Key Points
SOX7 expression is silenced in common myeloid malignancies.
SOX7 interacts directly with β-catenin and regulates the Wnt pathway in acute myeloid leukemia.
Abstract
SOX7 belongs to the SOX (Sry-related high-mobility group [HMG] box) gene family, a group of transcription factors containing in common a HMG box domain. Its role in hematologic malignancies and, in particular, acute myeloid leukemia (AML) is completely unknown. Here, we showed that SOX7 expression was regulated by DNA hypermethylation in AML but not in acute lymphoblastic leukemia or normal bone marrow cells. In cell lines (KG1, ML2, and K562) and in primary CD34+ AML samples, SOX7 expression could be induced by the DNA demethylating agent 5-aza-2′-deoxycytidine. Overexpression of SOX7 in K562 cells inhibited cell proliferation, with cell cycle delay in S/G2/M phases and reduced clonogenic activity. Apoptosis was unaffected. Ectopic expression of SOX7 in K562 and THP-1 cells, as well as primary CD33+CD34+ AML cells, abrogated leukemia engraftment in xenogeneic transplantation. SOX7 expression inhibited the Wnt/β-catenin pathway through direct protein binding to β-catenin, and the antileukemia effects of SOX7 in THP-1 cells were significantly reduced by deletion of its β-catenin binding site. The results provided unequivocal evidence for a novel tumor suppressor role of SOX7 in AML via a negative modulatory effect on the Wnt/β-catenin pathway.
Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous diseases with distinct cytogenetic and genetic features. Aberrant DNA methylation plays an important role in leukemogenesis, and distinctive methylation profiles have been reported in cytogenetically defined AML.1,2 Recurrent mutations in epigenetic regulator protein, including DNMT3A and TET2, were recently reported and associated with inferior clinical prognosis.3,4 In fact, the treatment outcome of AML has been largely unsatisfactory, underscoring the need for new pathogenetic models that may provide targets for therapeutic intervention.
The SOX (Sry-related high-mobility group [HMG] box) genes belong to a family of transcription factors containing an HMG box domain that shares >60% homology to that in SRY.5 SRY (sex-determining region on the Y chromosome) was the first member in the family.6 The HMG box domain is associated with DNA binding and bending and protein interaction and transportation across the nuclear membrane. Most SOX proteins also contain functional domains outside the HMG box at the carboxyl and amino terminal regions, which are highly conserved among members of the same group. At present, more than 20 SOX genes in human and mouse genomes have been identified.7
Information about SOX gene function was derived mostly from gene targeting in mice and position cloning for human syndromes.8 SOX proteins are involved in diverse embryonic processes, including male sex determination6 ; neural,9 skeletal,10 and hair development11 ; definitive endoderm development12,13 ; cardiogenesis14 ; angiogenesis15 ; and, more recently, hematopoiesis.16 They are also expressed in different human cancer cell lines, including those of brain, lung, skin, gynecologic, and colonic origins.17 The pathogenetic pathways whereby SOX gene expression leads to human cancers are poorly understood; their role in AML is completely unknown.
SOX7 belongs to SOX gene subgroup F, which also includes the closely related genes SOX17 and SOX18. In Xenopus and zebrafish, sox7, together with sox18, has been implicated in cardiogenesis14 and vasculogenesis.18,19 Recently, Sox7 was demonstrated to be an essential regulator of early hematopoiesis and development of hematopoietic stem cells16 and was associated with tumorigenesis in human cancers.20 Deregulation of SOX7 was observed in various neoplasms, including prostate and colorectal cancer in which SOX7 expression was silenced by DNA hypermethylation.21 In prostate cancer, deletion was found in ∼50% of cases at the short arm of chromosome 8 (8p23.1) where SOX7 gene normally resides.22
In this study, we examined the hitherto unknown expression pattern, regulation, and pathogenetic mechanism of SOX7 in human myeloid malignancies. SOX7 gene was uniquely silenced in myeloid malignancies due to promoter methylation. Forced expression of SOX7 significantly reduced leukemia cell proliferation and leukemia initiation in a humanized mouse model, in association with reduced Wnt/β-catenin signaling. The results might provide us with novel insights into the mechanisms of AML leukemogenesis.
Materials and methods
Materials and cell lines
Unless otherwise specified, the sources of materials and cell lines are tabulated in supplemental Table 1, available on the Blood Web site. Collection of bone marrow (BM) and peripheral blood from patients and healthy donors, as well as umbilical cord blood (UCB) samples, has been approved by the institutional review board in accordance to the Declaration of Helsinki.
SOX gene expression
Total RNA was extracted from the mononuclear cell (MNC) fraction of BM or peripheral blood from normal donors and leukemia patients and of cell lines using the TRIzol reagent or RNeasy kit, treated with RNase-free DNase, and reverse-transcribed (RT) using the Superscript II Reverse Transcriptase with oligo(dT) primers. Expression of SOX genes were examined by gene-specific polymerase chain reaction (PCR) (supplemental Table 2). The precursor B-cell acute lymphoblastic leukemia (ALL) cell line Nalm20 that consistently expresses SOX7 (see below) was included in every experiment, and SOX7 expression in each sample was normalized with respect to Nalm20.
Plasmid preparation
Plasmids used in this study are tabulated in supplemental Table 3. Preparation of green fluorescent protein (GFP)-SOX and GFP-SOX7Δ is described in detail in supplemental Table 4.
5-AdC treatment
Primary AML and leukemia cell lines (KG-1, ML-2, K562, and THP-1) were seeded in a 6-well culture dish at 0.5 to 1.0 × 106 cells per milliliter. 5-Aza-2′-deoxycytidine (5-AdC; 2 μM in a 1:1 acetic acid/double-distilled H2O solution) was added to the culture medium. Fresh 5-AdC or vehicle containing medium was replenished every 24 hours.
Bisulfite modification, MS- PCR, and bisulfite sequencing
Genomic DNA from primary leukemic samples or cell lines was extracted, dissolved in Tris-EDTA buffer, stored at –20°C, and bisulfite treated during the experiment. Protocols of methylation-specific (MS)-PCR and bisulfite sequencing are described in supplemental Methods.
TOPFlash dual-luciferase assay
THP-1 and MV4-11 cells (2 × 106 cells) were nucleofected with 0.5 μg of either pOT-FLASH or pOF-FLASH plus 0.1 μg of pRL-CM, using Cell Line Nucleofector Kit V and L. pMax-GFP, pMax-GFP-SOX7, or pMax-GFP-SOX7Δ vector (1 μg) was conucleofected, and the cells were incubated overnight. Firefly luciferase activity from TOPFlash or FOPFlash was normalized to Renilla luciferase activity, and results were expressed as corrected TOPFlash minus FOPFlash.
Immunologic methods
To examine SOX7 and β-catenin protein expression, proteins extracted from K562 and THP-1 cell lines were separated with a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel, transferred to nitrocellulose membrane, and blotted with rabbit polyclonal anti-SOX7 or mouse monoclonal anti-β-actin antibodies. Mouse monoclonal antibody against total β-catenin was used for immunoprecipitation. The membranes were visualized by the chemiluminescent substrate method.
Virus packaging and transduction
Production of lentivirus and the transduction protocols are described in supplemental Methods.
In vitro cellular assays
A seminaphtorhodafluor (SNARF)-1 assay was performed by incubating 0.5 × 106 K562 cells with 1 μM SNARF-1 for 5 minutes, after which they were washed, resuspended in culture medium, and evaluated by flow cytometry 2 days later. Apoptosis assays were performed 48 hours after viral transduction. Colony-forming and thymidine-incorporation assays are described in the legends of respective figures.
Xenogeneic transplantation
Primary leukemia cells and K562 cells were suspended at various cell numbers in 200 μL of Hanks balanced salt solution supplemented with 2% fetal bovine serum. They were transplanted intrafemorally (primary leukemia cells) or via IV injection (K562 and THP-1 cells) into sublethally irradiated (250 cGy) NOD/SCID mice (NOD.CB17-Prkdcscid/J strain from The Jackson Laboratory, Bar Harbor, ME) at 6 to 8 weeks of age. Monoclonal anti-murine CD122 antibody, raised from the TM-β1 hybridoma cell line,23 was injected into peritoneum at 200 μg per mouse 1 day before transplantation. The studies were approved by the Committee of the Use of Live Animals for Teaching and Research of the University of Hong Kong.
Gene expression profiling
K562, ML2, and NB4 cells expressing GFP or GFP-SOX7 were sorted by fluorescence-activated cell sorting (FACS), and the total RNA of the cells was extracted with TRIzol reagent (Life Technologies). Extracted total RNA was purified using QIAGEN RNeasy MiniElute Cleanup Kit and quantified (Agilent 2100 Bioanalyzer). RNA labeling, array hybridization (human GeneChip; Affymetrix), staining, and scanning were performed according to the manufacturer’s protocol (Affymetrix). Array data were analyzed using GeneSpring GX v.11 (Agilent Technologies). The effect of SOX7 overexpression on gene expression patterns was compared by the signal ratio of SOX7-GFP to GFP. Genes consistently upregulated by more than 1.5-fold in all 3 cell lines were selected for further analysis and validated by quantitative PCR (supplemental Table 2) with SYBR Green assay (Applied Biosystems). Array data have been deposited in the Gene Expression Omnibus database (accession number GSE67817).
Statistical analysis
Comparisons between groups of data were evaluated by 2-tailed paired or unpaired Student t test for numerical data (SPSS, Chicago, IL). All statistical data were presented as mean ± standard error of the mean unless stated otherwise. A P value <.05 was considered statistically significant.
Results
SOX7 was silenced in myeloid malignancies
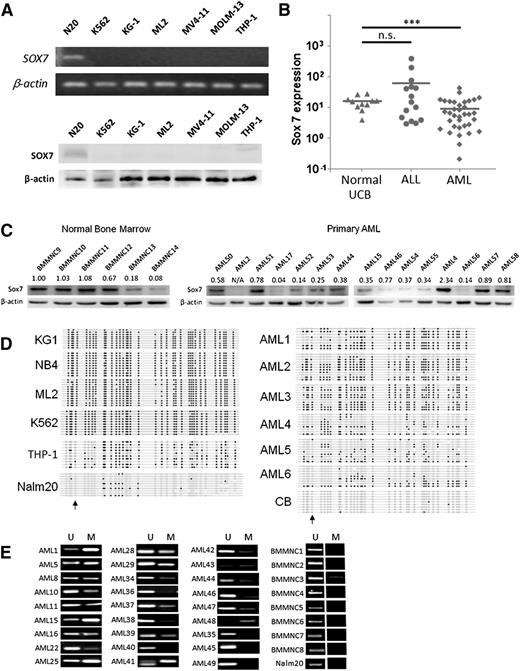
In an initial screen for SOX gene expression in AML, we examined the expression of 19 SOX genes in primary BM MNCs (BMMNCs) from patients with AML (n = 23), chronic myelogenous leukemia (CML; n = 13), myelodysplastic syndrome (MDS; n = 16), and ALL (n = 23), as well as in BMMNCs from healthy donors (n = 12). Cell lines derived from AML (NB4, KG1, and ML2), T-cell ALL (Jurkat and HPB-ALL), and precursor B-cell ALL (Nalm20) were also evaluated. Among the other SOX genes, SOX7 gene showed an interesting pattern: it was robustly expressed in most cases of normal BMMNCs and precursor B-cell ALL but consistently silenced in myeloid malignancies (supplemental Figure 1). Differential SOX7 gene and protein expression in myeloid and lymphoid leukemia cell lines was demonstrated by RT-PCR and western blot analysis (Figure 1A). Only precursor B-cell ALL, and not AML cell lines (K562, KG-1, ML2, MV4-11, MOLM-13, and THP-1), expressed SOX7. The results were further validated in primary AML myeloblasts by quantitative real-time RT-PCR: SOX7 was significantly downregulated in AML compared to CD34+ cells from normal UCB and ALL (Figure 1B). The protein expression of SOX7 was further examined in normal BM and AML samples. With an arbitrary cutoff of 0.5 (band-intensity ratio of Sox7 to β-actin, normalized by the ratio of BMMNC9), 4 of 6 normal BM samples (compared to only 6 of 15 AML samples) expressed SOX7 at the protein level (Figure 1C) (P = .014, χ2 test), supporting the proposition that SOX7 expression is preferentially silenced in AML at both the transcript and protein levels.
SOX7 expression was downregulated in myeloid malignancies. (A) SOX7 transcript (upper) and protein (lower) were downregulated in AML cell lines (K562, KG-1, ML2, MV4-11, MOLM-13, and THP-1) as compared to Nalm20 (N-20). (B) Expression of SOX7 transcript was measured by quantitative real-time RT-PCR in AML (n = 35), normal UCB CD34+ cells (n = 11), and ALL (n = 13), which was carried out by SYBR Green method in a StepOnePlus thermal cycler (Applied Biosystems, Foster City, CA). SOX7 was significantly downregulated in AML samples. The bars show the mean of expression, and comparisons were evaluated by unpaired Student t test. ***P < 0.001, n.s., not significant. (C) SOX7 protein was more commonly expressed in normal BM samples compared to primary AML samples. (D) Bisulfite sequencing on leukemia cell lines (left). Robust hypermethylation on SOX7 gene was observed in all myeloid leukemia cell lines but not in the precursor B-cell ALL cell line (Nalm20). DNA methylation could also be demonstrated in AML samples 1 to 6 but not in normal UCB CD34+ cells (right). Black dots, white dots, and arrows indicate methylated and unmethylated cytosine in CpG and the position of transcription start site, respectively. (E) DNA methylation of SOX7 in 27 AML and 8 normal BM samples and in the ALL cell line Nalm20 was demonstrated by MS-PCR. M, methylated allele; U, unmethylated allele.
SOX7 expression was downregulated in myeloid malignancies. (A) SOX7 transcript (upper) and protein (lower) were downregulated in AML cell lines (K562, KG-1, ML2, MV4-11, MOLM-13, and THP-1) as compared to Nalm20 (N-20). (B) Expression of SOX7 transcript was measured by quantitative real-time RT-PCR in AML (n = 35), normal UCB CD34+ cells (n = 11), and ALL (n = 13), which was carried out by SYBR Green method in a StepOnePlus thermal cycler (Applied Biosystems, Foster City, CA). SOX7 was significantly downregulated in AML samples. The bars show the mean of expression, and comparisons were evaluated by unpaired Student t test. ***P < 0.001, n.s., not significant. (C) SOX7 protein was more commonly expressed in normal BM samples compared to primary AML samples. (D) Bisulfite sequencing on leukemia cell lines (left). Robust hypermethylation on SOX7 gene was observed in all myeloid leukemia cell lines but not in the precursor B-cell ALL cell line (Nalm20). DNA methylation could also be demonstrated in AML samples 1 to 6 but not in normal UCB CD34+ cells (right). Black dots, white dots, and arrows indicate methylated and unmethylated cytosine in CpG and the position of transcription start site, respectively. (E) DNA methylation of SOX7 in 27 AML and 8 normal BM samples and in the ALL cell line Nalm20 was demonstrated by MS-PCR. M, methylated allele; U, unmethylated allele.
SOX7 silencing in AML by promoter CpG island methylation
SOX7 gene is located at the short arm of chromosome 8 (8p23.1), and deletion was found in ∼50% of prostate cancer.22 However, fluorescence in situ hybridization did not reveal SOX7 deletion in myeloblasts from either CD34+- or CD34−-purified primary AML samples (supplemental Figure 2A). We analyzed in silico the genomic sequence of SOX7 spanning −1700 to +4500 bps of transcription start site and identified 2 cytosine guanine dinucleotide (CpG) islands at the 5′ regulatory and the coding region (supplemental Figure 2B). CpGs were densely clustered around the transcription/ translation start site. Bisulfite sequencing of the genome region from −43 to +250 bps of transcription start site demonstrated CpG island hypermethylation in NB4, KG1, ML2, K562, and THP-1 cell lines and in primary AML samples but not in Nalm20 and normal UCB in which SOX7 was robustly expressed (Figure 1D). MS-PCR demonstrated promoter methylation in 22 of 27 primary AML samples, in 1 of 8 normal BMMNCs, and none in Nalm20 (Figure 1E).
SOX7 expression in myeloid malignancies could be induced by DNA demethylation
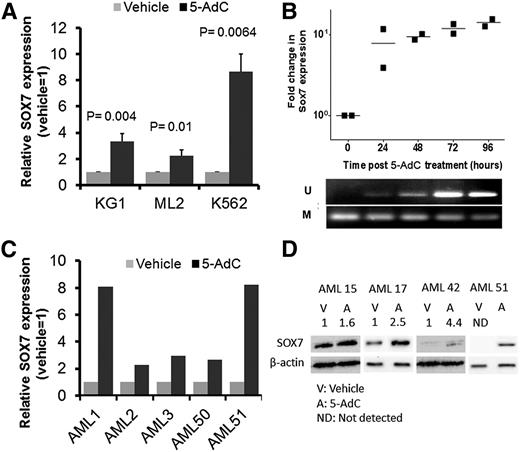
To elucidate the regulatory role of DNA methylation in SOX7 expression, we examined SOX7 expression in KG1, ML2, and K562 cells, as well as in CD34+ myeloblasts from primary AML samples upon treatment with DNA demethylating agent 5-AdC (2.0 μM) for 96 hours. 5-AdC treatment induced SOX7 expression in all 3 cell lines as compared to vehicle control (1:1 acetic acid/double-distilled H2O at 0.05% vol/vol) (Figure 2A). 5-AdC treatment of KG1 at different time points showed that SOX7 expression, as referenced to the baseline at time 0, was correlated with DNA demethylation (Figure 2B). The apparent smaller response to 5-AdC in Figure 2A was at least partially accounted for by an increase in SOX7 expression upon treatment with the vehicle control itself (supplemental Figure 3) that acted as the reference based on which SOX7 expression response was calculated. As for primary CD34+ AML myeloblasts, the expression of SOX7 was consistently induced upon 5-AdC treatment (Figure 2C). SOX7 protein expression was also induced upon demethylation (Figure 2D).
SOX7 expression could be induced by DNA demethylation. (A) SOX7 transcript expression induced by DNA demethylating agent 5-AdC in AML cell lines KG1, ML2, and K562. Cells were treated with vehicle (control) or 5-AdC for 96 hours, and SOX7 expression in vehicle was arbitrarily set as 1. (B) SOX7 expression in KG-1 cells treated with 5-AdC was evaluated every 24 hours by quantitative real-time RT-PCR. SOX7 expression at 0 hours was normalized to 1 (top). Upregulation of SOX7 expression was temporally correlated with its promoter demethylation as shown by MS-PCR (bottom). (C) SOX7 transcript expression induced by 5-AdC in 5 primary AML samples. (D) 5-AdC induced SOX7 protein in 4 primary AML samples. The intensity ratios of SOX7 to β-actin (normalized to the vehicle control) are indicated above the western blot image.
SOX7 expression could be induced by DNA demethylation. (A) SOX7 transcript expression induced by DNA demethylating agent 5-AdC in AML cell lines KG1, ML2, and K562. Cells were treated with vehicle (control) or 5-AdC for 96 hours, and SOX7 expression in vehicle was arbitrarily set as 1. (B) SOX7 expression in KG-1 cells treated with 5-AdC was evaluated every 24 hours by quantitative real-time RT-PCR. SOX7 expression at 0 hours was normalized to 1 (top). Upregulation of SOX7 expression was temporally correlated with its promoter demethylation as shown by MS-PCR (bottom). (C) SOX7 transcript expression induced by 5-AdC in 5 primary AML samples. (D) 5-AdC induced SOX7 protein in 4 primary AML samples. The intensity ratios of SOX7 to β-actin (normalized to the vehicle control) are indicated above the western blot image.
SOX7 expression in K562 cells reduced proliferation
To examine the effects of SOX7 expression on leukemogenesis, K562 cells were lentivirally transduced with GFP-tagged SOX7 or GFP control plasmid and sorted by FACS on the basis of GFP expression. SOX7 expression was confirmed by western blot analysis (Figure 3A). Forced SOX7 expression in K562 significantly reduced cellular proliferation as shown by the reduction in cell number (Figure 3B), 3[H] thymidine incorporation (Figure 3C), and retention of SNARF-1 staining in culture (Figure 3D). It also significantly decreased G1 and increased S and G2/M phases of cell cycle (Figure 3E, upper image). SOX7 expression had no effect on apoptosis as enumerated by 7-aminoactinomycin D−/annexin V+ population (Figure 3E, lower image). We further evaluated the effects of SOX7 expression on the clonogenic activity of K562 cells. Expression of GFP or GFP-SOX7 in the colonies was confirmed by green fluorescence (Figure 3F), and SOX7 expression significantly reduced the clonogenic activity of K562 cells as shown by the reduced number of colonies (Figure 3G). Differentiation, on the basis of CD11b expression, was not affected (supplemental Figure 4A). 5-AdC treatment of K562 and THP-1 cells also significantly reduced their proliferation and clonogenic activity. The changes were associated with demethylation of SOX7 promoter (supplemental Figure 4B).
SOX7 expression in K562 cell line inhibited cell proliferation. (A) Overexpression of SOX7 protein in K562 was confirmed by western blot analysis. (B) SOX7 expression reduced the rate of cell growth in culture. Cell number was enumerated on days 0, 3, and 6 on the basis of trypan blue exclusion (n = 3 separate experiments). (C) SOX7 expression significantly reduced 3[H] thymidine uptake (n = 3 separate experiments). The 3[H] thymidine-incorporation assay was performed in which 0.1 × 106 transduced K562 cells were incubated in 200 μL of 3[H]thymidine-containing culture medium at 0.025 mCi/mL for 18 hours. Cells were applied on a Cytostar-T scintillating microplate (Perkin Elmer, Waltham, MA) using vacuum suction, dried at 55°C in an incubator overnight, and scintillant was thereafter added to the microplate. The counts-per-minute (cpm) reading was recorded using TopCount NXT (Perkin Elmer). (D) SOX7 expression significantly reduced the rate of cell division as shown by SNARF-1 staining (n = 4 separate experiments). Flow cytometric histograms on day 0 (white area) and day 2 (green area) were superimposed (inset). (E) GFP-SOX7 expression (upper) delayed cell cycle progression in S and G2/M phases (n = 3 separate experiments). Comparisons between GFP and GFP-SOX7: G1, P = .004; S/G2/M, P = .011. SOX7 expression (lower) had no effect on apoptosis on the basis of 7-aminoactinomycin D (7AAD)/annexin V staining. Representative records of 2 separate experiments. (F-G) SOX7 expression reduced the clonogenic activity of K562 cells. GFP and GFP-SOX7 expression was confirmed by the presence of green fluorescence (F), and SOX7 expression significantly reduced colony frequency (G) (n = 4 separate experiments). Clonogenic assay was performed by plating transduced K562 cells onto 35-mm tissue culture plates in triplicates of 1.0 × 103 cells/mL in MethoCult GF H4230 supplemented with 10% fetal bovine serum (STEMCELL Technologies, Vancouver, BC, Canada). Colonies were enumerated on day 12 after culture. Scale bars represent 1.0 mm.
SOX7 expression in K562 cell line inhibited cell proliferation. (A) Overexpression of SOX7 protein in K562 was confirmed by western blot analysis. (B) SOX7 expression reduced the rate of cell growth in culture. Cell number was enumerated on days 0, 3, and 6 on the basis of trypan blue exclusion (n = 3 separate experiments). (C) SOX7 expression significantly reduced 3[H] thymidine uptake (n = 3 separate experiments). The 3[H] thymidine-incorporation assay was performed in which 0.1 × 106 transduced K562 cells were incubated in 200 μL of 3[H]thymidine-containing culture medium at 0.025 mCi/mL for 18 hours. Cells were applied on a Cytostar-T scintillating microplate (Perkin Elmer, Waltham, MA) using vacuum suction, dried at 55°C in an incubator overnight, and scintillant was thereafter added to the microplate. The counts-per-minute (cpm) reading was recorded using TopCount NXT (Perkin Elmer). (D) SOX7 expression significantly reduced the rate of cell division as shown by SNARF-1 staining (n = 4 separate experiments). Flow cytometric histograms on day 0 (white area) and day 2 (green area) were superimposed (inset). (E) GFP-SOX7 expression (upper) delayed cell cycle progression in S and G2/M phases (n = 3 separate experiments). Comparisons between GFP and GFP-SOX7: G1, P = .004; S/G2/M, P = .011. SOX7 expression (lower) had no effect on apoptosis on the basis of 7-aminoactinomycin D (7AAD)/annexin V staining. Representative records of 2 separate experiments. (F-G) SOX7 expression reduced the clonogenic activity of K562 cells. GFP and GFP-SOX7 expression was confirmed by the presence of green fluorescence (F), and SOX7 expression significantly reduced colony frequency (G) (n = 4 separate experiments). Clonogenic assay was performed by plating transduced K562 cells onto 35-mm tissue culture plates in triplicates of 1.0 × 103 cells/mL in MethoCult GF H4230 supplemented with 10% fetal bovine serum (STEMCELL Technologies, Vancouver, BC, Canada). Colonies were enumerated on day 12 after culture. Scale bars represent 1.0 mm.
The link between SOX7 and β-catenin in AML
To evaluate the mechanisms whereby SOX7 expression reduced leukemia growth, its effects on the Wnt/β-catenin pathway (which plays a pivotal role in leukemogenesis) were evaluated.24 We nucleofected K562 cells with β-catenin together with either pMAX-GFP, pMAX-GFP-SOX7, or pMAX-GFP-SOX7Δ (deletion of SOX7 β-catenin binding site) plasmid (Figure 4A).25 Defective β-catenin binding of SOX7Δ was confirmed by coimmunoprecipitation (Figure 4B), suggesting that SOX7 may regulate Wnt activity through direct β-catenin binding. Interestingly, the level of β-catenin protein was unchanged in presence of SOX7 or SOX7Δ. Furthermore, their subcellular location in K562 cells was also unchanged. In particular, β-catenin was expressed predominantly in the cytoplasm in all cases, and GFP-SOX7 and GFP-SOX7Δ proteins were expressed in the nucleus (supplemental Figure 5). Differential binding of SOX7 and SOX7Δ to β-catenin was also seen in THP-1 cells in which β-catenin was expressed endogenously (supplemental Figure 6). Furthermore, although SOX7 expression significantly reduced β-catenin signaling, the effects of SOX7Δ were reduced, supporting the proposition that SOX7 inhibits Wnt signaling by direct binding to β-catenin in K562, THP-1, and MV4-11 cells (Figure 4C-E). To examine the direct linkage of SOX7 overexpression and the Wnt/β-catenin pathway, we transfected β-catenin and wild-type SOX7, SOX7Δ, or GFP (control) into K562 cells. Importantly, only SOX7, but not SOX7Δ or GFP, transfection significantly reduced the nonphosphorylated (active) form of β-catenin, providing a mechanistic basis for the reduced TOPFlash activity upon SOX7 overexpression in AML (Figure 4F). To correlate changes in β-catenin signaling and leukemia growth, overexpression of β-catenin was shown to reverse the antileukemia effects of SOX7 expression (Figure 4G). However, despite modest suppression of β-catenin signaling upon SOX7Δ expression (Figure 4C-E), the latter had no effect on leukemia growth in vitro, suggesting that a critical level of β-catenin suppression was required.
SOX7 inhibited Wnt/β-catenin activity in K562 cells. (A) DNA (upper) and protein alignment (lower) of SOX7 and SOX7-deleted β-catenin binding site (SOX7Δ). (B) K562 cells nucleofected with β-catenin and either GFP, GFP-SOX7, or GFP-SOX7Δ. Immunoprecipitation (IP) was performed using anti-β-catenin antibody, and both the input lysate and the pull-downs were subjected to western blot analysis using anti-SOX7 antibody. Whole cell lysates of the K562 were also subjected to western blot analysis using anti-β-catenin and SOX7 antibodies, with β-actin as a loading control. GFP-SOX7Δ showed diminished binding to β-catenin as compared to SOX7. SOX7 expression downregulated Wnt/β-catenin activity in K562 (C), THP-1 (D), and MV4-11 cells (E), as enumerated by TOPFlash, normalized with respect to GFP-transduced cells. Deletion of β-catenin binding site in SOX7Δ significantly reduced the inhibitory effects of SOX7 on β-catenin activity. TOPFlash activity in GFP-transduced cells was arbitrarily set as 1 (K562, n = 3; THP-1, n = 4; MV4-11, n = 3 separate experiments). (F) SOX7 overexpression in K562 significantly reduced the active form (nonphosphorylated) of β-catenin, whereas SOX7Δ overexpression had no effect. (G) SOX7 overexpression in THP-1 cells significantly reduced the cellular proliferation, and the effect could be reversed by β-catenin expression. SOX7Δ expression with or without β-catenin had no significant effect.
SOX7 inhibited Wnt/β-catenin activity in K562 cells. (A) DNA (upper) and protein alignment (lower) of SOX7 and SOX7-deleted β-catenin binding site (SOX7Δ). (B) K562 cells nucleofected with β-catenin and either GFP, GFP-SOX7, or GFP-SOX7Δ. Immunoprecipitation (IP) was performed using anti-β-catenin antibody, and both the input lysate and the pull-downs were subjected to western blot analysis using anti-SOX7 antibody. Whole cell lysates of the K562 were also subjected to western blot analysis using anti-β-catenin and SOX7 antibodies, with β-actin as a loading control. GFP-SOX7Δ showed diminished binding to β-catenin as compared to SOX7. SOX7 expression downregulated Wnt/β-catenin activity in K562 (C), THP-1 (D), and MV4-11 cells (E), as enumerated by TOPFlash, normalized with respect to GFP-transduced cells. Deletion of β-catenin binding site in SOX7Δ significantly reduced the inhibitory effects of SOX7 on β-catenin activity. TOPFlash activity in GFP-transduced cells was arbitrarily set as 1 (K562, n = 3; THP-1, n = 4; MV4-11, n = 3 separate experiments). (F) SOX7 overexpression in K562 significantly reduced the active form (nonphosphorylated) of β-catenin, whereas SOX7Δ overexpression had no effect. (G) SOX7 overexpression in THP-1 cells significantly reduced the cellular proliferation, and the effect could be reversed by β-catenin expression. SOX7Δ expression with or without β-catenin had no significant effect.
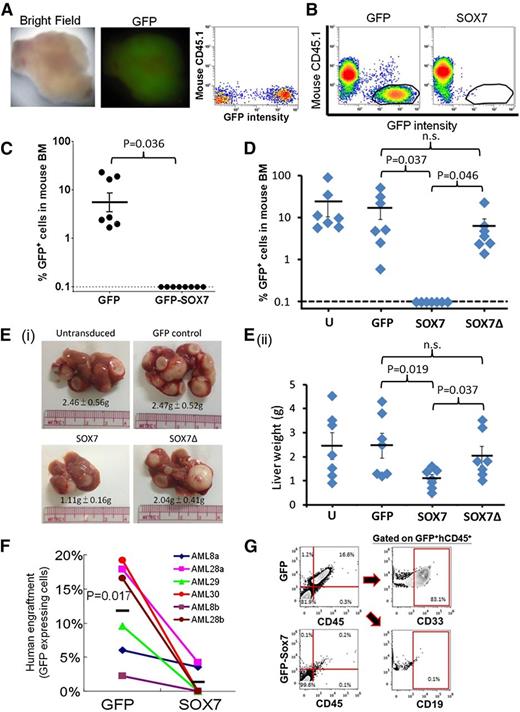
SOX7 expression reduced the repopulating potential of K562 cells and primary AML samples in immunodeficient NOD/SCID mice
We next examined the effect of SOX7 expression on the repopulating potential of IV injected K562 cells in sublethally irradiated NOD/SCID mice. In all experiments, GFP-transduced K562 cells engrafted in the recipient animals 6 weeks after transplant, and 3 of 7 mice developed myeloid sarcoma expressing GFP (Figure 5A). SOX7 expression completely abolished the repopulating potential of K562 cells (Figure 5B-C), and SOX7-transduced K562 cells did not develop myeloid sarcoma in mice. These observations could be recapitulated in THP-1 cells. Remarkably, the inhibitory effects on cellular proliferation (supplemental Figure 6C) and repopulating potential in BM (Figure 5D) and liver (Figure 5E) were significantly ameliorated in SOX7Δ-transduced THP-1 cells. To confirm the effects of SOX7 expression in primary AML myeloblasts, we isolated 0.2 to 0.5 × 106 CD33+CD34+ cells from AML patients and lentivirally transduced them with either GFP or GFP-SOX7 constructs. Transduced cells were selected by GFP expression (ranging from 10 to 100 × 103 cells) and transplanted via the intrafemoral route. GFP-expressing AML myeloblasts from different samples engrafted at different time points (range, 9-12 weeks) after transplantation. SOX7 expression consistently and significantly reduced leukemia-initiating cell (LIC) activity of these samples at equivalent cell doses (Figure 5F). To exclude the possibility that GFP expression of lentivirally transduced cells might be aberrantly silenced, recipient-mouse BM cells were stained with anti-human CD45 antibody. All engrafting GFP+ cells coexpressing human CD45 and GFP− cells were invariably negative for human CD45 (Figure 5G). Furthermore, the engrafting human CD45+ cells expressed only CD33 (and not CD19), an immunophenotype characteristic of the leukemic identity.
SOX7 expression reduced the repopulating potential of K562 and THP-1 cells and primary CD33+CD34+ AML samples in immunodeficient NOD/SCID mice. (A) A representative of the myeloid sarcomas detected in 3 of 7 mice transplanted with K562 cells (left). The excised tumor shows GFP positivity (middle). The GFP intensity in the sarcoma was detected by flow cytometry (right). (B) K562 cells engrafted in mouse BM were enumerated by flow cytometry as mouse CD45.1−/ GFP+ population (left, circled) and completely abolished by SOX7 expression (right; n = 3 separate experiments based on a total of 7 and 8 mice transplanted with GFP and GFP-SOX7 transduced cells, respectively). (C) A summary of GFP+ population in 3 sets of the experiment in panel B. SOX7 expression completely abolished the repopulating potential of K562 in mice. (D) SOX7 overexpression in THP-1 cells completely abolished engraftment in mouse BM, whereas SOX7Δ expression had no significant effect. (E) (i) Mice transplanted with untransduced, GFP-expressing, and SOX7Δ-expressing THP-1 cells resulted in leukemic deposits in liver, whereas in those with SOX7 overexpression, leukemia deposition was significantly less. (ii) Individual data on liver weights are shown. Mice were euthanized at 6 to 8 weeks. (F) Six CD33+CD34+ AML samples from 4 patients were lentivirally transduced with GFP or GFP-SOX7 and transplanted into irradiated NOD/SCID mice with equal cell doses. BM was aspirated at 6 and 9 weeks and harvested 12 weeks after transplantation. SOX7 expression nearly abolished leukemia engraftment. (G) Mouse BM cells stained by PC5-conjugated human CD45 antibody and subjected to flow cytometry show correlation between GFP and CD45 expression. In all xenotransplantation studies, successful leukemia engraftment was defined by the presence of GFP+, human CD45+, and murine CD45.1− cells in the recipient BM enumerated by flow cytometry. h, human.
SOX7 expression reduced the repopulating potential of K562 and THP-1 cells and primary CD33+CD34+ AML samples in immunodeficient NOD/SCID mice. (A) A representative of the myeloid sarcomas detected in 3 of 7 mice transplanted with K562 cells (left). The excised tumor shows GFP positivity (middle). The GFP intensity in the sarcoma was detected by flow cytometry (right). (B) K562 cells engrafted in mouse BM were enumerated by flow cytometry as mouse CD45.1−/ GFP+ population (left, circled) and completely abolished by SOX7 expression (right; n = 3 separate experiments based on a total of 7 and 8 mice transplanted with GFP and GFP-SOX7 transduced cells, respectively). (C) A summary of GFP+ population in 3 sets of the experiment in panel B. SOX7 expression completely abolished the repopulating potential of K562 in mice. (D) SOX7 overexpression in THP-1 cells completely abolished engraftment in mouse BM, whereas SOX7Δ expression had no significant effect. (E) (i) Mice transplanted with untransduced, GFP-expressing, and SOX7Δ-expressing THP-1 cells resulted in leukemic deposits in liver, whereas in those with SOX7 overexpression, leukemia deposition was significantly less. (ii) Individual data on liver weights are shown. Mice were euthanized at 6 to 8 weeks. (F) Six CD33+CD34+ AML samples from 4 patients were lentivirally transduced with GFP or GFP-SOX7 and transplanted into irradiated NOD/SCID mice with equal cell doses. BM was aspirated at 6 and 9 weeks and harvested 12 weeks after transplantation. SOX7 expression nearly abolished leukemia engraftment. (G) Mouse BM cells stained by PC5-conjugated human CD45 antibody and subjected to flow cytometry show correlation between GFP and CD45 expression. In all xenotransplantation studies, successful leukemia engraftment was defined by the presence of GFP+, human CD45+, and murine CD45.1− cells in the recipient BM enumerated by flow cytometry. h, human.
Potential target genes of SOX7 by gene expression profiling
To identify potential target genes of SOX7 in myeloid leukemia, the GFP-SOX7 and GFP plasmids were lentivirally transduced into 3 myeloid leukemia cell lines (K562, ML2, and NB4), and changes in gene expression were evaluated by microarray. Fifteen genes (including SOX7) exhibited upregulation by more than 1.5-fold upon SOX7 expression in all 3 cell lines, and selected genes were successfully validated by quantitative PCR and conventional RT-PCR (supplemental Table 5; supplemental Figure 7). SOX5 and SOX6 expression was significantly upregulated, suggesting that transcriptional transactivation is associated with SOX7 expression. Furthermore, Sprouty 3, a negative modulator of receptor tyrosine kinase signaling,26 was significantly upregulated, providing an important lead for studying the tumor suppressor function of SOX7. The functions of other genes upregulated upon SOX7 expression, with particular reference to tumor suppression, are currently unclear. Intriguingly, no gene was consistently downregulated by SOX7 overexpression.
Discussion
In this study, we demonstrated that SOX7 was silenced in myeloid malignancies including AML, CML, and MDS but was robustly expressed in ALL and in normal BM and UCB. Gene silencing was associated with promoter methylation and could be reversed by treatment with a demethylating agent. Both 5-AdC treatment and forced expression of SOX7 significantly reduced cellular proliferation and clonogenicity. Wnt/β-catenin signaling was significantly reduced and the effect was mediated at least partially by direct binding of SOX7 to β-catenin but not by protein degradation and subcellular localization of β-catenin. LIC activity in K562 and THP-1 cells, as well as in primary AML CD34+CD33+ myeloblasts, was significantly impaired. Therefore, our observation underscored the hitherto undescribed tumor suppressor function of SOX7 in AML and shed new light on the pathogenesis of this disease.
First, SOX7 expression in AML cell lines and primary samples could be induced 24 hours after 5-AdC treatment and temporally correlated with SOX7 promoter demethylation, implicating a direct demethylation of SOX7 promoter by 5-AdC. In fact, primary AML samples that were less methylated in the SOX7 CpG promoter region also had a higher, albeit variable, SOX7 protein expression compared to those with a higher level of methylation (supplemental Figure 8). Regulation of SOX7 expression by promoter CpG island methylation has also been reported in MDS27 and colorectal cancers.28 In contrast, promoter methylation at this genomic region was not detectable in primary CML samples (A.Y.H.L., unpublished data) in which SOX7 gene expression was also silenced (supplemental Figure 1), suggesting that promoter methylation at a more upstream genomic region or alternative mechanisms of gene regulation may operate in other myeloid malignancies.
Second, SOX7 expression by lentiviral transduction inhibited cell proliferation, clonogenicity, and leukemia propagation of K562 and THP-1, as well as of primary AML myeloblasts, supporting the tumor suppressor role of SOX7 in human AML. The effects were associated with cell cycle delay in S and G2/M phases. Apoptosis was not affected, suggesting that SOX7 expression specifically induced antiproliferative, but not general cytotoxic, effects. The level of SOX7 expression in leukemia cell lines was quantitatively comparable to endogenous SOX7 expression in normal CD34+ UCB cells (supplemental Figure 9). In colon cancers, by contrast, SOX7 expression was associated with both reduced cellular proliferation and increased apoptosis.28 Importantly, SOX7 expression abolished AML repopulation (a surrogate of LIC activity) when limiting cell doses of SOX7-transduced CD34+CD33+ myeloblasts were transplanted intrafemorally into NOD/SCID mice. Collectively, the data suggested that SOX7 expression had a tumor suppressor effect on both the LICs and downstream myeloblasts.
Third, we demonstrated for the first time a link between SOX7 and Wnt/β-catenin signaling in human leukemia. In human and murine myeloid malignancies, β-catenin is required for leukemic transformation of hematopoietic stem and progenitor cells and self-renewal of primitive leukemia stem cells.24,29 Therefore, the negative regulation of Wnt/β-catenin signaling by SOX7 expression provided a mechanistic link between SOX7 silencing, Wnt/β-catenin signaling, and leukemogenesis. SOX7 expression specifically reduced the active form of β-catenin by direct binding, and deletion of a β-catenin binding site in SOX7 significantly ameliorated its leukemia suppressive effect. β-catenin inhibition by SOX7 has also been recently reported in endometrial,30 colorectal,28 and prostate cancers.21 However, the function of SOX7 in lymphoid malignancies has not been investigated. The gene was expressed in ALL and may play a distinct role in leukemogenesis. In fact, knockdown of SOX7 significantly reduced leukemia initiation of ALL in xenotransplantation (H.W. and A.Y.H.L., unpublished data). In this context, a high level of canonical Wnt/β-catenin activity has been shown to inhibit leukemia growth and sensitize the leukemia cells to the cytotoxicity of precursor B-cell ALL,31 providing an important foundation for further research into the mechanistic link between SOX7, Wnt/β-catenin pathway, and leukemogenesis in this disease.
Lastly, gene expression profiling of 3 distinct myeloid leukemia cell lines overexpressing SOX7 was performed to examine the common target genes of SOX7 in myeloid leukemia. SOX7 overexpression resulted predominantly in activation, but not repression, of gene expression, consistent with the exclusive presence of the transactivation domain.32 Genes that have been induced by SOX7 expression in solid cancers33 were not identified in the present study, suggesting that SOX7 expression in myeloid leukemia might induce distinct gene expression profile and tumor suppression. SOX5 and SOX6 expression was upregulated, supporting the proposition that SOX7 could transactivate other SOX members in myeloid leukemia. In particular, the transcriptional function of SOX6 depends on its partnering with other proteins, including SOX5 and β-catenin,34 and more recent data have suggested that it might have a tumor suppressor effect in esophageal cancers.35 In fact, SOX6 expression was also silenced in the myeloid leukemia cell lines tested, as well as in half the AML and all the MDS and CML samples (supplemental Figure 1), suggesting a possible tumor suppressor function in myeloid malignancies. It is intriguing that the set of genes with unanimous upregulation upon SOX7 expression in all 3 leukemia lines was limited, underscoring the diversity of downstream signaling pathways in different leukemias and the lack of a common mechanistic pathway to tumor suppression. In addition to SOX6 and SOX5, Sprouty has been shown to modulate signaling of receptor tyrosine kinase, providing an important lead for future research of the tumor suppression function of SOX7 in AML.26
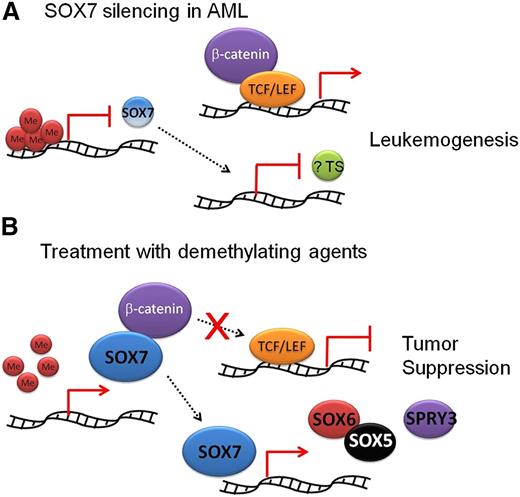
These observations have portrayed a novel pathogenetic role of SOX7 silencing in AML (Figure 6). SOX7 expression is normally silenced in AML by promoter CpG island methylation. Because SOX7 binds to β-catenin and modulates its activity, at least in some AML, unopposed Wnt/β-catenin signaling upon SOX7 gene silencing may result in leukemogenesis. Heterogeneity among AML, with some cases expressing a significant level of SOX7, was evident. Contamination by nonleukemic cells was unlikely because these samples were obtained by density-gradient centrifugation of blood or BM and comprised primarily blast population. Whether these SOX7-expressing AML samples represent a subgroup with distinct clinicopathologic and prognostic features should be further ascertained in larger cohorts of patients with uniform treatments. SOX7 promoter methylation has been correlated with inferior prognosis in MDS27 and lung cancers.36 Furthermore, deletion of β-catenin binding site in SOX7 did not completely reverse its inhibitory effects on Wnt/β-catenin signaling, suggesting that an alternative mechanism or mechanisms independent of direct β-catenin binding may exist. Previous studies have demonstrated that SOX17 bound T-cell factor (TCF) transactivators and promoted their proteasomal degradation,37 and that SOX6 could recruit histone deacetylase 1 to β-catenin and suppress its transcription.38 SOX-Neuro could also suppress β-catenin activity by competing TCF-binding site with TCF transactivator and stabilizing the TCF/Groucho repressor complex.39 Lastly, whether SOX7 expression in AML patients treated with hypomethylating agents may be used as a surrogate predictor of response should be further evaluated.
Proposed tumor suppressor role of SOX7 in AML. (A) SOX7 expression is normally silenced in AML by promoter CpG island hypermethylation. Wnt/β-catenin signaling is unopposed. SOX7 silencing might be associated with downregulation of other tumor suppressors (?TS). Both SOX7 suppression and Wnt/β-catenin activity might result in leukemogenesis. (B) When SOX7 was expressed, β-catenin was bound and Wnt/β-catenin signaling was checked. SOX7 might induce expression of other tumor suppressors, including SOX6 and Sprouty 3 (SPRY3). These changes were associated with an antileukemia effect on AML.
Proposed tumor suppressor role of SOX7 in AML. (A) SOX7 expression is normally silenced in AML by promoter CpG island hypermethylation. Wnt/β-catenin signaling is unopposed. SOX7 silencing might be associated with downregulation of other tumor suppressors (?TS). Both SOX7 suppression and Wnt/β-catenin activity might result in leukemogenesis. (B) When SOX7 was expressed, β-catenin was bound and Wnt/β-catenin signaling was checked. SOX7 might induce expression of other tumor suppressors, including SOX6 and Sprouty 3 (SPRY3). These changes were associated with an antileukemia effect on AML.
The array data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE67817).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The work was supported by the FACS Core Facility of the Li Ka Shing Faculty of Medicine, The University of Hong Kong, the Hong Kong Blood Cancer Foundation, and the General Research Fund (HKU771611); and by the Lady Tata Memorial Trust (T.K.F.). A.Y.H.L. is the Li Shu Fan Medical Foundation Professor in Haematology at the University of Hong Kong and received funding from its endowment.
Authorship
Contribution: C.H.M., H.W., T.K.F., and A.F. performed the experiments, analyzed the data, and wrote the manuscript; C.Y.C and T.S.K.W. performed some of the experiments; T.T. provided the anti-CD122 antibody required for xenotransplantation; C.W.E.S. and Y.L.K. analyzed the data and wrote the manuscript; N.N. and C.H. performed some of the experiments; and A.Y.H.L designed the research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anskar Y. H. Leung, Department of Medicine, Queen Mary Hospital, Room K418, K Block, Pok Fu Lam Rd., Hong Kong; e-mail: ayhleung@hku.hk.
References
Author notes
C.H.M., T.K.F., and H.W. contributed equally to this study.

![Figure 3. SOX7 expression in K562 cell line inhibited cell proliferation. (A) Overexpression of SOX7 protein in K562 was confirmed by western blot analysis. (B) SOX7 expression reduced the rate of cell growth in culture. Cell number was enumerated on days 0, 3, and 6 on the basis of trypan blue exclusion (n = 3 separate experiments). (C) SOX7 expression significantly reduced 3[H] thymidine uptake (n = 3 separate experiments). The 3[H] thymidine-incorporation assay was performed in which 0.1 × 106 transduced K562 cells were incubated in 200 μL of 3[H]thymidine-containing culture medium at 0.025 mCi/mL for 18 hours. Cells were applied on a Cytostar-T scintillating microplate (Perkin Elmer, Waltham, MA) using vacuum suction, dried at 55°C in an incubator overnight, and scintillant was thereafter added to the microplate. The counts-per-minute (cpm) reading was recorded using TopCount NXT (Perkin Elmer). (D) SOX7 expression significantly reduced the rate of cell division as shown by SNARF-1 staining (n = 4 separate experiments). Flow cytometric histograms on day 0 (white area) and day 2 (green area) were superimposed (inset). (E) GFP-SOX7 expression (upper) delayed cell cycle progression in S and G2/M phases (n = 3 separate experiments). Comparisons between GFP and GFP-SOX7: G1, P = .004; S/G2/M, P = .011. SOX7 expression (lower) had no effect on apoptosis on the basis of 7-aminoactinomycin D (7AAD)/annexin V staining. Representative records of 2 separate experiments. (F-G) SOX7 expression reduced the clonogenic activity of K562 cells. GFP and GFP-SOX7 expression was confirmed by the presence of green fluorescence (F), and SOX7 expression significantly reduced colony frequency (G) (n = 4 separate experiments). Clonogenic assay was performed by plating transduced K562 cells onto 35-mm tissue culture plates in triplicates of 1.0 × 103 cells/mL in MethoCult GF H4230 supplemented with 10% fetal bovine serum (STEMCELL Technologies, Vancouver, BC, Canada). Colonies were enumerated on day 12 after culture. Scale bars represent 1.0 mm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/25/10.1182_blood-2014-06-580993/4/m_3928f3.jpeg?Expires=1767750492&Signature=fNWnagIfTJpgA6q-vgwaj0gLkb06ejtcbZaGM7dGpbaUAVpdHATerWGjxQ5eEnDtpuRyxLHGdMW0ywXb-GXBLR6rDN~TgTFe3~qxfIw-X~9HFX3rDNL52NXNdHMHQLqI0Cd3U0n5p~YuXaVBQcEXhqn36W9yhNfR5AG-tjpk7j0ygv4lGcI2XTVrMIjwaFrPhI2vzD4uWHiPZ8UzJVPoD05WdVYDkbhrAI1DQxi7AKxlEUP71bp9la0eQlQnQHdGvFJVySxnBhxd0BfRduY3hFgzh0Auf9W1patrRKH2D2Ej29-CYLPnvs0hgTwPLQGIxe7CGTwZk8Wh4L2W-QMeaQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)