Key Points
miR-139-3p and miR-199a-3p, induced by ICL-induced damage, respectively, cause a loss and gain of hematopoietic progenitors.
miR-199a-3p is an onco-microRNA (onco-miR) causing AML in a Cebpa-deficient mouse model. Target genes of miR-199a-3p include PRDX6, RUNX1, and SUZ12.
Abstract
Interstrand crosslinks (ICLs) are toxic DNA lesions that cause severe genomic damage during replication, especially in Fanconi anemia pathway-deficient cells. This results in progressive bone marrow failure and predisposes to acute myeloid leukemia (AML). The molecular mechanisms responsible for these defects are largely unknown. Using Ercc1-deficient mice, we show that Trp53 is responsible for ICL-induced bone marrow failure and that loss of Trp53 is leukemogenic in this model. In addition, Ercc1-deficient myeloid progenitors gain elevated levels of miR-139-3p and miR-199a-3p with age. These microRNAs exert opposite effects on hematopoiesis. Ectopic expression of miR-139-3p strongly inhibited proliferation of myeloid progenitors, whereas inhibition of miR-139-3p activity restored defective proliferation of Ercc1-deficient progenitors. Conversely, the inhibition of miR-199a-3p functions aggravated the myeloid proliferation defect in the Ercc1-deficient model, whereas its enforced expression enhanced proliferation of progenitors. Importantly, miR-199a-3p caused AML in a pre-leukemic mouse model, supporting its role as an onco-microRNA. Target genes include HuR for miR-139-3p and Prdx6, Runx1, and Suz12 for miR-199a-3p. The latter genes have previously been implicated as tumor suppressors in de novo and secondary AML. These findings show that, in addition to TRP53-controlled mechanisms, miR-139-3p and miR-199a-3p are involved in the defective hematopoietic function of ICL-repair deficient myeloid progenitors.
Introduction
Bone marrow failure (BMF) syndromes are characterized by impaired hematopoiesis, leading to single- or multilineage cytopenia.1 Although the underlying causes of BMF syndromes are heterogeneous, they share the elevated risk to progress toward leukemia.2 Ineffective repair of DNA damage is one of the major causes of BMF and leukemic transformation.3 Fanconi anemia (FA) is the most frequently inherited cause of BMF.4 Defective interstrand crosslink (ICL) repair is a major hallmark of FA.5 Mutations in at least 16 genes encoding FA proteins have been described.5-9
Structure-specific endonuclease complexes include Fanconi-associated nuclease 1, MUS81-EME1, and xeroderma pigmentosum, complementation group F (FA, complementation group Q [FANCQ])-ERCC1, which are recruited to ICL lesions.5 SLX4 (FA, complementation group P) guides FANCQ-ERCC1 to the sites of ICL damage, where it functions as the essential endonuclease required for repair.5,10-12 Mutation or loss of either FA, complementation group P, ERCC1, or FANCQ leads to the disruption of the FANCQ-ERCC1 endonuclease activity, impairs ICL-repair, and increases sensitivity to crosslinking agents, showing the relevance of these mutations for FA.6,13,14
ERCC1 is active in both nucleotide excision repair and ICL repair.15 Like the Fancd2- and Fanca-deficient models, Ercc1-deficient mice have reduced numbers of hematopoietic stem and progenitor cells (HSPCs).16-19 The observation that XPA knockout mice, which are exclusively nucleotide excision repair-deficient, did not show the accelerated bone marrow (BM) exhaustion suggested that defective ICL repair was predominantly responsible for this phenotype. Indeed, Ercc1-deficient hematopoietic progenitor cells showed the hypersensitivity to the DNA crosslinking agent mitomycin C (MMC) that is characteristic of ICL-repair deficiency.19 The underlying DNA-damage response (DDR) pathways responsible for the depletion and oncogenic transformation of ICL-deficient hematopoietic progenitors are still largely unknown. Recent studies have shown that proinflammatory cytokines such as tumor necrosis factor-α contribute to the BMF in FA20 and that inhibition of p38MAPK may ameliorate this effect.21
Here, we used Ercc1-deficient mice to identify mechanisms that contribute to ICL-induced BMF and leukemic transformation. We show that the deletion of Trp53 but not Cdkn2a alleviates the loss of lineage negative (lin−) Sca1+ c-Kit+ (LSK) BM cells in Ercc1-deficient mice. TRP53 acted as a major gatekeeper preventing leukemic transformation of Ercc1-deficient BM cells, corroborating studies in clinical FA.22 In addition, we identified two microRNAs (miRNAs), miR-139-3p and miR-199a-3p that were expressed at enhanced levels in common myeloid progenitors (CMPs) from Ercc1-deficient mice compared with control littermates and which exert strikingly opposite effects on the proliferation of myeloid progenitors. These miRNAs were also expressed at an elevated level in CD34+ BM cells from FA patients. Importantly, miR-199a-3p caused acute myeloid leukemia (AML) in a preleukemic mouse transplantation model, establishing its role as an onco-microRNA (onco-miR).
Materials and methods
Mice
The Cdkn2a+/−,23 Trp53+/−,24 Ercc1+/*292, and Ercc1+/− mice have been previously described.25 Ercc1 mice were generated in a F1 mixed background of C57BL/6 and FVB/n. To generate Cebpacre/fl mice, Cebpafl/fl were crossed with Cebpa-cre mice.26,27
For transplantation experiments with Ercc1-deficient cells, 12- to 14-week-old recipient F1 mice (FVB/n × C57BL/6) were irradiated (9 Gy), and transplanted by tail-vein injection with 1 × 107 total BM cells and 1 × 105 spleen cells. For other transplantation experiments, 6- to 8-week-old C57BL/6 recipient mice (The Jackson Laboratory) were irradiated (8.5 Gy) and tail-vein injected with retrovirally-transduced HSPCs (1-5 × 105 cells per mouse). Leukemia cells isolated from BM were injected into irradiated (5 Gy) 6- to 8-week-old C57BL/6 mice. All animal experiments were approved by the Erasmus Medical Center Animal Welfare/Ethics Committee.
Retroviral infection and colony assays
Murine stem cell virus (MSCV) barcoded miRNA vectors for constitutive overexpression of miRNAs and virus particles were generated as described previously.28 The 32D cells and BM-derived HSPCs were infected with MSCV-BC-miRNA virus using RetroNectin (Takara Bio, Inc) according to the manufacturer’s instructions with a multiplicity of infection of 0.5. Colony assays were performed as described.27 In brief, 10 000 MSCV-transduced cells (based on enhanced green fluorescent protein [EGFP] expression) or 50 000 total BM cells, per mL per 35 mm dish were plated in triplicate in methyl cellulose medium (MethoCult M3234, Stemcell Technologies), containing human colony-stimulating factor 3 (CSF3) (0.1 µg/mL), or mouse CSF2 (0.1 µg/mL), or human erythropoietin (4 mU/mL) plus transferrin (0.3 mM), hemin (0.2 mM) and mouse stem cell factor (0.1 µg/mL), and puromycin (1.5 μg/mL, only for transduced cells). Colonies containing 50 cells or more were scored on day 7 of culture. For miRNA inhibitory experiments, HSPCs were transfected with fluorescent-tagged miRCURY locked nucleic acid (LNA) inhibitors (Exiqon) with DharmaFECT 1 (Thermo Scientific). Research was conducted in accordance with the Declaration of Helsinki.
See supplemental Materials on the Blood Web site for quantitative proteomics, whole exome sequencing (WES) and data analysis, Luciferase reporter assays, Luminex experiments, antibodies, cell staining flow cytometry and cytospins, gene- and miRNA profiling quantitative polymerase chain reaction (PCR), and statistics and patient samples.
Results
Exhaustion of Ercc1-/*292 HSPCs is caused by TRP53-dependent rather than CDKN2A-dependent mechanisms
Ercc1 knockout mice are severely runted, weigh only about 20% compared with their normal littermates, and die at around 3 weeks of age.25,29 A premature stop codon at position 292 of mouse Ercc1 (Ercc1*292) causes a carboxyl-terminal deletion of 7 amino acids of ERCC1, which impairs dimerization with xeroderma pigmentosum, complementation group F.25 The life span of Ercc1-/*292 mice is approximately 22 weeks, and their hematopoietic phenotype is comparable to that of Ercc1 knockout mice.18 The BM of Ercc1-/*292 mice contains decreased numbers of myeloid progenitors that are strongly hampered in their ability to proliferate in colony assays.18 Both TRP53 and CDKN2A have been shown to regulate the DDR in HSPCs.22,30 Ataxia telangiectasia mutated (ATM)-deficient hematopoietic stem cells (HSCs) undergo reactive oxygen species (ROS)-induced genotoxic stress that evokes a DDR involving the upregulation of p16Ink4a 30. Because ICLs induce ATM-related activity,30 we asked whether CDKN2a, either through p16INK4a or through the TRP53-regulatory effects of p19ARF, contribute to the phenotype of Ercc1-deficient HSPCs. Therefore, we investigated how disruption of these individual loci affects Ercc1-/*292 HSPC maintenance. At 3 weeks of age, Ercc1-/*292 BM contained less than 50% of LSKs compared with wild-type (WT) (Figure 1A). Furthermore, the colony-forming capacity of Ercc1-/*292 myeloid and erythroid progenitor cells was markedly reduced (Figure 1B-D). Deletion of Cdkn2a did not affect this phenotype (Figure 1A-D). In contrast, homozygous deletion of Trp53 in Ercc1-/*292 mice restored the fraction of LSK cells and granulocyte-macrophage colony-forming units (CFU)-CSF2 (Figure 1E-H). These results showing that TRP53-driven, rather than CDKN2A-dependent DDR mechanisms cause ICL-induced loss of HSPCs corroborate studies in clinical FA.22 Under conditions of CSF3 and stem cell factor/erythropoietin stimulation, a partial rescue of CFU-CSF3 or erythroid burst forming unit colony growth was seen (Figure 1G-H).
Loss of Trp53 but not of Cdkn2a, restores HSPC content in Ercc1-/*292 mice. (A, E) LSK frequencies in lin− fractions of indicated mouse genotypes at 3 weeks of age are shown. (B-D and F-H) CFUs per 5 × 104 unfractionated BM cells in the presence of indicated growth factors relative to the WT controls are shown. All bars represent the mean and standard deviations (SD) of n ≥3 mice. The significance was calculated by the Student t tests (asymptotic significance [2-tailed]), *P < .05].
Loss of Trp53 but not of Cdkn2a, restores HSPC content in Ercc1-/*292 mice. (A, E) LSK frequencies in lin− fractions of indicated mouse genotypes at 3 weeks of age are shown. (B-D and F-H) CFUs per 5 × 104 unfractionated BM cells in the presence of indicated growth factors relative to the WT controls are shown. All bars represent the mean and standard deviations (SD) of n ≥3 mice. The significance was calculated by the Student t tests (asymptotic significance [2-tailed]), *P < .05].
Affected pathways in Ercc1-/*292 HSPCs
To investigate how Ercc1-deficiency affects BM HSPCs at the molecular level, we performed gene expression profiling (GEP) and proteomics. Ingenuity Downstream Effect Analysis (IDEA) of differentially expressed genes (supplemental Table 1; supplemental Figure 1A) showed decreased activity of networks required for hematopoietic development and leukocyte differentiation in Ercc1-/*292 LSK BM cells at weeks 20 to 22 of age compared with control cells (supplemental Figure 1B). Differentially expressed DDR genes included P53 and DNA damage regulated 1, cyclin-dependent kinase inhibitor 1a (Cdkn1a, p21), and Xiap-associated factor 1, all TRP53-induced genes (supplemental Table 1).
LSK cells are enriched in HSCs that have a low cycling activity. Because committed hematopoietic progenitors are highly proliferative, the consequences of defective ICL-repair are expected to be more severe in these cells. We therefore also analyzed lineage-depleted BM cells that contain HSCs and progenitor cells. GEP identified a set of differentially expressed transcripts in Ercc1-/*292 lin− cells relative to controls (supplemental Figure 1C; supplemental Table 2). IDEA showed that pathways involved in cell death, the regulation of ROS, cell-cycle control, hematopoiesis, and DNA repair are more prominently deregulated in Ercc1-/*292 lin− cells compared with LSKs (supplemental Figure 1B vs 1D). Expression of TRP53 target genes, eg, p21 and Gadd45b, was increased in Ercc1-/*292 lin− cells, indicating an activated DDR, which is in agreement with the previously reported γH2AX accumulation in Ercc1-deficient cells31 (supplemental Table 2). Quantitative proteome analysis on Ercc1-/*292 and control HSPCs showed that 457 proteins were significantly differentially expressed (supplemental Table 3). Notably, a high correlation between the changes in messenger RNA and protein level was observed (supplemental Figure 1E). IDEA on these protein data showed that the main deregulated biological functions identified by proteomics are largely similar to those detected by GEP, ie, pathways involved in cell death and the regulation of ROS (supplemental Figure 1F).
miRNA expression in Ercc1-/*292 HSPCs
Because miRNAs play key roles in cellular stress responses, including the regulation of TRP53-dependent pathways,32,33 we interrogated which miRNAs are differentially expressed in LSK and lin− cells from Ercc1-/*292 mice. From a panel of 365 miRNAs tested by TaqMan quantitative PCR in Ercc1-/*292 LSKs isolated at week 20 of age, none were significantly altered in its expression relative to control LSKs. In contrast, in lin−Ercc1-/*292 cells, expression of 4 miRNAs (miR-139-3p, miR-199a-3p, miR-34a-5p, and miR-342-5p) was significantly elevated relative to WT lin− controls (twofold to fourfold, P < .05) (Figure 2A). To further specify which myeloid progenitor subsets express these miRNAs, we analyzed fluorescence-activated cell sorter-purified CMPs, granulocyte macrophage progenitors, and megakaryocyte-erythroid progenitors. In Ercc1-/*292 CMPs, the levels of miR-34a-5p, miR-139-3p, and miR-199a-3p were elevated relative to controls, whereas miR-342-5p was not expressed (Figure 2B). In Ercc1-/*292 granulocyte macrophage progenitors, miR-139-3p was not expressed and only the level of miR-199a-3p was significantly elevated (Figure 2B). In Ercc1-/*292 megakaryocyte-erythroid progenitors, only the expression of miR-342-5p was significantly induced (Figure 2B).
Differentially expressed miRNAs in Ercc1-/*292 cells. (A) The expression of miRNAs and U6 in lin− BM cells was determined by quantitative PCR in quadruplicate. The expression of indicated miRNAs relative to U6 and normalized to WT controls is depicted. The error bars represent the SD of 5 mice. (B) CMP (top), GMP (middle), and MEP (bottom). The expression of indicated miRNAs relative to U6 and normalized to WT is depicted for different hematopoietic progenitor subpopulations isolated from the BM of 20-week-old mice. The error bars represent SD of 3 measurements. (C) The expression of indicated miRNAs relative to U6 in lin− BM cells of 3-week-old Ercc1-/*292;Trp53 WT mice and Ercc1-/*292;Trp53−/−, and relative to WT controls is shown. All bars represent the mean and SD of n ≥ 3 mice. (D) The expression of miR-139-3p and miR-199a-3p relative to U6 in WT lin− cells and normalized to the control condition without MMC, is depicted for the indicated MMC concentrations. The error bars represent SD of 3 measurements. In all panels, the significance was calculated with the Mann-Whitney U test (asymptotic significance [2-tailed], *P < .05]. NE, not expressed; NS, not significant.
Differentially expressed miRNAs in Ercc1-/*292 cells. (A) The expression of miRNAs and U6 in lin− BM cells was determined by quantitative PCR in quadruplicate. The expression of indicated miRNAs relative to U6 and normalized to WT controls is depicted. The error bars represent the SD of 5 mice. (B) CMP (top), GMP (middle), and MEP (bottom). The expression of indicated miRNAs relative to U6 and normalized to WT is depicted for different hematopoietic progenitor subpopulations isolated from the BM of 20-week-old mice. The error bars represent SD of 3 measurements. (C) The expression of indicated miRNAs relative to U6 in lin− BM cells of 3-week-old Ercc1-/*292;Trp53 WT mice and Ercc1-/*292;Trp53−/−, and relative to WT controls is shown. All bars represent the mean and SD of n ≥ 3 mice. (D) The expression of miR-139-3p and miR-199a-3p relative to U6 in WT lin− cells and normalized to the control condition without MMC, is depicted for the indicated MMC concentrations. The error bars represent SD of 3 measurements. In all panels, the significance was calculated with the Mann-Whitney U test (asymptotic significance [2-tailed], *P < .05]. NE, not expressed; NS, not significant.
At week 3 of age, only the levels of miR-34a-5p and miR-342-5p were significantly elevated in Ercc1-/*292 lin− cells (Figure 2C). Crossings of Ercc1-/*292 mice with Trp53−/− mice revealed that the expression of miR-34a-5p and miR-342-5p was fully TRP53 dependent (Figure 2C). In contrast, expression of miR-139-3p and miR-199a-3p was not detectable at this earlier time point. Apparently, these miRNAs are only induced when cells with DNA damage due to unrepaired ICLs accumulate over a longer time frame (Figure 2A,C). In support of a direct relationship between ICLs and these miRNAs, the ICL-inducing agent MMC (10 µM, 24 hours) induced the expression of miR-199a-3p and miR-139-3p in normal lin− cells (Figure 2D).
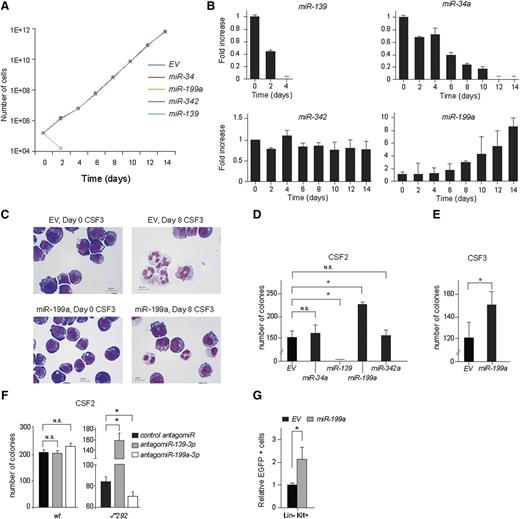
Opposite roles of miR-139 and miR-199a in myeloid progenitor exhaustion and expansion
To determine their role in myeloid cell survival, proliferation, and differentiation, we ectopically expressed the miRNAs from barcoded retroviral vectors (MSCV) in the murine cell line 32D expressing human CSF3R (32D-CSF3R).28,34 In interleukin (IL)-3–containing expansion medium, miR-139 expressing 32D-CSF3R cells died rapidly, whereas no alteration in cell survival and proliferation was seen in cells expressing miR-34a, miR-199a, or miR-342 (Figure 3A). When these cells were mixed in a 1:1 ratio with MSCV-empty vector (EV) transduced control cells and transferred to CSF3-containing differentiation medium, miR-139 again inhibited proliferation (Figure 3B), whereas miR-34a and miR-342 had little or no effect (Figure 3B). In contrast, miR-199a expressing 32D-CSF3R cells had gained a competitive growth advantage in this setting (Figure 3B). In addition, we noted an increased percentage of blast-like cells in miR-199–overexpressing 32D cell cultures (37% miR-199 vs 21% EV control) and a decreased percentage of fully differentiated cells (23% miR-199 vs 40% EV control), whereas the percentage of intermediate differentiated cells was comparable to EV control cells at day 8 of CSF3 treatment (Figure 3C). This suggests that miR-199a inhibits differentiation and/or enhances proliferation/self-renewal of myeloid progenitors.
Expression of miR-139 and miR-199a alters the balance of HSPC loss and expansion. (A) Murine 32D cells were infected with MSCV-BC vectors containing either, miR-139, miR-199a, miR-34a, miR-342, or no miRNA (EV) as control; 32D cells expressing miRNAs were expanded in IL-3 containing medium and the number of cells at indicated time points is plotted. (B) Equal numbers of 32D cells expressing miRNAs were mixed with control EV-expressing 32D cells and switched to CSF3-containing medium. Cell samples were taken at indicated time points and genomic DNA was isolated. The abundance of the different barcodes relative to the EV barcode signal and normalized to day 0 is depicted. Representative data of 3 independent experiments are shown. The error bars represent SD of 3 measurements. (C) 32D-CSF3R cells were switched from IL-3 to CSF3-containing medium on t = 0. Micrographs show the morphology of 32D-CSF3R cells on indicated time points of CSF3 treatment. Size bars indicate 10 µm. (D) CSF2-colony assays of 1 × 104 plated lin cells transduced with different miRNA expressing viruses. The number of CFU consisting of more than 50 cells after 7 days of growth relative to EV control is depicted. Data are from 2 independent experiments performed in triplicate and represent the mean and SD of 3 plates counted. (E) CSF3-colony assays were performed with 2 × 104 lin− cells transduced with miR-199a expressing viruses. The number of CFU consisting of more than 50 cells per colony is depicted. Data represent the mean and SD of 3 plates counted. (F) Same as (C). The number of CFU of 1 × 104 mouse Ercc1-/*292 and WT lin− cells transfected with LNA antagomiR against miR-139-3p and miR-199a-3p are shown. Data represent the mean and SD of 3 plates counted. (G) Lin− cells transduced with MSCV–miR-199a or MSCV-EV and mixed with untransduced WT lin− cells in a 1:10 ratio were transplanted in irradiated recipient mice (n = 8 per group). The change in lin−Kit+ (LK, progenitors) fraction in the BM 10 weeks posttransplantation and relative to the EV control is shown (*P < .05). The error bars represent SD of n = 8 mice. In panels (C-E), the significance was calculated with the Student t test (2-tailed, *P < .05).
Expression of miR-139 and miR-199a alters the balance of HSPC loss and expansion. (A) Murine 32D cells were infected with MSCV-BC vectors containing either, miR-139, miR-199a, miR-34a, miR-342, or no miRNA (EV) as control; 32D cells expressing miRNAs were expanded in IL-3 containing medium and the number of cells at indicated time points is plotted. (B) Equal numbers of 32D cells expressing miRNAs were mixed with control EV-expressing 32D cells and switched to CSF3-containing medium. Cell samples were taken at indicated time points and genomic DNA was isolated. The abundance of the different barcodes relative to the EV barcode signal and normalized to day 0 is depicted. Representative data of 3 independent experiments are shown. The error bars represent SD of 3 measurements. (C) 32D-CSF3R cells were switched from IL-3 to CSF3-containing medium on t = 0. Micrographs show the morphology of 32D-CSF3R cells on indicated time points of CSF3 treatment. Size bars indicate 10 µm. (D) CSF2-colony assays of 1 × 104 plated lin cells transduced with different miRNA expressing viruses. The number of CFU consisting of more than 50 cells after 7 days of growth relative to EV control is depicted. Data are from 2 independent experiments performed in triplicate and represent the mean and SD of 3 plates counted. (E) CSF3-colony assays were performed with 2 × 104 lin− cells transduced with miR-199a expressing viruses. The number of CFU consisting of more than 50 cells per colony is depicted. Data represent the mean and SD of 3 plates counted. (F) Same as (C). The number of CFU of 1 × 104 mouse Ercc1-/*292 and WT lin− cells transfected with LNA antagomiR against miR-139-3p and miR-199a-3p are shown. Data represent the mean and SD of 3 plates counted. (G) Lin− cells transduced with MSCV–miR-199a or MSCV-EV and mixed with untransduced WT lin− cells in a 1:10 ratio were transplanted in irradiated recipient mice (n = 8 per group). The change in lin−Kit+ (LK, progenitors) fraction in the BM 10 weeks posttransplantation and relative to the EV control is shown (*P < .05). The error bars represent SD of n = 8 mice. In panels (C-E), the significance was calculated with the Student t test (2-tailed, *P < .05).
Similar results were obtained in primary CFU-CSF2 and CFU-CSF3 colony cultures. Whereas ectopic expression of miR-34a and miR-342 did not alter, and miR-139 completely abrogated colony outgrowth, the CFU-CSF2 colony formation capacity of miR-199a– expressing BM cells was increased by 1.5-fold (P < .05) (Figure 3D). Ectopic miR-199a expression also increased the CSF3-stimulated colony-forming capacity (Figure 3E), indicating that ectopic miR-199a expression did not induce autonomous proliferation of 32D cells. miR-199a–expressing 32D cells died rapidly when switched to medium without IL-3 (data not shown).
Next, we used LNA antagomiRs to explore how the inhibition of miR-139-3p and miR-199a-3p affects the outgrowth of Ercc1-/*292 CFU-CSF2. Treatment of Ercc1-/*292 lin− cells with miR-199a-3p LNA antagomiR reduced the number of CFU-CSF2 by ∼20% (Figure 3F). Conversely, miR-139-3p LNA antagomiR rescued the colony-forming capacity of Ercc1-/*292 progenitors. In contrast, both antagomiRs did not affect CFU-CSF2 colony formation from normal BM (Figure 3F).
Finally, we assessed the effects of miR-199a expression on HSPC expansion in vivo, by transplanting MSCV–miR-199a and MSCV-EV infected lin− cells mixed with noninfected lin− cells in mice. Myeloid progenitor cells expressing miR-199a significantly increased in numbers over nontransduced controls at 10 weeks posttransplantation, whereas cells transduced with MSCV-EV were maintained at a similar frequency compared with input (Figure 3G). In summary, these data show that ectopic expression of miR-139-3p inhibits and miR-199a-3p enhances the outgrowth of normal myeloid progenitors, both in vitro and in vivo, and that antagonizing their endogenous expression in Ercc1-deficient cells restores (anti–miR-139-3p) or further aggravates (anti–miR-199a-3p) the ICL-induced phenotype.
Identification of miR-139-3p and miR-199a-3p targets
miRNAs regulate gene expression by inhibition of translation and/or destabilization of target transcripts.35-37 Of the downregulated transcripts and proteins in Ercc1-/*292 lin− cells compared with controls, 7 genes contain predicted and evolutionary well-conserved binding sites for either miR-139-3p or miR-199a-3p in their 3′-untranslated regions (UTRs) (TargetScan; www.targetscan.org) (supplemental Tables 2 and 3). The RNA-binding protein HuR was the only identified target of miR-139-3p in Ercc1-/*292 HSPCs. Six genes, Prdx6, Suz12, Pon2, Fubp1, Calu, and Runx1, contain miR-199a-3p recognition sites. To test whether miR-139-3p and miR-199a-3p directly control the expression of these genes by binding to the predicted miRNA binding sites, we cloned these 3′-UTR regions downstream of a luciferase reporter. Ectopic expression of miR-139 and miR-199a caused a 30% to 65% reduction of luciferase activity of all target 3′-UTR fragments tested (Figure 4A). Mutation of the predicted miRNA binding sites abolished the inhibition of luciferase activity, confirming the predicted target sites as major determinants for miR-139-3p and miR-199a-3p–mediated regulation (Figure 4A).
MiR-139-3p, miR-199a-3p, and their targets are aberrantly expressed in FA. (A) Luciferase reporter assays containing the 3′-UTR sequences of indicated targets of miR-139-3p or miR-199a-3p with WT or mutated miRNA binding sites. Luciferase activities relative to the EV control are shown. Error bars represent the SD of 3 experiments. The significance was calculated with the Student t test (2-tailed) (*P < .05). (B) Expression of miR-139-3p and miR-199a-3p normalized to U6 in BM CD34+ cells isolated from healthy individuals (nBM) and FA patients. Data are plotted relative to the average of nBM. Patients with severe BMF are indicated with a circle. The bars show the median relative expression of each group. The significance was calculated with the Mann-Whitney U test (asymptotic significance [2-tailed], *P< .05). (C) Transcript levels of indicated miR-139-3p or miR-199a-3p targets, TP53, and ACTB in low-density mononuclear BM cells from 21 FA patients and 11 healthy volunteers (normal) are shown. Data were taken from the FA Transcriptome Consortium database (Gene Expression Omnibus, 2009 and publicly available as GSE16334). The bars indicate the mean relative expression of each group. The significance was calculated with the non-paired Student t test (asymptotic significance [2-tailed], *P < .05). nBM, normal BM.
MiR-139-3p, miR-199a-3p, and their targets are aberrantly expressed in FA. (A) Luciferase reporter assays containing the 3′-UTR sequences of indicated targets of miR-139-3p or miR-199a-3p with WT or mutated miRNA binding sites. Luciferase activities relative to the EV control are shown. Error bars represent the SD of 3 experiments. The significance was calculated with the Student t test (2-tailed) (*P < .05). (B) Expression of miR-139-3p and miR-199a-3p normalized to U6 in BM CD34+ cells isolated from healthy individuals (nBM) and FA patients. Data are plotted relative to the average of nBM. Patients with severe BMF are indicated with a circle. The bars show the median relative expression of each group. The significance was calculated with the Mann-Whitney U test (asymptotic significance [2-tailed], *P< .05). (C) Transcript levels of indicated miR-139-3p or miR-199a-3p targets, TP53, and ACTB in low-density mononuclear BM cells from 21 FA patients and 11 healthy volunteers (normal) are shown. Data were taken from the FA Transcriptome Consortium database (Gene Expression Omnibus, 2009 and publicly available as GSE16334). The bars indicate the mean relative expression of each group. The significance was calculated with the non-paired Student t test (asymptotic significance [2-tailed], *P < .05). nBM, normal BM.
miR-139-3p, miR-199a-3p, and their targets are deregulated in human FA BM cells
Because defective ICL repair is a major hallmark of FA,5 we assessed the expression of miR-139-3p and miR-199a-3p in CD34+ BM progenitor cells from 8 FA patients (supplemental Table 4). Similar to the Ercc1-/*292 lin− cells, miR-139-3p expression was higher in CD34+ cells from FA patients (who exhibited no signs for malignant transformation) compared with healthy individuals (median = 14.5-fold [P < .05]) (Figure 4B). Expression of miR-199a-3p was significantly higher in FA patients with a severe BMF compared with controls (median = 15.7-fold [P < .05]) than in patients with mild BMF (Figure 4B; supplemental Table 4), suggesting a correlation between the severity of ICL-induced damage and the expression of miR-199a-3p in FA-BM.
RNA-seq analysis on BM CD34+ cells from an FA patient with severe BMF showed that the targets of miR-139-3p and miR-199a-3p identified in the Ercc1-deficient mouse model were also downregulated in this patient compared with normal CD34+ cells (supplemental Figure 2). Conversely, expression of GADD45B and CDKN1A, two TP53-controlled DDR genes, was significantly increased in FA BM CD34+ cells (supplemental Figure 2). To corroborate these results in a larger group of patients, we analyzed the FA Transcriptome Consortium database (Gene Expression Omnibus, 2009, publicly available as GSE16334), containing the transcriptome data of low-density mononuclear BM cells from 21 FA patients and 11 healthy donors.38 In this data set, expression of the target genes of miR-139-3p and miR-199a-3p was again significantly lower in FA patients relative to healthy controls (Figure 4C), whereas the DDR-induced genes GADD45B and CDKN1A were upregulated (Figure 4C).
Loss of Trp53 uncovers the leukemogenic nature of Ercc1 deficiency
Having established that the genetic deletion of Trp53 rescued Ercc1-/*292 HSPCs from exhaustion, we investigated whether the loss of one Trp53 allele promotes the leukemic transformation of Ercc1-deficient HSPCs. From 51 mice transplanted with Ercc1-/*292;Trp53+/− HSPCs, 41 died with an average latency of 28 weeks with symptoms of leukemia, including enlarged liver, spleen, or thymus. In contrast, mice transplanted with Ercc1+/*292;Trp53+/− or Ercc1+/−;Trp53+/− survived without symptoms (Figure 5A). Of the 32 leukemic mice that could be fully analyzed, all had symptoms of acute leukemia. With few exceptions, these leukemic blast cells expressed T-cell receptor α/β rearrangements, and CD4 and CD8, indicative of T-cell leukemia (data not shown).
Development of leukemia in Ercc1-/*292;Trp53+/− transplanted mice. (A) Kaplan–Meier plots showing overall survival of Ercc1-/*292;Trp53+/−, Ercc1+/−Trp53+/−, and Ercc1+/*292Trp53+/−. (B) Heat map showing consistent patterns of CNV. The CNVs are shown for each leukemia sample (indicated by a 9-digit number, eg, 11-11713-01) along the genome. Autosomal chromosomes (1 to 19) are indicated on the left. Blue: loss of genetic material; Red: gain of genetic material; and White: retention of genetic material. (C) LOH along the genomes from isolated leukemias. In all samples, chromosome 11 harbors partial or complete LOH. Trp53 is located within regions affected by LOH, leading to homozygous deletions. Blue: LOH; Yellow: retention of heterozygosity; and White: no informative single nucleotide polymorphisms. (D) Expression of miR-139-3p and miR-199a-3p normalized to U6 in cells isolated from Ercc1-/*292 leukemias or WT lin− controls. Bars show the mean relative expression of each group. Significance was calculated with the Mann-Whitney U test (asymptotic significance [2-tailed], *P < .05).
Development of leukemia in Ercc1-/*292;Trp53+/− transplanted mice. (A) Kaplan–Meier plots showing overall survival of Ercc1-/*292;Trp53+/−, Ercc1+/−Trp53+/−, and Ercc1+/*292Trp53+/−. (B) Heat map showing consistent patterns of CNV. The CNVs are shown for each leukemia sample (indicated by a 9-digit number, eg, 11-11713-01) along the genome. Autosomal chromosomes (1 to 19) are indicated on the left. Blue: loss of genetic material; Red: gain of genetic material; and White: retention of genetic material. (C) LOH along the genomes from isolated leukemias. In all samples, chromosome 11 harbors partial or complete LOH. Trp53 is located within regions affected by LOH, leading to homozygous deletions. Blue: LOH; Yellow: retention of heterozygosity; and White: no informative single nucleotide polymorphisms. (D) Expression of miR-139-3p and miR-199a-3p normalized to U6 in cells isolated from Ercc1-/*292 leukemias or WT lin− controls. Bars show the mean relative expression of each group. Significance was calculated with the Mann-Whitney U test (asymptotic significance [2-tailed], *P < .05).
To identify somatic insertions/deletions (indels), 17 leukemia samples and germ-line control DNAs isolated from the brains of 6 donor mice were subjected to WES. Because the chromosomal damage caused by ICLs is repaired by error-prone nonhomologous end-joining rather than homologous recombination, leukemia genomes from patients deficient in ICL-repair frequently contain gross genetic aberrations.39,40 We developed an algorithm (M.A.S., R.H., and P.J.M.Valk, manuscript submitted 2015), allowing for the identification of these abnormalities based on WES data sets. Different patterns of copy number variations (CNVs) were seen, such as gains on chromosomes 4, 5, 11, and 15 and losses on chromosomes 3, 6-8, 13, 16, and 19 (Figure 5B). However, the copy number neutral loss of heterozygosity (LOH) of chromosome 11 caused complete loss of Trp53 in all leukemia samples analyzed (Figure 5B-C). Expression of miR-199a-3p was markedly increased (3.14-fold, P < .05) in Ercc1-/*292 leukemia cells and miR-139-3p was not detectable (Figure 5D). miR-199a-3p is expressed from 3 genes, miR-199a1, miR-199a2, and miR-199b, localized on mouse chromosome 9, chromosome 1, and chromosome 2, respectively. miR-139 is located on mouse chromosome 7. However, neither of these loci harbored genomic abnormalities in the tumors analyzed (n = 12).
miR-199a contributes to the development of AML
Because the leukemias arising in this model are mostly of T-cell origin, we sought to investigate the consequences of enhanced miR-199a-3p in a mouse model prone to developing AML. We reasoned that a Cebpa conditional knockout model is especially suitable to investigate the leukemogenic potential of miR-199a. Cebpa knockout mice develop leukemia only when secondary oncogenic events are introduced in hematopoietic stem and progenitors.41,42 Moreover, CEBPA expression is significantly decreased in FA (Figure 4C). We used the CebpaCre/fl mouse model in which the expression of Cre-recombinase is driven by the Cebpa promoter.27 The Cre enzyme recombines the floxed Cebpa allele, resulting in the loss of Cebpa mainly in myeloid precursors. This leads to a differentiation block at the CMP level.41 Fetal liver CebpaCre/fl HSPCs were transduced with MSCV-EGFP control virus, MSCV-EGFP–miR-199a, or -miR-106, a non-oncogenic miRNA that induces expansion of HSPCs,28,43 and transplanted in irradiated recipients (Figure 6A). All mice transplanted with miR-199a transduced HSPCs (n = 5) developed leukemia within 60 to 200 days, whereas no malignancies occurred in mice transplanted with control cells (EV, n = 9 or miR-106, n = 4) (Figure 6B). Transplantation of the leukemic cells in secondary recipients caused leukemia with a shortened latency (60 to 150 days) and more than 95% blasts in the BM (Figure 6B, supplemental Table 5). The complete loss of Cebpa expression in EGFP-positive leukemia cells was confirmed by Cebpa locus specific PCR and Cebpa expression analysis of leukemia samples (data not shown). Leukemic mice had splenomegaly, severe anemia, high percentages (70% to 90%) of blast cells in the BM (Figure 6C; supplemental Table 5), and leukemic infiltration in peripheral blood, liver, and spleen (Figure 6D-E). All leukemia samples expressed c-Kit and/or Sca1 (Figure 6F). They lacked markers for terminal myelo-monocytic differentiation (CD11b, GR1), T cells (CD4, CD8), B-cells (CD19), or erythroid cells (TER-119) (data not shown).
Forced expression of miR-199a drives leukemogenesis in mice. (A) Schematic overview of the HSC transplantation experiment. HSPCs were isolated from CebpaCre/fl fetal livers and infected with MSCV-GFP-miRNA or with MSCV-GFP-EV control viruses. Recipient mice were lethally irradiated (8.5 Gy) and transplanted with transduced cells by tail-vein injection. (B) Cumulative survival of mice transplanted with lin− cells expressing GFP, with miR-199a (n = 5) (P < .0005 compared with EV n = 9), miR-106 (n = 4) (a non-oncogenic miRNA that promotes myeloid progenitor expansion),28,43 GFP only, or secondary recipients of miR-199a leukemia cells (n = 7) (miR-199 compared with EV P < .0005, and secondary transplants compared with primary tumors P < .03). Statistical significance was calculated with the log-rank Mantel–Cox test. (C) Splenomegaly and anemic femurs and tibiae isolated from miR-199a–transplanted leukemic mice. (D) Fluorescence-activated cell sorter plots showing the percentage of GFP-expressing AML cells in the BM, blood, liver, and spleen of the leukemic mice. (E) Micrographs showing the morphology of leukemic blasts in the different hematologic organs (BM, blood, and spleen). Black bar indicates 10 µm. (F) Flow cytometric analysis of GFP-positive BM cells. GFP-positive AML cells from mouse 2 are an example of a stem cell-like phenotype (c-Kit, Sca1 double-positive), whereas the AML cells from mouse 3 have a progenitor-like (c-Kit high, Sca1 low) phenotype. GFP, green fluorescent protein; LTR, long terminal repeat; SSC-A, side scatter channel-A.
Forced expression of miR-199a drives leukemogenesis in mice. (A) Schematic overview of the HSC transplantation experiment. HSPCs were isolated from CebpaCre/fl fetal livers and infected with MSCV-GFP-miRNA or with MSCV-GFP-EV control viruses. Recipient mice were lethally irradiated (8.5 Gy) and transplanted with transduced cells by tail-vein injection. (B) Cumulative survival of mice transplanted with lin− cells expressing GFP, with miR-199a (n = 5) (P < .0005 compared with EV n = 9), miR-106 (n = 4) (a non-oncogenic miRNA that promotes myeloid progenitor expansion),28,43 GFP only, or secondary recipients of miR-199a leukemia cells (n = 7) (miR-199 compared with EV P < .0005, and secondary transplants compared with primary tumors P < .03). Statistical significance was calculated with the log-rank Mantel–Cox test. (C) Splenomegaly and anemic femurs and tibiae isolated from miR-199a–transplanted leukemic mice. (D) Fluorescence-activated cell sorter plots showing the percentage of GFP-expressing AML cells in the BM, blood, liver, and spleen of the leukemic mice. (E) Micrographs showing the morphology of leukemic blasts in the different hematologic organs (BM, blood, and spleen). Black bar indicates 10 µm. (F) Flow cytometric analysis of GFP-positive BM cells. GFP-positive AML cells from mouse 2 are an example of a stem cell-like phenotype (c-Kit, Sca1 double-positive), whereas the AML cells from mouse 3 have a progenitor-like (c-Kit high, Sca1 low) phenotype. GFP, green fluorescent protein; LTR, long terminal repeat; SSC-A, side scatter channel-A.
Discussion
In this study, we used Ercc1-deficient mice as a model to identify mechanisms involved in ICL-driven BMF and leukemic progression. Similar to the observations in FA patients,22 we found that Trp53 is crucially involved in the exhaustion of HSPCs in Ercc1-/*292 mice and that deletion of Trp53 restored the colony-forming capacity and the LSK content of Ercc1-/*292 HSPCs. In contrast, this was not achieved by the deletion of the Cdkn2a locus, which encodes the cyclin-dependent kinase inhibitor p16INK4a.44 Although being a major effector of BMF caused by the loss of ATM protein,30 p16INK4a thus appears dispensable for ICL-induced DDR in HSPCs. Cdkn2a also encodes p19ARF, which sequesters the TRP53 degradation protein MDM2. Hence, the lack of effects of Cdkn2a deletion on myeloid colony formation of Ercc1-/*292 HSPCs also argues against a dominant role of p19ARF/MDM2-controlled TRP53 stability in HSPCs.
The loss of Trp53 was a common genetic abnormality in the Ercc1-/*292 leukemia genomes. This is in line with studies showing that deletion of Trp53 cooperates with the loss of Fancc or Fancd2 in tumorigenesis.45,46 TP53 mutations are rare in FA progression toward myelodysplastic syndrome/AML.40 However, this does not exclude that TP53 activities or epigenetic silencing of critical target genes of TP53 may occur in these patients.
In addition, we found that miR-139-3p and miR-199a-3p, two miRNAs with a poorly characterized function in hematopoiesis, play opposite roles in the expansion of ICL repair-deficient HSPCs. We identified HuR as a major target of miR-139-3p, and blocking of miR-139-3p activity by antagomiRs partly restored myeloid colony formation from Ercc1-/*292 BM cells, showing its importance for the loss of HSPCs during ICL-stress. How HuR, an RNA-binding protein, protects ICL-damaged HSPCs from exhaustion is currently unknown. Previous studies have shown that HuR has a broad pro-survival function in hematopoietic progenitors by controlling the expression of Bcl-2, Bcl-xl, Survivin, Caspase-9, Noxa, and Puma.47,48 In contrast, the expression of miR-199a-3p antagonized the inhibitory effects of miR-139-3p expression in Ercc1-deficient HSPCs. Notably, miR-199a expression in 32D cells causes a proliferative advantage in response to CSF3, but not IL-3, suggesting that miR-199a inhibits myeloid differentiation kinetics, thereby causing an enhanced proliferation/self-renewal rate of 32D cells.
CEBPA mutations have not been found in FA, but its expression is significantly reduced (Figure 4C). Forced expression of miR-199a-3p in Cebpa-deficient HSPCs gave rise to a transplantable leukemia in mice, establishing its role as an onco-miR and suggestive of a tumor suppressor function of its major target gene(s), Pon2, Prdx6, Suz12, and Runx1. PON2 and PRDX6 protect cells from oxidative stress49,50 and their repression may contribute to the excessive ROS levels in ICL-deficient progenitors.51 SUZ12 is a component of the polycomb repressor complex 2 (PRC2) involved in the silencing of multiple genes, including HOX genes.52 Indeed, expression of the PRC2 target HOXA11 was increased in FA samples, whereas expression of HOXA5, which is not controlled by PRC2,53 was not changed (supplemental Figure 3). miR-199a–mediated repression of Suz12 may, at least partly, explain the increased expansion of progenitors without impairing their differentiation capacity, in line with what has been reported in a heterozygous Suz12 (Suz12Plt8/wt) model.54 Additionally, miR-199a-3p–induced downregulation of Runx1, a transcription factor critical for normal hematopoiesis55 may contribute to leukemogenesis. Intriguingly, RUNX1 has been shown to protect HSPCs from oncogenic insults via a fail-safe mechanism involving BMI-1 that neutralizes oncogenic RAS signaling.56 Other targets of miR-199a-3p that have been implicated in leukemogenesis, ie, mTOR and CD4457-59 were not found in our study. We cannot exclude that these targets were missed for technical reasons, eg, the limited sensitivity of the GEP microarray and proteomics strategy used. Of note, miR-199a-3p has recently been shown to compete with pluripotency factors in the reprogramming of mouse embryonic fibroblasts to induced pluripotent stem cells.60 A difference between this and our study is that miR-199a-3p processing in mouse embryonic fibroblasts depended on TRP53, contrary to what was observed in Ercc1-deficient leukemia cells (Figure 2C). Finally, decreased expression of hsa-miR-181c was reported in EBV-transformed B cells and in peripheral blood mononuclear cells from FA patients relative to healthy subjects.61 We did not observe decreased hsa-miR-181c expression in Ercc1-deficient HSPCs, which may be due to differences between species or cell types investigated.
In conclusion, we have presented evidence for a model in which ICL-stress causes a loss of HSPCs through the combinatorial actions of both Trp53 and miR-139-3p (Figure 7). The elevated expression of miR-199a-3p also seen at this stage is insufficient to compensate for the exhaustion of HSPCs, but when expression of Trp53 and miR-139-3p is low or absent, miR-199a-3p expression promotes HSPC proliferation. Furthermore, miR-199a-3p acts as an onco-miR that promotes the development of AML in the Cebpa-deficient mice. Whereas these findings have implications for understanding leukemic progression of FA, the role of miR-139a and miR-199a may not be restricted to FA. For instance, a preliminary screen in myelodysplastic syndrome and AML revealed that miR-199a-3p is also expressed in a subset of these patients. This may open therapeutic avenues to reduce stem cell exhaustion and leukemic transformation, eg, by selective use of antagomiRs.
Model of how miRNAs control the balance between cell loss, cell survival and expansion, and leukemogenesis during BM stress. During homeostasis, miR-139-3p and miR-199a-3p are expressed at detectable levels in hematopoietic progenitors. The activity of TRP53, miR-139-3p, and miR-199a-3p is induced by ICLs. TRP53 and miR-139 causes the loss of HSPCs, whereas elevated levels of miR-199a-3p only partially compensate cell loss by stimulating HSPC proliferation. Targets of miR-139-3p and miR-199a-3p, Suz12, Runx1, Pon2, and Prdx6 and HuR are downregulated at this stage. Trp53 and miR-139-3p are lost in leukemias, whereas miR-199a-3p is highly expressed. miR-199a-3p is an onco-miR and drives oncogenic transformation of HSPCs toward leukemia.
Model of how miRNAs control the balance between cell loss, cell survival and expansion, and leukemogenesis during BM stress. During homeostasis, miR-139-3p and miR-199a-3p are expressed at detectable levels in hematopoietic progenitors. The activity of TRP53, miR-139-3p, and miR-199a-3p is induced by ICLs. TRP53 and miR-139 causes the loss of HSPCs, whereas elevated levels of miR-199a-3p only partially compensate cell loss by stimulating HSPC proliferation. Targets of miR-139-3p and miR-199a-3p, Suz12, Runx1, Pon2, and Prdx6 and HuR are downregulated at this stage. Trp53 and miR-139-3p are lost in leukemias, whereas miR-199a-3p is highly expressed. miR-199a-3p is an onco-miR and drives oncogenic transformation of HSPCs toward leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge the personnel of the Erasmus Medical Center Experimental Animal Center for mouse care, Dr E. Rombouts, P. van Geel, and N. Spierenburg-Papazian for assistance with flow cytometry and cell sorting, Dr R. Beekman and Dr E. Bindels for experimental assistance, and E. Simons for assistance with the preparation of figures. Prof Dr B. Schumacher, Cologne Cluster of Excellence in Cellular Stress Responses in Aging-Associated Diseases, University of Cologne, Germany is gratefully acknowledged for providing Trp53- and Cdkn2a-deficient mouse models. The authors also thank Prof Dr D. Tenen for the floxed Cebpa mice. Some results are based upon data generated by The Cancer Genome Atlas managed by the National Cancer Institute and National Human Genome Research Institute (http://cancergenome.nih.gov).
This study was supported by the Dutch Cancer Society (KWF Kankerbestrijding). P.M.H.v.S. was supported by a ZonMw E-Rare grant. S.E.S. was supported by a KiKa Children Cancer Free grant.
Authorship
Contribution: M.F.A. and J.R.H. performed experiments and analyzed data; H.W.J.d.L., P.M.H.v.S., J.V.-O., and Y.C. performed experiments; R.H. and M.A.S. processed and analyzed next generation sequencing data; A.H.d.R., G.M.C.J., and P.A.v.V., designed and performed proteomics and analyzed proteomics data; S.E.S. and M.B.B. isolated and investigated BM samples from FA patients; M.v.L., I.P.T., and S.J.E. supervised the project; and M.F.A., J.R.H., I.P.T., and S.J.E. designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stefan J. Erkeland, Department of Hematology, Erasmus University Medical Center, Cancer Institute, Wytemaweg 80, 3015 CN, Rotterdam, The Netherlands; e-mail: s.erkeland@erasmusmc.nl; and Ivo P. Touw, Department of Hematology, Erasmus University Medical Center, Cancer Institute, Wytemaweg 80, 3015 CN, Rotterdam, The Netherlands; e-mail: i.touw@erasmusmc.nl.
References
Author notes
M.F.A. and J.R.H. contributed equally to this study.

![Figure 1. Loss of Trp53 but not of Cdkn2a, restores HSPC content in Ercc1-/*292 mice. (A, E) LSK frequencies in lin− fractions of indicated mouse genotypes at 3 weeks of age are shown. (B-D and F-H) CFUs per 5 × 104 unfractionated BM cells in the presence of indicated growth factors relative to the WT controls are shown. All bars represent the mean and standard deviations (SD) of n ≥3 mice. The significance was calculated by the Student t tests (asymptotic significance [2-tailed]), *P < .05].](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/25/10.1182_blood-2014-11-612507/4/m_3937f1.jpeg?Expires=1767780679&Signature=4~Q1bmcfKD8ip7FcZzca2jv5E4V0VpdCviIebwiev0GfysXln9O26DsX8NE~Z~2peJz1g-CJy-R6bztmN1vw35putZG-F9oPJ9IL1W7CsjQWSMtzm0CWmGIUD~-LTG2F3tsZ4GmQ~IJJeKAjVkNtE7PgzLA49z2-tuYrXUXZikCS6W82lg6DiSZIqb5J~i8i1k2Rlo1wGRVjulPqGvOLoJKLhSLwTIl277EdZglDDWL~m3ADPqlKL36FFRWjQfO595wZ70OnBLxDa6p7v9Ej5er4-A75Cu164u~IzptbLuLSxjlNwWEQyShOEBLpHjJwEK-gUc9XZ-DsCiGNgcR9MA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Differentially expressed miRNAs in Ercc1-/*292 cells. (A) The expression of miRNAs and U6 in lin− BM cells was determined by quantitative PCR in quadruplicate. The expression of indicated miRNAs relative to U6 and normalized to WT controls is depicted. The error bars represent the SD of 5 mice. (B) CMP (top), GMP (middle), and MEP (bottom). The expression of indicated miRNAs relative to U6 and normalized to WT is depicted for different hematopoietic progenitor subpopulations isolated from the BM of 20-week-old mice. The error bars represent SD of 3 measurements. (C) The expression of indicated miRNAs relative to U6 in lin− BM cells of 3-week-old Ercc1-/*292;Trp53 WT mice and Ercc1-/*292;Trp53−/−, and relative to WT controls is shown. All bars represent the mean and SD of n ≥ 3 mice. (D) The expression of miR-139-3p and miR-199a-3p relative to U6 in WT lin− cells and normalized to the control condition without MMC, is depicted for the indicated MMC concentrations. The error bars represent SD of 3 measurements. In all panels, the significance was calculated with the Mann-Whitney U test (asymptotic significance [2-tailed], *P < .05]. NE, not expressed; NS, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/25/10.1182_blood-2014-11-612507/4/m_3937f2.jpeg?Expires=1767780679&Signature=FQIuIA~eWhggzEvYV0OcM0-gA2ecpPZYCd6SpbeaOpWJzJQGBwoLAa6OBzDxXKLA0XsjkE2nyJRTYczPXPABntlVbwfgPW0yka4UKdzSjjbqbGbxMW~FCXVkz-An-6Yxy9NIHx1CQIh~pVKGzC1KS0Ls9klVsXIX2sJ7mBC14~n61ekXPmL6mRSrVZkTRPcd79NSzOy-XuXoqbJABrqwPCEYJGRIFjWg0znFAcNEzKzvIMxSG8fWaDfT0QSKxyjN~UvqBOEKbwUn0psQMT~2Z-EhtLTJce2UL5oBJxPsN-krVT2Ra-VF7TKM4q~4hUd9u-~X3H7SfCQ4~Ac6T-yQgw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. MiR-139-3p, miR-199a-3p, and their targets are aberrantly expressed in FA. (A) Luciferase reporter assays containing the 3′-UTR sequences of indicated targets of miR-139-3p or miR-199a-3p with WT or mutated miRNA binding sites. Luciferase activities relative to the EV control are shown. Error bars represent the SD of 3 experiments. The significance was calculated with the Student t test (2-tailed) (*P < .05). (B) Expression of miR-139-3p and miR-199a-3p normalized to U6 in BM CD34+ cells isolated from healthy individuals (nBM) and FA patients. Data are plotted relative to the average of nBM. Patients with severe BMF are indicated with a circle. The bars show the median relative expression of each group. The significance was calculated with the Mann-Whitney U test (asymptotic significance [2-tailed], *P< .05). (C) Transcript levels of indicated miR-139-3p or miR-199a-3p targets, TP53, and ACTB in low-density mononuclear BM cells from 21 FA patients and 11 healthy volunteers (normal) are shown. Data were taken from the FA Transcriptome Consortium database (Gene Expression Omnibus, 2009 and publicly available as GSE16334). The bars indicate the mean relative expression of each group. The significance was calculated with the non-paired Student t test (asymptotic significance [2-tailed], *P < .05). nBM, normal BM.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/25/10.1182_blood-2014-11-612507/4/m_3937f4.jpeg?Expires=1767780679&Signature=twf3AOa3y0Z2XYVnI0zdMFGr2L1II7mvQFK-IXIOlsat--HHbeT4fsuXBT7OeHYN21YZ3mtfaAo4y4C~rmDnksPnz7DIQ9ckGBes7epu0xJLUVd7a3dgZTfZxEVf0XU6--jX9MTS~jhACKGB4aZerY01fe8~MtVyQnoZWzL3PJrsL0pskcHM1qvOs2x0-SsEVbzkCeOpBPVPN~F00C119CBEEB19fZy9MerztV55Rr1RE0dVJ4fPTB524ji4GFbW8vvxH5kblUQ69RcU5LzInw-QUoMdzAo7mZ~dfzXtDiz0X~4jYc9IiDqRTaFwry0GRTQE-ILljuFzp2ee4Hbgfw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Development of leukemia in Ercc1-/*292;Trp53+/− transplanted mice. (A) Kaplan–Meier plots showing overall survival of Ercc1-/*292;Trp53+/−, Ercc1+/−Trp53+/−, and Ercc1+/*292Trp53+/−. (B) Heat map showing consistent patterns of CNV. The CNVs are shown for each leukemia sample (indicated by a 9-digit number, eg, 11-11713-01) along the genome. Autosomal chromosomes (1 to 19) are indicated on the left. Blue: loss of genetic material; Red: gain of genetic material; and White: retention of genetic material. (C) LOH along the genomes from isolated leukemias. In all samples, chromosome 11 harbors partial or complete LOH. Trp53 is located within regions affected by LOH, leading to homozygous deletions. Blue: LOH; Yellow: retention of heterozygosity; and White: no informative single nucleotide polymorphisms. (D) Expression of miR-139-3p and miR-199a-3p normalized to U6 in cells isolated from Ercc1-/*292 leukemias or WT lin− controls. Bars show the mean relative expression of each group. Significance was calculated with the Mann-Whitney U test (asymptotic significance [2-tailed], *P < .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/25/10.1182_blood-2014-11-612507/4/m_3937f5.jpeg?Expires=1767780679&Signature=1mMxG-UFMfqB9KqumgiMVkLbhqTGkticeIvKWzfEX4TSDpRduBK3rUVC-HXMmENa16aChlsvjxjIHPFNFEmk~RG4UWyS8HZ50U1DlpbDCOx4A87qiDoTOgTowAa4Ashz3489SBk1~RqBCLr2xYi9HyAQOvGDhLDDu7PPUVa9bJFufkXlEiHMs3FxqnHbZD0rVTk06xyk1kycl~k~DOS4HU~3SfXm43xTQcWpwKIX4jLOcjhvZ8XQV7rXeYLOa8ACTfc3~jHUgiJcWbbhz-gQnRqgv3jxUCCHx93mMw4f6zgdRkAQj84C~UABbB4DHtWO~GIUa~kt6GUFlsUNel6FeA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal