Abstract
Cytogenetically normal acute myeloid leukemia (CN-AML) is a heterogeneous disease with regard to genetic alterations and clinical outcome. Recent sequencing studies categorized the growing number of recurrently mutated genes into different functional groups, e.g. myeloid transcription factors, tumor suppressors, signal transducers, chromatin modifiers, cohesin-complex and spliceosome-complex. We set out to characterize mutations in genes linked to epigenetic regulation during the progression of CN-AML. Besides genes directly involved in chromatin modification (i.e. DNMT3A, TET2, MLL, ASXL1, KDM6A, KDM2A, NSD1 and EZH2), we also studied mutations in WT1 and IDH1/2 since they are known to inhibit TET2 function.
Targeted sequencing of 46 genes related to leukemia (mean coverage >500x) was performed on matched diagnostic, remission and relapse samples of 50 patients with CN-AML (median age: 66, range: 21-89). We called somatic variants at diagnosis or at relapse and filtered for mutations with translational consequences, excluding known error-prone genes and common germline polymorphisms (dbSNP 138; MAF>=1%).
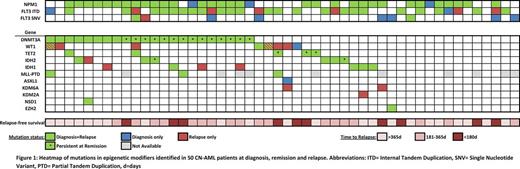
At diagnosis, 36/50 patients (72%) carried a total of 48 mutations in epigenetic regulators (Figure 1). The majority of patients harbored a single mutation affecting this functional group, while 2 or 3 mutations were observed in 9 and 1 patient(s), respectively. The median variant allele frequency (VAF) of the mutations was 42% (range: 22-98%), indicating that mutations in epigenetic regulators are early events and are present in the founding clone. Of the 48 mutations detected at diagnosis, only 2 were lost at relapse, highlighting the stability of these lesions during disease progression. Moreover, in 12/50 patients (24%), mutations in epigenetic regulators were acquired at relapse. All but one of these patients already had a mutation in another epigenetic regulator at diagnosis. We did not identify patients who acquired DNMT3A, TET2 or ASXL1 mutations during disease progression. However, mutations in WT1, IDH1, and KDM6A were gained in several patients at relapse. In 4/13 cases, the gained mutations were already detectable at low levels at diagnosis (median VAF: 2.9%, range: 0.3-6%, mean coverage at the investigated sites: 629x, range: 85-1625x).
We also evaluated the presence of these mutations in remission: In 18 out of 36 (50%) patients, some of the mutations affecting DNMT3A (n=14), TET2 (n=3) or IDH2 (n=2) were present at a VAF >5% (median: 22%, range: 9-75%) in cytomorphologically defined complete remission, suggesting the persistence of pre-leukemic clones with limited response to chemotherapy. Longer relapse-free survival was observed in patients with DNMT3A mutations that did not persist at remission (np-DNMT3A) in comparison to patients with persisting DNMT3A mutations (p-DNMT3A). Remarkably, the latter group was enriched for patients that also harbored FLT3 internal tandem duplications (ITDs) (10/14 versus 1/8; Fisher's exact test, p=0.02). The vast majority of p-DNMT3A showed alterations of R882, whereas mutations at other positions of DNMT3A tended to be undetectable in remission. When including the NPM1 status, only 1/8 patient with np-DNMT3A was triple mutated, compared to 11/14 patients with p-DNMT3A, suggesting that co-occurrence of DNMT3A, FLT3- ITD and NPM1 c is associated with p-DNMT3A (p=0.006).
In summary, we show that a high proportion of patients (72%) with relapsing CN-AML is affected by mutations in genes linked to epigenetic regulation. The stability of these mutations between diagnosis and relapse in combination with their acquisition during disease progression, as well as the frequent persistence of DNMT3A, TET2 and IDH2 mutations during remission underscore the necessity for new therapeutic approaches. The striking association of DNMT3A R882 mutations with NPM1 c and FLT3 -ITD suggest a unique mechanism of oncogenic collaboration. Persistence of DNMT3A R882 mutations may indicate a fertile ground for relapse. Further studies will be required to clarify whether the actual relapse arises from a preleukemic clone harboring only the founder mutation or from residual leukemia cells containing several genetic lesions.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal