Key Points
Electron microscopy and hydrogen-deuterium exchange establish the C1 domain as the major binding site for the VWF D′D3 domain on FVIII.
Additional sites implicated in the FVIII-VWF interaction are located within the a3 acidic peptide and the A3 and C2 domains of FVIII.
Abstract
Association with the D′D3 domain of von Willebrand factor (VWF) stabilizes factor VIII (FVIII) in the circulation and maintains it at a level sufficient to prevent spontaneous bleeding. We used negative-stain electron microscopy (EM) to visualize complexes of FVIII with dimeric and monomeric forms of the D′D3 domain. The EM averages show that FVIII interacts with the D′D3 domain primarily through its C1 domain, with the C2 domain providing a secondary attachment site. Hydrogen-deuterium exchange mass spectrometry corroborated the importance of the C1 domain in D′D3 binding and implicates additional surface regions on FVIII in the interaction. Together, our results establish that the C1 domain is the major binding site on FVIII for VWF, reiterate the importance of the a3 acidic peptide in VWF binding, and suggest that the A3 and C2 domains play ancillary roles in this interaction.
Introduction
The high-affinity, noncovalent association of factor VIII (FVIII) with von Willebrand factor (VWF) in the circulation protects FVIII from otherwise rapid clearance.1,2 Quantitative insufficiency of VWF, characteristic of type 1 and type 3 von Willebrand disease (VWD), as well as impaired FVIII binding by VWF, observed in type 2N VWD, result in a secondary deficiency of FVIII and a bleeding diathesis.1,3,4 During biosynthesis, VWF monomers (∼260 kDa) form disulfide-bonded dimers through their C-terminal cystine knot (CK) domains. Upon formation of homotypic disulfide bonds between juxtaposed N-terminal D′D3 domains, these dimers form mature VWF multimers that range in size from 500 kDa to >20 MDa. The structural determinants for FVIII binding have been localized to the D′D3 domain by proteolytic fragmentation and by analysis of naturally occurring type 2N VWD mutations.5,6 Consistent with these findings, recombinant D′D3 domains have been shown to be sufficient to elevate levels of endogenous FVIII in VWF-deficient mice.7 The reciprocal binding site for VWF on FVIII was initially localized broadly to its light chain.8 Subsequent studies demonstrated that the a3 acidic peptide region (E1649-R1689)9 and a restricted fragment thereof (K1673-R1689),10 as well as sulfation on residue Y1680 within this region, are important for VWF binding.11 In addition, mutation analysis and antibody competition studies have implicated the C1 and C2 domains in VWF binding.9,12-16 To better understand the interaction between FVIII and VWF, we visualized FVIII-D′D3 complexes by electron microscopy (EM) and used hydrogen-deuterium exchange mass spectrometry (HDX-MS) to identify structural perturbations in FVIII upon D′D3 binding.
Materials and methods
B domain–deleted FVIII and FVIII-Fc fusion protein (rFVIIIFc), and monomeric and dimeric forms of VWF D′D3 domain, were expressed in HEK 293 cells and purified as described in supplemental Methods, found on the Blood Web site. Experimental methods for EM and HDX-MS are also described in the supplemental Methods.
Results and discussion
Electron microscopy of FVIII in complex with D′D3 domains
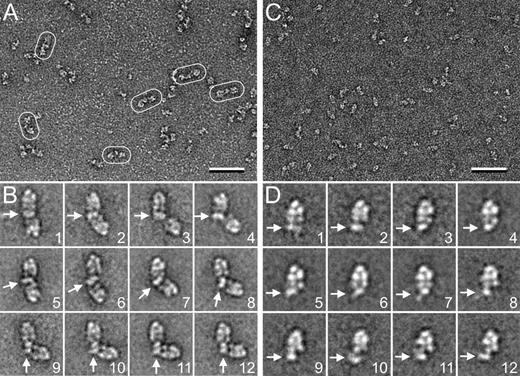
Negatively stained FVIII in complex with dimeric D′D3 showed substantial structural variability (Figure 1A), but many particles appeared to consist of 2 peripheral densities connected by a smaller central density (Figure 1A, circled). Class averages (Figure 1B and supplemental Figure 1) revealed the peripheral densities as FVIII, and thus the central, elongated density as the dimeric D′D3 (Figure 1B, arrows). The angle between the 2 FVIII molecules varies dramatically (supplemental Video 1), and the dimeric D′D3 can also be positioned differently with respect to the 2 FVIII molecules. In panels 1 to 4 of Figure 1B, the long axis of the dimeric D′D3 is aligned with the 2 C domains of the FVIII on top (indicated by horizontal arrows), but only in some cases is it also aligned with the C domains of the second FVIII (panels 1 and 2 vs panels 3 and 4). In the remaining panels of Figure 1B, the long axis of the dimeric D′D3 is positioned either diagonally (panels 5-8) or perpendicular (panels 9-12) to the 2 C domains of the FVIII on top (indicated by the direction of the arrows).
EM analysis of FVIII in complex with dimeric and monomeric D′D3. (A) Representative raw image of dimeric FVIII-D′D3 in negative stain. Some of the complexes are circled. The scale bar represents 50 nm. (B) Selected class averages of dimeric FVIII-D′D3 complex illustrating that the angle between the 2 FVIII molecules varies greatly. White arrows indicate the density representing the dimeric D′D3 domain. The side length of individual panels is 75.1 nm. (C) Representative raw image of monomeric FVIII-D′D3 in negative stain. The scale bar represents 50 nm. (D) Selected class averages of monomeric FVIII-D′D3 complex illustrating that structural heterogeneity persists in this complex. White arrows indicate the density representing the monomeric D′D3 domain. The side length of individual panels is 33.4 nm.
EM analysis of FVIII in complex with dimeric and monomeric D′D3. (A) Representative raw image of dimeric FVIII-D′D3 in negative stain. Some of the complexes are circled. The scale bar represents 50 nm. (B) Selected class averages of dimeric FVIII-D′D3 complex illustrating that the angle between the 2 FVIII molecules varies greatly. White arrows indicate the density representing the dimeric D′D3 domain. The side length of individual panels is 75.1 nm. (C) Representative raw image of monomeric FVIII-D′D3 in negative stain. The scale bar represents 50 nm. (D) Selected class averages of monomeric FVIII-D′D3 complex illustrating that structural heterogeneity persists in this complex. White arrows indicate the density representing the monomeric D′D3 domain. The side length of individual panels is 33.4 nm.
To distinguish whether the observed heterogeneity resulted from structural variability of the 2 D′D3 domains in the dimeric construct or from different ways in which FVIII interacts with a D′D3 domain, we analyzed FVIII in complex with monomeric D′D3. The particles appeared more homogeneous (Figure 1C), but class averages revealed that structural variability persisted (Figure 1D and supplemental Figure 2). In particular, the density representing monomeric D′D3 (indicated by arrows) varies in shape and location relative to FVIII (supplemental Video 2). The difference in shape may be caused by different orientations or by partial loss of density upon averaging. Importantly, although in some averages D′D3 appears to interact with both C domains of FVIII (Figure 1D, panels 1-7), in other averages it appears to interact with only 1 C domain (Figure 1D, panels 8-12). In the conventional orientation used to display FVIII, the molecule has the shape of a reversed “R,” with the C1 domain at the bottom left of the molecule and the C2 domain at the bottom right (supplemental Figure 3). Our averages thus suggested that the C1 domain is the primary binding site for D′D3 and that the C2 domain is a secondary binding site. To confirm this conclusion and to further characterize the interaction between D′D3 and FVIII, we performed HDX-MS.
Hydrogen-deuterium exchange mass spectrometry
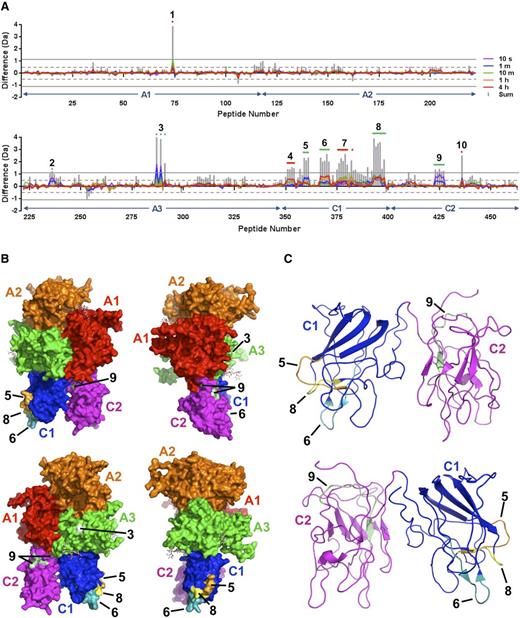
Because of its ready availability and an affinity for VWF that is comparable with that of FVIII, pharmaceutical-grade rFVIIIFc was used for HDX-MS.17 HDX-MS was performed with rFVIIIFc alone and in complex with monomeric D′D3. Stringent criteria were used to determine whether differential deuterium exchange was statistically significant: for a given peptide, the mass difference must exceed ±0.5 Da for at least 1 time point, and the sum of mass differences over all time points must exceed ±1.1 Da.18 Based on these criteria, 10 distinct regions of differential deuterium exchange were initially identified within the FVIII component (Figure 2A). Region 1 was identified in the heavy chain of FVIII, within the A1 domain and encompassing residue C248, which forms a disulfide bond with C329. Although differential deuterium uptake at this site cannot be ruled out, given the distance from known VWF interaction sites, the observed mass difference (supplemental Figure 4B, peptide 74) could be more readily rationalized by intramolecular hydrogen transfer upon generation of protein CysS• radicals during disulfide bond photolysis, as has been observed for immunoglobulins.20 The remaining 9 regions were located within the light chain: 2 in A3, 5 in C1, and 2 in C2. Each cluster of overlapping peptides was further analyzed to establish logical consistency of their respective overlap patterns and to narrow down the sequences to which perturbations in deuterium exchange could clearly be attributed (supplemental Figures 5 through 13). Three regions (4, 7, and 10) were eliminated from further consideration on the basis of irreconcilable peptide overlap patterns. Of the remaining 6 regions, 2 could be unambiguously assigned in the A3 domain: one in the a3 acidic peptide (Region 2, V1670-D1678; Figure 2A and supplemental Figure 5) and one in the A3 domain proper (Region 3; V1857-N1861 and E1875: Figure 2A-B and supplemental Figure 6). Three regions were unambiguously assigned in the C1 domain: Region 5 (W2062-S2069; Figure 2A-C and supplemental Figure 8), Region 6 (T2086-S2095; Figure 2A-C and supplemental Figure 9), and Region 8 (S2157-L2166; Figure 2A-C and supplemental Figure 11). One region was assigned in the C2 domain and corresponded to the C2-A1 domain interface (Region 9; Q2235-T2244; Figure 2A-C and supplemental Figure 12).
Mapping of the D′D3 domain-binding site on FVIII by HDX-MS. (A) HDX difference plots illustrating localized differential deuterium uptake by the FVIII component of rFVIIIFc in the presence and absence of the monomeric D′D3 construct for the FVIII heavy chain (upper panel) and light chain (lower panel). The x-axis indicates the 463 peptides derived from the B domain–deleted FVIII sequence ordered from the N to C terminus based on their respective midpoints, and the y-axis indicates the calculated mass difference caused by differential deuterium uptake for peptides derived from FVIII in the D′D3-bound and D′D3-free states. FVIII domain boundaries are indicated with arrows. Individually colored curves correspond to the average mass difference values calculated for data acquired at the time points indicated. Vertical gray bars represent the sum of differences across all time points for a given peptide. Positive values for x indicate peptides for which deuterium uptake is reduced in the presence of the D′D3 domain. Peptides and overlapping clusters of peptides exhibiting statistically significant perturbations in deuterium uptake according to criteria defined by Houde et al18 are numbered and indicated with colored horizontal bars or dots, with green indicating those for which noncontradictory peptide overlap patterns could be derived and red indicating those for which this was not possible within the given constraints for statistical significance. (B) Surface representations of the FVIII structure (PDB ID: 2R7E19 ) with individual domains colored as indicated. Numbered areas correspond to sites of differential deuterium uptake identified in (A). (C) Ribbon representations of the FVIII C1 and C2 domains with areas corresponding to differential deuterium uptake numbered as in (B).
Mapping of the D′D3 domain-binding site on FVIII by HDX-MS. (A) HDX difference plots illustrating localized differential deuterium uptake by the FVIII component of rFVIIIFc in the presence and absence of the monomeric D′D3 construct for the FVIII heavy chain (upper panel) and light chain (lower panel). The x-axis indicates the 463 peptides derived from the B domain–deleted FVIII sequence ordered from the N to C terminus based on their respective midpoints, and the y-axis indicates the calculated mass difference caused by differential deuterium uptake for peptides derived from FVIII in the D′D3-bound and D′D3-free states. FVIII domain boundaries are indicated with arrows. Individually colored curves correspond to the average mass difference values calculated for data acquired at the time points indicated. Vertical gray bars represent the sum of differences across all time points for a given peptide. Positive values for x indicate peptides for which deuterium uptake is reduced in the presence of the D′D3 domain. Peptides and overlapping clusters of peptides exhibiting statistically significant perturbations in deuterium uptake according to criteria defined by Houde et al18 are numbered and indicated with colored horizontal bars or dots, with green indicating those for which noncontradictory peptide overlap patterns could be derived and red indicating those for which this was not possible within the given constraints for statistical significance. (B) Surface representations of the FVIII structure (PDB ID: 2R7E19 ) with individual domains colored as indicated. Numbered areas correspond to sites of differential deuterium uptake identified in (A). (C) Ribbon representations of the FVIII C1 and C2 domains with areas corresponding to differential deuterium uptake numbered as in (B).
Conclusions
Despite the structural variability observed by EM among complexes of FVIII with both monomeric and dimeric D′D3, involvement of the C1 domain was a common feature of all complexes. Furthermore, HDX-MS analysis revealed multiple sites of structural perturbation within the C1 domain upon binding of monomeric D′D3. In general, FVIII missense mutations known to impair VWF binding, and thereby giving rise to mild or moderate hemophilia A, cluster within the C1 domain, and approximately half of these are located within or near regions identified by HDX-MS (supplemental Figure 14). Less pronounced perturbations were observed in the a3 acidic peptide, consistent with its established role in VWF binding, as well as in the A3 domain proper and C2 domain, suggesting a secondary role for these sites in the FVIII-D′D3 interaction.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Damian Houde, Steven Berkowitz, and Andrew Weiskopf for helpful discussions. T.W. is an investigator with the Howard Hughes Medical Institute.
This research was funded in part by Biogen Idec.
Authorship
Contribution: P.-L.C., G.M.B.-A., R.T.P., J.D.K., and T.W. designed the experiments; P.-L.C., G.M.B.-A., E.S.C., M.G.C., J.D.K., and T.W. performed research and analyzed data; and P.-L.C., G.M.B.-A., J.D.K., and T.W. wrote the manuscript.
Conflict-of-interest disclosure: G.M.B.-A., E.S.C., R.T.P., and J.D.K. are employees of and hold equity interest in Biogen Idec.
Correspondence: Thomas Walz, Department of Cell Biology, Harvard Medical School, 240 Longwood Ave, Boston, MA 02115; e-mail: twalz@hms.harvard.edu; John D. Kulman, Biogen, 250 Binney St, Cambridge, MA 02142; e-mail: john.kulman@biogen.com.
References
Author notes
P.-L.C. and G.M.B.-A. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal