZFNs in cultured CD34+ cells ex vivo. We collected BM and enriched CD34+ cells by bead-based positive selection from “steady state” animals that were not primed with granulocyte colony-stimulating factor or stem cell factor. Following overnight culture at 37°C, ZFNs targeting the 5′ region of the macaque CCR5 open reading frame (Figures 2 and 3A) were delivered via electroporation of ZFN mRNA. Cells were then incubated overnight at 30°C (Figure 1).20 By western blot, ZFN protein was evident 1 day after electroporation, but was absent at further time points, suggesting that this gene delivery strategy results in a short burst of nuclease expression (Figure 3B). Importantly, this transient window of expression resulted in robust cleavage at the targeted CCR5 locus (Figure 3C). These results suggest that our mRNA electroporation delivery strategy is capable of eliciting transient ZFN protein expression and significant disruption of the on-target CCR5 locus.
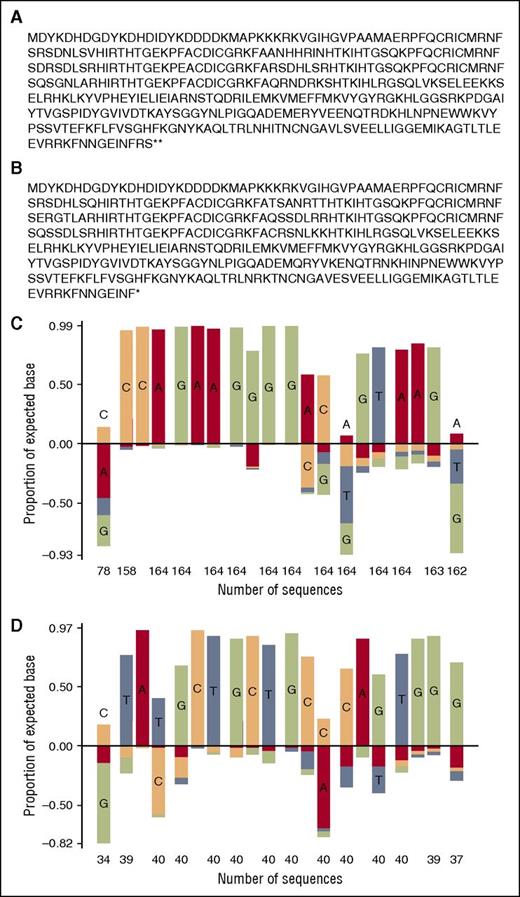
Amino acid sequence and target site preference for macaque CCR5 ZFNs. Full sequences of (A) ZFN-L and (B) ZFN-R are shown. A target site selection assay was performed on (C) ZFN-L and (D) ZFN-R, yielding the indicated position-frequency matrix plot. At each position in the plot, the frequency of the intended target base is shown as a positive value, whereas frequencies of unintended bases are plotted below the x-axis.
Amino acid sequence and target site preference for macaque CCR5 ZFNs. Full sequences of (A) ZFN-L and (B) ZFN-R are shown. A target site selection assay was performed on (C) ZFN-L and (D) ZFN-R, yielding the indicated position-frequency matrix plot. At each position in the plot, the frequency of the intended target base is shown as a positive value, whereas frequencies of unintended bases are plotted below the x-axis.
The CCR5 locus in macaque HSPCs is efficiently targeted by macaque-specific CCR5 (mCCR5) ZFNs ex vivo. (A) Schematic of the CCR5 locus showing TM, CL, and EL domains. Single, double, and triple bars indicate sites of a previously published human CCR5 ZFN target region, macaque CCR5 ZFN target region, and the human Δ32 mutation, respectively. The lower schematic shows the macaque CCR5 coding sequence, highlighting the target sites for the ZFN pair 32106 (ZFN-L) and 32118 (ZFN-R). (B) At the indicated times after electroporation of macaque CCR5 ZFN mRNA into macaque HSPCs, cells were collected and western blots were performed for ZFN and actin protein. (C) Representative Cel I nuclease assay demonstrating robust targeting of mCCR5 in HSPCs following delivery of mCCR5 ZFN mRNA. The percentage of disrupted mCCR5 alleles, as measured by densitometry, is indicated below each lane. CL, cytoplasmic loop; EL, extracellular loop; GFP, green fluorescent protein; Mock, control cells that were not electroporated; TM, transmembrane.
The CCR5 locus in macaque HSPCs is efficiently targeted by macaque-specific CCR5 (mCCR5) ZFNs ex vivo. (A) Schematic of the CCR5 locus showing TM, CL, and EL domains. Single, double, and triple bars indicate sites of a previously published human CCR5 ZFN target region, macaque CCR5 ZFN target region, and the human Δ32 mutation, respectively. The lower schematic shows the macaque CCR5 coding sequence, highlighting the target sites for the ZFN pair 32106 (ZFN-L) and 32118 (ZFN-R). (B) At the indicated times after electroporation of macaque CCR5 ZFN mRNA into macaque HSPCs, cells were collected and western blots were performed for ZFN and actin protein. (C) Representative Cel I nuclease assay demonstrating robust targeting of mCCR5 in HSPCs following delivery of mCCR5 ZFN mRNA. The percentage of disrupted mCCR5 alleles, as measured by densitometry, is indicated below each lane. CL, cytoplasmic loop; EL, extracellular loop; GFP, green fluorescent protein; Mock, control cells that were not electroporated; TM, transmembrane.
CCR5 ZFN transplants are feasible and safe
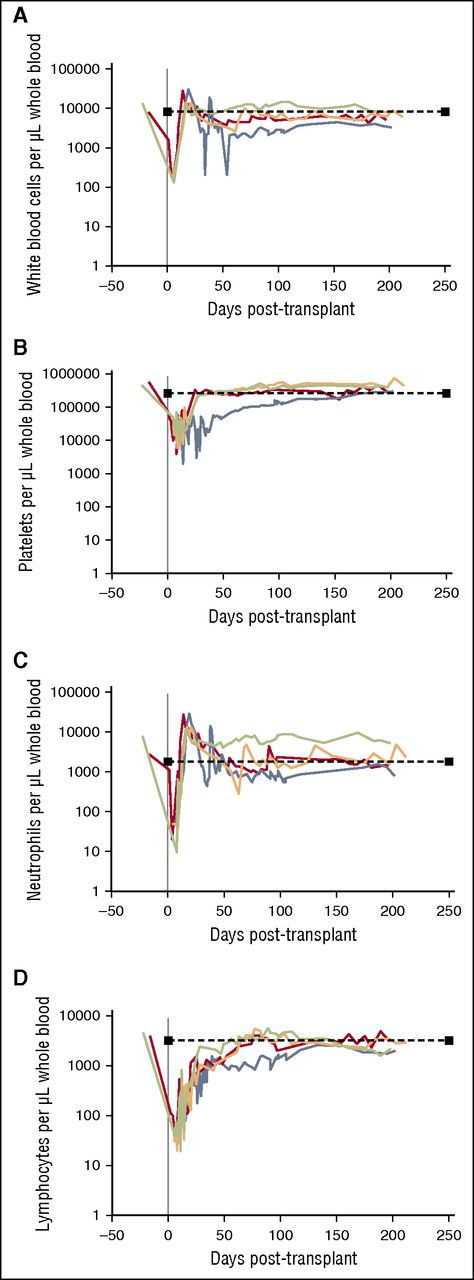
To evaluate the general safety and tolerability of CCR5-modified macaque CD34+ HSPCs in vivo, we transplanted 4 pigtailed macaques with autologous, ZFN mRNA-electroporated CD34+ HSPCs. Our protocol proceeded similarly, and cell infusion products were comparable to our previously published lentivirus-mediated gene therapy transplants (see “Materials and methods,” Figure 1, and supplemental Figure 1).16 At longitudinal time points immediately prior to and up to 200 days following conditioning and cell infusion, we monitored white blood cell (WBC), platelet, neutrophil, and lymphocyte recovery in each transplanted animal (Figure 4). Our conditioning regimen, consisting of 1020 cGy TBI, led to marked depletion of each of these counts in the days following cell infusion. Nadir was followed by a rebound of WBC and neutrophil counts within 3 weeks, with cell counts returning to within the normal range by 6 months posttransplant (Figure 4A,C). Platelet counts recovered within 4 weeks in 3 out of 4 animals, with animal ID A11210 requiring a longer period for recovery but still reaching normal levels within 200 days (Figure 4B). Finally, lymphocyte recovery, which should rebound relatively slowly, was observed within 10 to 11 weeks in 3 out of 4 animals; again, A11210 demonstrated a slower but sufficient recovery (Figure 4D). The kinetics of peripheral blood recovery in these animals was similar to animals transplanted with lentivirus-transduced HSPCs.21 We conclude that the infusion of autologous, CCR5 ZFN-electroporated HSPCs into macaques receiving myeloablative TBI is feasible and well tolerated.
Hematopoietic recovery following infusion of ZFN mRNA-electroporated HSPCs. At the indicated time points post-transplant, peripheral blood was drawn from animal IDs A11217 (red), A11210 (blue), Z12161 (orange), and Z12220 (green). Automated differential counts were used to quantify (A) WBCs, (B) platelets, (C) neutrophils, and (D) lymphocytes. Dashed lines represent normal values for each count.
Hematopoietic recovery following infusion of ZFN mRNA-electroporated HSPCs. At the indicated time points post-transplant, peripheral blood was drawn from animal IDs A11217 (red), A11210 (blue), Z12161 (orange), and Z12220 (green). Automated differential counts were used to quantify (A) WBCs, (B) platelets, (C) neutrophils, and (D) lymphocytes. Dashed lines represent normal values for each count.
CCR5 disruption is detected in ZFN-electroporated CD34+ cell infusion products
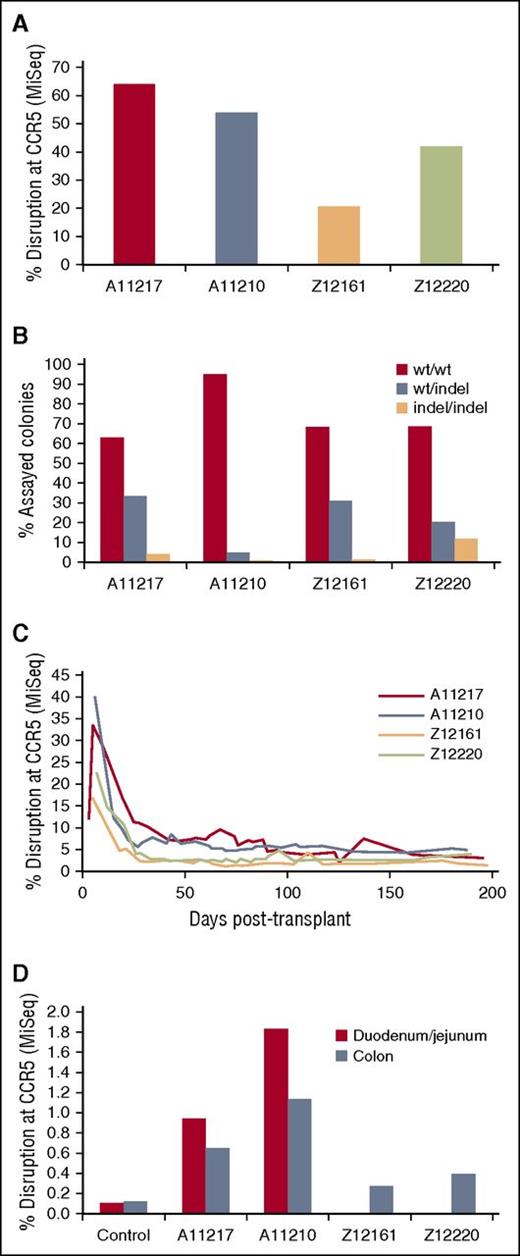
We next measured CCR5 disruption in the autologous CD34+ cell infusion product using Illumina MiSeq. Bulk CCR5 disruption in ex vivo cultured CD34+ cells was maintained over a 5-day culture period, suggesting that no overt toxicity was associated with this approach (data not shown). Peak disruption for each of the 4 transplanted animals ex vivo ranged between 20.77% and 64.03% of sequenced CCR5 alleles (Figure 5A). To address the frequency of monoallelic vs biallelic CCR5 targeting by our ZFNs, we performed colony-forming assays on the ZFN-treated HSPCs. Because individual colonies should arise from a single cell, we reasoned that deep sequencing of single colonies would allow us to classify wild-type (WT) and indel genotypes from each colony as WT/WT, WT/indel, or indel/indel. We detected indel/indel colonies in between 0.54% and 11.84% of the sequenced colonies from each animal (Figure 5B). Together, these data demonstrate that electroporation of ZFN-encoding mRNA into CD34+ HSPCs is capable of inducing indels in greater than two-thirds of all CCR5 alleles in the bulk population, resulting in up to 11% biallelic modification.
Efficient disruption of CCR5 ex vivo and detection of CCR5-modified cells in vivo. (A-B) A small aliquot of electroporated HSPCs from 4 animals transplanted with mCCR5 ZFNs was maintained ex vivo to quantify bulk and biallelic disruption of CCR5. (A) Total gDNA isolated from ex vivo cultured HSPCs; displayed is the peak disruption during a 5-day liquid culture time course. (B) Biallelic disruption was measured in colony forming assays as described in “Materials and methods.” (C) Peripheral blood was collected from each animal at the indicated time points post-transplant, and gDNA was isolated from hemolysed total leukocytes. (D) GI biopsies were collected from the indicated animals at ∼190 days post-transplant and total gDNA was isolated. Duodenum/Jejunum biopsies were not collected from Z12161 and Z12220 due to insufficient size of the animals. CCR5 disruption was measured from each gDNA sample by Illumina MiSeq. “Control” are representative values from 1 of 3 CCR5 WT animals from which GI tissues were collected following transplant with lentivirus-transduced cells. All values are significantly increased relative to the controls at the P < .05 level of significance (Bonferroni-corrected).
Efficient disruption of CCR5 ex vivo and detection of CCR5-modified cells in vivo. (A-B) A small aliquot of electroporated HSPCs from 4 animals transplanted with mCCR5 ZFNs was maintained ex vivo to quantify bulk and biallelic disruption of CCR5. (A) Total gDNA isolated from ex vivo cultured HSPCs; displayed is the peak disruption during a 5-day liquid culture time course. (B) Biallelic disruption was measured in colony forming assays as described in “Materials and methods.” (C) Peripheral blood was collected from each animal at the indicated time points post-transplant, and gDNA was isolated from hemolysed total leukocytes. (D) GI biopsies were collected from the indicated animals at ∼190 days post-transplant and total gDNA was isolated. Duodenum/Jejunum biopsies were not collected from Z12161 and Z12220 due to insufficient size of the animals. CCR5 disruption was measured from each gDNA sample by Illumina MiSeq. “Control” are representative values from 1 of 3 CCR5 WT animals from which GI tissues were collected following transplant with lentivirus-transduced cells. All values are significantly increased relative to the controls at the P < .05 level of significance (Bonferroni-corrected).
CCR5-disrupted CD34+ cells support long-term engraftment in blood and tissues
To measure the persistence of the modified CD34+ cells in vivo following cell infusion, we longitudinally monitored CCR5 disruption in peripheral blood from the 4 animals transplanted with ZFN-electroporated cells. Peripheral blood samples were collected 1 to 2 times per week, and gDNA was isolated from total leukocytes for sequencing analysis by Illumina MiSeq. We observed high levels of gene disruption in vivo during the first month following cell infusion, likely due to the co-infusion of CCR5-targeted progenitor and other short-term repopulating cells (Figure 5C). These early engraftment kinetics were nearly identical to lentivirus-transplanted (CCR5 WT) animals.21 Between days 40 and 200 posttransplant, levels of CCR5 disruption in peripheral blood stabilized at ∼3% to 5% in each animal, relative to an average 0.1% background level in lentivirus-transplanted (CCR5 WT) animals (data not shown). The persistence of CCR5-disrupted cells over more than 6 months indicates that gene-targeted HSPCs are capable of long-term engraftment in vivo. We further inquired whether CCR5-targeted cells homed to secondary lymphoid organs such as the gastrointestinal (GI) tract. Approximately 6 months posttransplant, each animal underwent colonic (lower GI) biopsies. Animal IDs A11217 and A11210 also underwent duodenal/jejunal (upper GI) biopsies; this material could not be collected from animal IDs Z12161 and Z12220 due to insufficient size. Following enzymatic dissociation of biopsy material, gDNA was prepared from single cell suspensions and CCR5 disruption was measured by MiSeq (Figure 5D). CCR5 disruption in all samples was significantly higher than background levels measured in samples from lentivirus-transplanted (CCR5 WT) control animals (P < .05 Bonferroni-corrected19 ). This finding is consistent with the ability of HSPC-derived cells to home to relevant secondary lymphoid organs, supporting the conclusion that CCR5-disrupted HSPCs are long lived and give rise to progeny which traffic to the GI tract.
Macaque CCR5 ZFNs are highly specific
A SELEX-based target site selection assay performed with the DNA binding domains of our mCCR5 ZFNs demonstrated selectivity for their intended targets (Figure 2). To experimentally determine the relative amount of gene disruption at off-target loci relative to CCR5, we quantified mutations both at CCR5 and at a set of candidate off-target sites chosen for their similarity to the consensus DNA binding sites for the ZFNs, using an Illumina MiSeq deep sequencing approach (supplemental Table 1). We first evaluated the specificity of the ZFNs in a macaque epithelial cell line, LLC-MK2. Gene disruption at CCR5 reached ∼28% when cells were recovered at 30°C, consistent with previous observations (supplemental Table 2).20 Two candidate off-target sites demonstrated modification above the background established in mock-electroporated LLC-MK2 cells: an uncharacterized locus on chromosome 10 and the CXCR3 locus on the X chromosome.
Next, we extended our ZFN off-target analyses to the relevant primary cells, CD34+ HSPCs. We evaluated the same panel of putative off-target loci in (1) ex vivo cultured HSPCs from the cell infusion product of 1 transplanted animal, and (2) posttransplant peripheral blood total leukocytes collected 5, 20, and 196 days following cell infusion (Table 1). Within the sensitivity of our assays, we did not observe disruption in these samples at any interrogated off-target locus (including the sites identified in LLC-MK2 cells). These data suggest that the ZFNs used here are highly specific for the CCR5 locus in primary cells.
Macaque CCR5 ZFN off-target activity in vitro and in vivo following delivery into HSPCs and infusion
| OT # . | Database . | Chrom . | Location . | Gene . | Exon/Intron . | ZFNBS . | In vitro 5-7 d GFP . | In vitro 5-7 d ZFN . | In vivo 6-11 d ZFN . | In vivo 189-198 d ZFN . |
|---|---|---|---|---|---|---|---|---|---|---|
| Intended | rheMac2 | chr2 | 90143599 | CCR5 | Exon | L_6_R | 0.11 ± 0.02 | 52.76 ± 9.29* | 33.55 ± 7.69* | 3.65 ± 0.39* |
| 1 | rheMac2 | chrX | 70535298 | CXCR3 | Exon | L_6_R | 0.03 ± 0.02 | 0.05 ± 0.02 | 0.47 ± 0.71 | 0.04 ± 0.03 |
| 2 | rheMac2 | chr14 | 74379519 | WNT11 | Intron | R_5_L | 0.03 ± 0.00 | 0.03 ± 0.01 | 0.02 ± 0.00 | 0.02 ± 0.01 |
| 3 | rheMac2 | chr10 | 30952397 | — | Intergenic | L_5_R | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.01 ± 0.00 |
| 4 | rheMac3 | chr20 | 2291152 | — | Unknown | R_6_L | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.01 ± 0.01 |
| 5 | rheMac2 | chr4 | 46918712 | — | Intergenic | L_5_R | 0.02 ± 0.00 | 0.01 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.00 |
| 6 | rheMac2 | chr15 | 2710971 | — | Intergenic | L_6_R | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.01 |
| 7 | rheMac2 | chr13 | 58922755 | — | Intergenic | R_5_L | 0.01 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| 8 | rheMac2 | chr10 | 32119972 | C20ORF112 | Intron | L_5_R | 0.05 ± 0.01 | 0.05 ± 0.00 | 0.06 ± 0.01 | 0.04 ± 0.01 |
| 9 | rheMac3 | chr10 | 94067013 | — | Unknown | R_6_L | 0.02 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.00 |
| 10 | rheMac2 | chr11 | 61360349 | C12orf56 | Intron | R_6_L | 0.07 ± 0.02 | 0.09 ± 0.02 | 0.08 ± 0.02 | 0.08 ± 0.01 |
| 11 | rheMac2 | chr9 | 79520402 | — | Intergenic | R_5_L | 0.28 ± 0.03 | 0.26 ± 0.08 | 0.30 ± 0.04 | 0.27 ± 0.02 |
| 12 | rheMac2 | chr8 | 109331649 | — | Intergenic | R_6_L | 0.04 ± 0.01 | 0.05 ± 0.03 | 0.07 ± 0.03 | 0.05 ± 0.00 |
| 13 | rheMac2 | chr8 | 126618925 | FER1L6 | Intron | R_5_L | 0.02 ± 0.01 | 0.01 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.00 |
| 14 | rheMac2 | chr16 | 39404073 | STXBP4 | Intron | L_5_R | 0.11 ± 0.03 | 0.17 ± 0.00 | 0.10 ± 0.01 | 0.08 ± 0.01 |
| 15 | rheMac2 | chr19 | 15545514 | — | Intergenic | L_5_R | 0.06 ± 0.01 | 0.05 ± 0.00 | 0.05 ± 0.02 | 0.05 ± 0.02 |
| 16 | rheMac3 | chr10 | 39382414 | — | Unknown | R_5_L | 0.02 ± 0.00 | 0.03 ± 0.02 | 0.02 ± 0.00 | 0.01 ± 0.00 |
| 17 | rheMac3 | chr19 | 13168773 | — | Unknown | L_5_R | 0.07 ± 0.03 | 0.40 ± 0.46 | 0.13 ± 0.04 | 0.23 ± 0.10 |
| 18 | rheMac2 | chr11 | 2692851 | CACNA1C | Intron | L_6_R | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.00 |
| OT # . | Database . | Chrom . | Location . | Gene . | Exon/Intron . | ZFNBS . | In vitro 5-7 d GFP . | In vitro 5-7 d ZFN . | In vivo 6-11 d ZFN . | In vivo 189-198 d ZFN . |
|---|---|---|---|---|---|---|---|---|---|---|
| Intended | rheMac2 | chr2 | 90143599 | CCR5 | Exon | L_6_R | 0.11 ± 0.02 | 52.76 ± 9.29* | 33.55 ± 7.69* | 3.65 ± 0.39* |
| 1 | rheMac2 | chrX | 70535298 | CXCR3 | Exon | L_6_R | 0.03 ± 0.02 | 0.05 ± 0.02 | 0.47 ± 0.71 | 0.04 ± 0.03 |
| 2 | rheMac2 | chr14 | 74379519 | WNT11 | Intron | R_5_L | 0.03 ± 0.00 | 0.03 ± 0.01 | 0.02 ± 0.00 | 0.02 ± 0.01 |
| 3 | rheMac2 | chr10 | 30952397 | — | Intergenic | L_5_R | 0.00 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.01 ± 0.00 |
| 4 | rheMac3 | chr20 | 2291152 | — | Unknown | R_6_L | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.01 ± 0.01 |
| 5 | rheMac2 | chr4 | 46918712 | — | Intergenic | L_5_R | 0.02 ± 0.00 | 0.01 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.00 |
| 6 | rheMac2 | chr15 | 2710971 | — | Intergenic | L_6_R | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.01 |
| 7 | rheMac2 | chr13 | 58922755 | — | Intergenic | R_5_L | 0.01 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.01 |
| 8 | rheMac2 | chr10 | 32119972 | C20ORF112 | Intron | L_5_R | 0.05 ± 0.01 | 0.05 ± 0.00 | 0.06 ± 0.01 | 0.04 ± 0.01 |
| 9 | rheMac3 | chr10 | 94067013 | — | Unknown | R_6_L | 0.02 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.00 |
| 10 | rheMac2 | chr11 | 61360349 | C12orf56 | Intron | R_6_L | 0.07 ± 0.02 | 0.09 ± 0.02 | 0.08 ± 0.02 | 0.08 ± 0.01 |
| 11 | rheMac2 | chr9 | 79520402 | — | Intergenic | R_5_L | 0.28 ± 0.03 | 0.26 ± 0.08 | 0.30 ± 0.04 | 0.27 ± 0.02 |
| 12 | rheMac2 | chr8 | 109331649 | — | Intergenic | R_6_L | 0.04 ± 0.01 | 0.05 ± 0.03 | 0.07 ± 0.03 | 0.05 ± 0.00 |
| 13 | rheMac2 | chr8 | 126618925 | FER1L6 | Intron | R_5_L | 0.02 ± 0.01 | 0.01 ± 0.00 | 0.02 ± 0.01 | 0.02 ± 0.00 |
| 14 | rheMac2 | chr16 | 39404073 | STXBP4 | Intron | L_5_R | 0.11 ± 0.03 | 0.17 ± 0.00 | 0.10 ± 0.01 | 0.08 ± 0.01 |
| 15 | rheMac2 | chr19 | 15545514 | — | Intergenic | L_5_R | 0.06 ± 0.01 | 0.05 ± 0.00 | 0.05 ± 0.02 | 0.05 ± 0.02 |
| 16 | rheMac3 | chr10 | 39382414 | — | Unknown | R_5_L | 0.02 ± 0.00 | 0.03 ± 0.02 | 0.02 ± 0.00 | 0.01 ± 0.00 |
| 17 | rheMac3 | chr19 | 13168773 | — | Unknown | L_5_R | 0.07 ± 0.03 | 0.40 ± 0.46 | 0.13 ± 0.04 | 0.23 ± 0.10 |
| 18 | rheMac2 | chr11 | 2692851 | CACNA1C | Intron | L_6_R | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.00 |
M nemestrina CD34+ cells were BTX electroporated with the mCCR5 ZFN (32106:32118) mRNA followed by a 30°C “cold shock” to increase ZFN activity. Cells were then transplanted back into the M nemestrina (in vivo) or cultured at 37°C (in vitro) for 5 days. Cells were collected at indicated time points and subjected to gDNA purification and sequencing analysis (MiSeq) to evaluate genome modification (% indels) at each of the potential target sites indicated. Data from 3 independent experiments were combined (n = 3) except the “in vitro, 5 to 7 days, ZFN” data set (n = 2). Shown are average ± standard deviation. ZFN-dependent disruption at the on-target CCR5 locus was significantly increased relative to GFP mRNA controls (*P ≤ .05). No off-target values were significantly different from the GFP mRNA control.
Chrom, chromosome; ZFNBS, zinc-finger nuclease binding site.
CCR5 ZFN-edited macaque HSPCs support multilineage engraftment
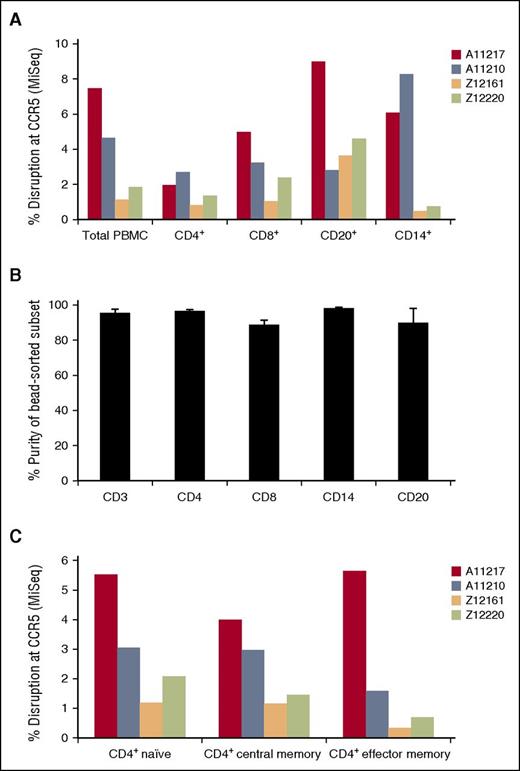
In addition to long-term engraftment of our gene-edited HSPCs, we asked whether these cells were capable of differentiating into CCR5-modified myeloid and lymphoid lineage cells. Following transplant and hematopoietic recovery in our cohort of 4 transplanted animals, we collected large-volume blood draws, and used bead-based positive selection to sort for CD4+, CD8+, CD20+, and CD14+ cells. We detected CCR5 disrupted cells in each subset, confirming that our gene-edited HSPCs supported multilineage engraftment (Figure 6A). Purities of each bead-sorted population were >90%, arguing against the contribution of contaminating lineages to these marking levels (Figure 6B). Moreover, a detailed analysis of the CD4+ T-cell subsets revealed gene disruption at CCR5 in both naïve and memory T cells (Figure 6C). Taken together, these data demonstrate CCR5 disruption across all assayed cell types, further suggesting modification of long-term repopulating HSPCs.
CCR5 modification is detected in lymphoid and myeloid lineages, including naïve and memory CD4+ T cells. Following hematopoietic recovery, large-volume peripheral blood draws were collected from 4 transplanted animals, and the indicated lymphoid and myeloid subsets were enriched. (A) Bead-based positive selection was used to isolate CD4+, CD8+, CD20+, and CD14+ subsets. CCR5 disruption in each subset was measured by Illumina MiSeq. (B) Purities of bead-separated subsets from (A) were measured by flow cytometry. Shown are the average and standard deviation for 3 out of 4 animals (an insufficient number of cells were available for this analysis from animal ID Z12161). (C) Fluorescence-activated cell sorting was used to isolate naïve, central memory, and effector memory CD4+ T-cell subsets, followed by quantification of CCR5 disruption as in (A). PBMC, peripheral blood mononuclear cells.
CCR5 modification is detected in lymphoid and myeloid lineages, including naïve and memory CD4+ T cells. Following hematopoietic recovery, large-volume peripheral blood draws were collected from 4 transplanted animals, and the indicated lymphoid and myeloid subsets were enriched. (A) Bead-based positive selection was used to isolate CD4+, CD8+, CD20+, and CD14+ subsets. CCR5 disruption in each subset was measured by Illumina MiSeq. (B) Purities of bead-separated subsets from (A) were measured by flow cytometry. Shown are the average and standard deviation for 3 out of 4 animals (an insufficient number of cells were available for this analysis from animal ID Z12161). (C) Fluorescence-activated cell sorting was used to isolate naïve, central memory, and effector memory CD4+ T-cell subsets, followed by quantification of CCR5 disruption as in (A). PBMC, peripheral blood mononuclear cells.
Mutation tracking suggests that ZFN-edited clones persist long-term
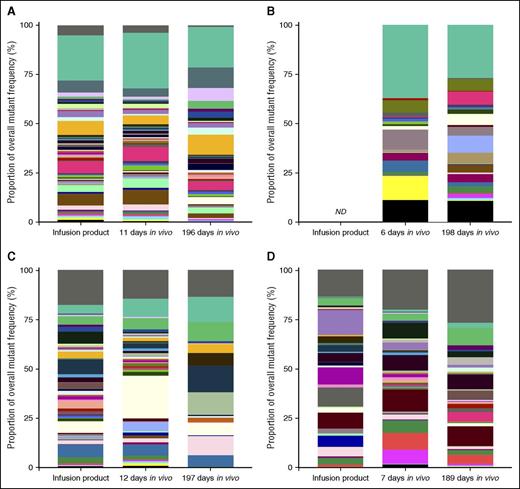
Finally, we leveraged our expertise in tracking of lentivirus integration sites in transduced CD34+ HSPCs22-24 to analyze the kinetics of individual CCR5 mutant cells over time. We reasoned that as individual CCR5 mutation sequences increased in complexity, they were more likely to represent unique gene-edited clones that could be followed in an analogous manner to a unique lentivirus integration site. Our mutant tracking bioinformatics pipeline (supplemental Methods) generated data that were consistent with the kinetics of total CCR5 disruption (Figure 5C; supplemental Figure 2A), and allowed us to calculate background levels of 1 bp, 2 bp, and >2 bp mutations at the CCR5 locus (supplemental Figure 2B). This facilitated analyses of significance of each mutation in ZFN-treated samples. Approximately two-thirds of mutations that were significantly enriched above background in our animals encoded frameshift mutations (supplemental Table 3). We quantified the number of mutants in each of the 4 transplanted animals: 89 to 340 mutations were found in the infusion product at levels significantly above background, 117 to 305 in the short-term in vivo time point, and 25 to 83 in the long-term in vivo time point (supplemental Table 3). Between 48.72% and 78.03% of the mutations were found in the infusion product and short-term engraftment time points from each animal, and between 69.23% and 97.59% of the mutations found in the long-term engraftment time point were found in at least one of the earlier time points (supplemental Table 3; Figure 7 and supplemental Figures 3 and 4). These data are consistent with our model that although a proportion of gene-edited cells are lost over time, a diverse population of gene-edited clones persists long-term in transplanted animals.
CCR5 mutations induced in CD34+ HSPCs ex vivo are detected in multiple post-transplant time points in vivo. Total gDNA was collected from ex vivo cultured CD34+ HSPCs from animal IDs A11217 (A), A11210 (B), Z12161 (C), and Z12220 (D), 5 to 6 days following electroporation (“infusion product”), or from total leukocytes collected from each animal 6 to 12 and 189 to 198 days following transplantation of these cells. CCR5 disruption was measured from total gDNA by Illumina MiSeq and the presence of the indicated mutants was tracked over time in each animal. The 50 most frequent mutants that were detected in more than one time point in a given animal (“recurrent mutants”) are shown in colored bars. Frequency of each mutant from the indicated animal/time point is displayed as a proportion of the total pool of mutants that were detected. In each graph, the top bars (dark gray) represent the sum of mutants that were detected in only one time point (“non-recurrent mutants”), and the second bars (teal) represent the sum of recurrent mutants that were not among the 50 most frequent. Single base pair point mutations were excluded from the analysis and infusion product from A11210 was not available for analysis. ND, not determined.
CCR5 mutations induced in CD34+ HSPCs ex vivo are detected in multiple post-transplant time points in vivo. Total gDNA was collected from ex vivo cultured CD34+ HSPCs from animal IDs A11217 (A), A11210 (B), Z12161 (C), and Z12220 (D), 5 to 6 days following electroporation (“infusion product”), or from total leukocytes collected from each animal 6 to 12 and 189 to 198 days following transplantation of these cells. CCR5 disruption was measured from total gDNA by Illumina MiSeq and the presence of the indicated mutants was tracked over time in each animal. The 50 most frequent mutants that were detected in more than one time point in a given animal (“recurrent mutants”) are shown in colored bars. Frequency of each mutant from the indicated animal/time point is displayed as a proportion of the total pool of mutants that were detected. In each graph, the top bars (dark gray) represent the sum of mutants that were detected in only one time point (“non-recurrent mutants”), and the second bars (teal) represent the sum of recurrent mutants that were not among the 50 most frequent. Single base pair point mutations were excluded from the analysis and infusion product from A11210 was not available for analysis. ND, not determined.
Discussion
ZFNs are the most extensively studied genome-editing platform in clinical trials, and have exhibited efficient delivery into CD4+ T cells, robust targeting of CCR5, and safety following infusion into autologous HIV+ patients.25 Here, we demonstrate that mRNA-encoding ZFNs that target macaque CCR5 can be efficiently delivered to macaque HSPCs ex vivo. Bulk CCR5 disruption rates of up to 64% are observed in targeted HSPCs, and colony forming assays show that up to 1 in 10 cells carries two disrupted alleles of CCR5. Following conditioning, these cells engraft into autologous animals, and are detectable in blood and tissues over 6 months after transplant, with no adverse hematologic phenomena observed. Off-target analyses suggest that these ZFNs are highly specific for cleavage at CCR5. Finally, we show that ZFN-modified cells are capable of long-term, multilineage engraftment. Our novel methodology to track gene-edited cells over time in vivo demonstrates that our approach induces hundreds of unique CCR5 mutants, a significant proportion of which persist over 6 months following transplantation.
As many as two-thirds of bulk CCR5 alleles were mutated in our CD34+ cell infusion products. In contrast to published mouse studies, we were able to test stem cell editing in a more clinically relevant setting using autologous stem cells, and studying multilineage and long-term repopulation.18 The high initial engraftment we observe, up to 40% in total peripheral blood leukocytes, is an unprecedented observation for gene-edited HSPCs in a large animal model. We did observe a decrease to ∼3% to 5% stable gene-edited cells at further time points post-transplant. Although we predict that the range of biallelic disruption in our infusion product (0.54% to 11.84%) would proportionally decrease in long-term engrafting HSCs in vivo, it is important to note that long-term HSCs carrying one disrupted allele of CCR5 may also provide clinical benefits.26 We are currently investigating the mechanism underlying the decline in the percentage of gene-edited NHP cells in vivo, which has been previously addressed in human cells.27,28 We believe it is a feature of the experimental conditions, for two reasons.29,30 First, we observed a similar magnitude reduction in the levels of marked cells in the peripheral blood immediately following transplant of NHPs with retrovirus-transduced HSPCs.31 Declines in the number of long-term engrafting, lentivirus-transduced HSCs were avoided in more recent studies when we used optimized gene transfer conditions for NHP HSPCs.16,30 Second, when modified human HSPCs are transplanted into NSG mice, only moderate decreases in gene-edited cells are observed32,33 (and data not shown), further supporting our hypothesis that the decline observed in our NHP studies likely represents differences between human and macaque CD34+ cell culturing and/or electroporation conditions. Previous findings from our group demonstrate that levels of lentivirus gene-marked cells stabilize at 4 to 6 months following infusion into autologous macaques, consistent with long-term engraftment.30,34 Compared with a recent cohort of animals transplanted with lentivirus-transduced cells,21 we did not observe acute toxicity or delayed hematopoietic reconstitution in our ZFN gene-editing cohort. Nevertheless, we cannot exclude the alternative model that our electroporation-based gene-delivery strategy may have impaired the long-term engraftment potential of a subset of infused cells. However, our data emphasize that when marking levels stabilized at 4 to 6 months, these levels tended to remain stable in multiple lineages, indicating long-term engraftment of modified, repopulating cells.
Our previously published methodology for tracking the unique integration site profiles of lentivirus-transduced cells was easily adapted to track CCR5 mutations induced by ZFN-dependent gene editing. We found that the diversity of unique mutations correlated with total levels of CCR5 mutation, when comparing the cell infusion product to the respective early and late post-transplant in vivo time points from each animal. These results suggest that the lower levels of long-term engrafting cells we detected were derived not from the residual output of a few long-lived progenitors, but rather from a smaller but still quite diverse pool of long-lived HSCs.
Deep sequencing data demonstrate that our macaque CCR5 ZFNs are highly specific for the on-target locus, an essential finding for the clinical translatability of this work. Past reports utilizing a ZFN pair targeting the 5′ region of human CCR5 detected low-level off-target cleavage at the CCR2 locus in primary cells.32,35 By contrast, our macaque CCR5 ZFN pair did not display any statistically significant off-target events in primary cells. Our improved methodology for evaluating the fidelity of ZFNs in silico (Figure 2), as well as the distinct target site used for macaque CCR5 vs human CCR5 (Figure 3), are the most likely reasons for our improved on:off target ratio. Interestingly, we observe increased off-target effects in a cultured cell line, in our case the rhesus macaque epithelial cell line LLC-MK2, relative to primary cells from our transplanted animals. These findings are consistent with recent reports suggesting that targeted nucleases’ accessibility to DNA is likely a function of many cell-type–specific parameters (eg, the chromatin structure at a given locus).36 In summation, our deep sequencing strategy allows us to accurately measure CCR5 disruption with increased sensitivity, and quantify safety and specificity of our ZFNs via examination of off-target mutation events.
A crucial finding from our data is that ZFN-driven CCR5 genome editing of HSPCs supports multilineage engraftment, suggesting that neither electroporation-dependent delivery of ZFN mRNA nor the genetic disruption of CCR5, impairs the ability of cells to differentiate into myeloid or lymphoid lineages. This finding has significant implications for the field of HSPC gene therapy/editing. There are many genetic disorders affecting the hematopoietic system that are currently being treated with HSPC gene therapy, including Wiskott–Aldrich syndrome,37 severe combined immunodeficiency,38 adenosine deaminase deficiency,39 and chronic granulomatous disease.40 Furthermore, HSPC gene therapy can contribute to therapeutic benefit in diseases of the peripheral and central nervous system through modification of glial cells (eg, in lysosomal storage diseases like adrenoleukodystrophy41 and metachromatic leukodystrophy42 ). Our NHP model has the potential to be useful in the study of a broad range of diseases in which gene-edited HSPCs might offer therapeutic benefit. For example, chronic granulomatous disease is representative of a class of disorders where targeted gene-editing approaches may be particularly warranted, due to the risk of leukemia associated with current viral vector-mediated gene therapy.43
The data presented here are particularly applicable to models of combination antiretroviral therapy (cART)-suppressed HIV infection, for which the NHP setting is uniquely suited.44 An important aspect of macaque models that distinguishes them from mouse xenografts is the ability to demonstrate infection resistance in multiple cell types, such as T cells, monocytes, dendritic cells, etc, and in relevant tissues that constitute viral reservoirs (sites that harbor replication competent provirus during suppressive cART). In the context of HIV eradication strategies, it will be essential to target infection-resistant cells to long-lived viral reservoirs in blood and secondary sites such as gut-associated lymphoid tissue, a well characterized reservoir for infected cells in patients on cART.45 We have observed engraftment of CCR5 gene-edited memory and naïve CD4+ T cells, and trafficking of HSPC-derived, CCR5-disrupted cells to the duodenum/jejunum and colon of our transplanted animals. These results suggest that CCR5 mutation is induced in infection-susceptible cells, and does not impair the ability of the cells to home to secondary lymphoid organs, consistent with previous experiments in NSG mice.32,46 Therefore, CCR5-edited NHPs can be directly applied to preclinically relevant models of HIV reservoir eradication.
In conclusion, we demonstrate for the first time the ability to model a targeted genome-editing approach for HSPCs in a clinically relevant large animal model. The maintenance of significant levels of multilineage gene-edited repopulating cells in our pigtailed macaque HSPC model has important implications for the field of HSPC gene therapy and genome editing. Our model more closely reflects the human clinical autologous transplant setting, and our results provide the basis to apply this model to evaluate genome-editing approaches for HIV and other human diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Grace Choi for help in preparing this manuscript and for creating the Figure 1 illustration; Veronica Nelson, Erica Curry, and Kelvin Sze for excellent support in our pigtailed macaque studies; Andrea Repetto, Willi Obenza, John McNevin, Cristina McAllister, and Sowmya Reddy for processing of macaque samples; Patrick Li, Emily Killingbeck, and Priscilla Cheung for Illumina MiSeq data analysis; Dmitry Guschin for SELEX studies; David Paschon and Xiangdong Meng for ZFN design; Sarah Hinkley, Anna Vincent, and Stephen Lam for ZFN gene assembly; Andreas Reik for initial ZFN testing; and Leon Flannery and Joel Ahrens for conducting GI biopsy procedures.
This study was supported by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (U19 AI096111 and R01 AI080326) and National Heart, Lung, and Blood Institute (R01 HL116217 and U19 HL129902). H.-P.K. is a Markey Molecular Medicine Investigator and received support as the inaugural recipient of the José Carreras/E. Donnall Thomas Endowed Chair for Cancer Research.
Authorship
Contribution: H.-P.K. is the principal investigator and designed and supervised the studies; C.W.P. designed and coordinated the experiments and optimized the ZFN delivery protocol with J.W.; J.W. assembled the MiSeq gene disruption data, which was generated by C.K.T., J.J.Y., and R.G.T.; K.K.N. conducted the autologous transplants; D.A.S. and Z.K.N. performed genome searches and developed software; O.H. and Z.K.N. developed the mutant tracking methodology; E.J.R. coordinated ZFN assembly and SELEX analysis; and M.C.H. and P.D.G. contributed substantially to the study conception and design, and critically reviewed the manuscript.
Conflict-of-interest disclosure: J.W., C.K.T., J.J.Y., R.G.T., D.A.S., E.J.R., M.C.H., and P.D.G. are full-time employees of Sangamo BioSciences, Inc. The remaining authors declare no competing financial interests.
Correspondence: Hans-Peter Kiem, Fred Hutchinson Cancer Research Center, PO Box 19024, Mail Stop D1-100, Seattle, WA 98109-1024; e-mail: hkiem@fredhutch.org.