Key Points
Fbxl10 is a bona fide oncogene in vivo.
Fbxl10 overexpression in HSCs induces mitochondrial metabolic activation and enhanced expression of Nsg2.
Abstract
We previously reported that deficiency for Samd9L, which was cloned as a candidate gene for −7/7q− syndrome, accelerated leukemia cooperatively with enhanced expression of a histone demethylase: F-box and leucine-rich repeat protein 10 (Fbxl10, also known as Jhdm1b, Kdm2b, and Ndy1). To further investigate the role of Fbxl10 in leukemogenesis, we generated transgenic (Tg) mice that overexpress Fbxl10 in hematopoietic stem cells (HSCs). Interestingly, Fbxl10 Tg mice developed myeloid or B-lymphoid leukemia with complete penetrance. HSCs from the Tg mice exhibited an accelerated G0/G1-to-S transition with a normal G0 to G1 entry, resulting in pleiotropic progenitor cell expansion. Fbxl10 Tg HSCs displayed enhanced expression of neuron-specific gene family member 2 (Nsg2), and forced expression of Nsg2 in primary bone marrow cells resulted in expansion of immature cells. In addition, the genes involved in mitochondrial oxidative phosphorylation were markedly enriched in Fbxl10 Tg HSCs, coupled with increased cellular adenosine 5′-triphosphate levels. Moreover, chromatin immunoprecipitation followed by sequencing analysis demonstrated that Fbxl10 directly binds to the regulatory regions of Nsg2 and oxidative phosphorylation genes. These findings define Fbxl10 as a bona fide oncogene, whose deregulated expression contributes to the development of leukemia involving metabolic proliferative advantage and Nsg2-mediated impaired differentiation.
Introduction
Appropriate patterns of epigenetic alterations in histone modifications are required to assure cell identity, and their deregulation can contribute to human diseases such as cancer.1 We previously generated knockout mice for Samd9L (which was cloned as a candidate gene for the −7/7q− syndrome frequently that is observed in myelodysplastic syndrome and acute leukemia patients) and demonstrated that mice deficient in Samd9L developed leukemia after a long latent period.2 In addition, retroviral insertional mutagenesis revealed that the onset of the disease was highly accelerated with upregulation of F-box and leucine-rich repeat protein 10 (Fbxl10) or ectopic virus integration 1 (Evi1).2 Clinically, an elevated expression of Fbxl10 is observed in patients with acute myeloid leukemia (AML) or acute lymphoid leukemia (ALL), seminoma, and pancreatic ductal adenocarcinoma.3-6 These findings prompted us to further investigate the role of Fbxl10 in leukemia development in vivo.
Fbxl10 belongs to the JmjC domain-containing histone demethylases, and contains an N-terminal JmjC domain, followed by a CXXC zinc finger domain, a plant homeodomain finger, an F-box, and 8 leucine-rich repeats.7 Fbxl10 preferentially demethylates the dimethylation of histone H3 at lysine 36 (H3K36me2), but not H3K36me1 and H3K36me3.6,8 Fbxl10 was also recently reported to mediate the monoubiquitination of the histone H2A at lysine 119 as a component of noncanonical polycomb-repressive complex 1 in embryonic stem (ES) cells.9,10 Several lines of evidence have so far suggested that Fbxl10 is a tumor accelerator. Overexpression of Fbxl10 in fibroblasts inhibited cellular senescence by repressing p16ink4a-p19Arf and p15ink4b loci in mouse as well as Rb and p53 loci in humans, resulting in cellular immortalization.6,11 In addition, wild-type (WT) Fbxl10 but not a demethylase activity-deficient mutant, accelerated the progression of pancreatic cancer in a mouse allograft model.5 Fbxl10 is highly expressed in lineage marker (Lin)−, Sca-1+, c-Kit+ (LSK) cells and Lin− undifferentiated bone marrow (BM) cells, and forced expression of Fbxl10 in hematopoietic stem cells (HSCs) retained high BM repopulation capacity.12 Furthermore, another study demonstrated that the demethylase enzymatic activity was required for leukemic transformation in a Hoxa9-Meis1–induced mouse BM transplantation model.13 These findings suggest that Fbxl10 contributes to the development of tumor in vivo.
Genomic and functional studies have identified 2 broad classes of mutations that cooperate during the development of acute leukemia.14,15 Class I mutations confer a proliferative and/or survival advantage of HSCs and progenitor cells. Class II mutations are associated with impaired hematopoietic differentiation.14,15 As a simplified model, it has been proposed that growth advantage together with impaired differentiation is required for the development of the acute leukemia phenotype. In this study, we report that Fbxl10 transgenic (Tg) mice that overexpress Fbxl10 in HSCs spontaneously develop leukemia involving these class I/class II mutation-mimetic properties.
Methods
Generation of Fbxl10 transgenic mice
Murine Fbxl10 complementary DNA (cDNA) with a Flag tag at the 3′ end was inserted at the ClaI site of the Ly-6A transgenic cassette (pLy-6A14) that encompasses the mouse Sca-1 gene.16,17 A fragment containing the Sca-1 promoter, Fbxl10 cDNA with Flag tag and Sca-1 3′ locus control regions was excised and microinjected into the pronuclei of C57BL/6N mice. All the mice were kept according to the guidelines of the Institute of Laboratory Animal Science, Hiroshima University.
Statistics
Mouse survival curves were constructed using the Kaplan-Meier methodology and compared by the log-rank test using the GraphPad Prism software. Other statistical analyses were performed using the Student t test, unless otherwise stated.
A complete and detailed description of methods can be found in the supplemental Methods (available on the Blood Web site).
Results
Fbxl10 transgenic mice spontaneously develop myeloid and B-lymphocytic leukemias
The Sca-1 gene promoter is activated in functional repopulating adult HSCs in mice and has been, so far, widely available for Tg research.16-19 To achieve Tg expression of Fbxl10 in mice, the Flag-tagged mouse Fbxl10 cDNA was inserted into the cloning site of the Ly6A/Sca-1 Tg cassette that allows high expression in the HSC compartment17 (supplemental Figure 1A). We confirmed a >10-fold upregulation of Fbxl10 messenger RNA (mRNA) in Sca-1+ cells in 2 independent lines (lines 3 and 4) (supplemental Figure 1B), both of which provided similar results in this study (thus, hereafter we refer to mice in both lines as Fbxl10 Tg mice). We found that the expression levels of Fbxl10 mRNA were higher in Tg cells than control cells in all of the compartments, especially in HSC-early progenitor fractions (supplemental Figure 1C). This is consistent with the result of the initial study reporting that the same Sca-1 promoter used in this study is vigorously active predominantly in murine HSCs.17 The overexpression of the Fbxl10 protein was confirmed by immunoprecipitation followed by western blot using an anti-Flag antibody (supplemental Figure 1D). Immunoblot of extracted histones obtained from sorted Sca-1+ cells and differentiated Sca-1− cells showed a strong reduction of dimethylated H3K36 (H3K36me2) levels in the Tg cells, indicating an increased catalytic function of Fbxl10 in mice (supplemental Figure 1E). One possible explanation for decreased H3K36me2 levels in Sca-1− cells may be that Fbxl10 protein was sustained in Sca-1− progenitor and other differentiated populations.
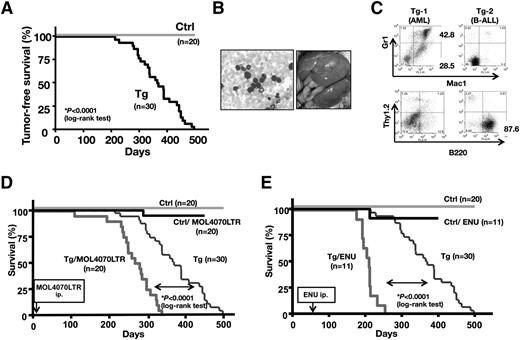
We followed 30 Tg and 20 control littermates over a 1.5-year period. Interestingly, all of the Fbxl10 Tg mice developed acute leukemias and died during the observation period (median survival = 367 days), whereas no hematopoietic disease was observed in the WT mice (Figure 1A). All moribund Tg mice showed leukocytosis and numerous circulating leukemic blasts in the peripheral blood (Figure 1B, left) and frequently displayed gross hepatosplenomegaly (Figure 1B, right). Leukemic cells from 14 of the 25 tested mice were abundantly positive for both Gr1 and Mac1 and were accordingly diagnosed as AML (Tg-1 in Figure 1C and supplemental Table 1). Seven mice were diagnosed as B-cell ALL (B-ALL), in which B220+ cells dominantly proliferated (Tg-2 in Figure 1C and supplemental Table 1). Regarding the remaining 4 leukemic mice, although surface marker analysis was not applicable, tumor tissues exhibited germline patterns in the TCRβ and IgH loci (supplemental Table 1). Therefore, we did not find any mice that had an expansion of Thy1.2 T-cell antigen-positive cells and/or rearrangements in TCRβ locus in the leukemic tissues, suggesting that Fbxl10 Tg mice exclusively developed myeloid and B-cell lineage leukemias. We intraperitoneally injected cells disaggregated from Tg leukemic spleens into immunodeficient NOD/SCID mice. The recipient mice exhibited splenomegaly as did the Fbxl10 Tg mice, indicating that leukemic cells were transplantable (supplemental Figure 2).
Transgenic expression of Fbxl10 in mice develop leukemia. (A) Kaplan-Meier survival plots for the Fbxl10 Tg (Tg) (n = 30) and control WT (Ctrl) (n = 20) mice. (B) Representative peripheral blood smear (left) and macroscopic appearance (right) of a leukemic Tg mouse. Proliferation of blast cells and massive hepatosplenomegaly are shown. (C) Flow cytometric profiles in the leukemic tissues of 2 Tg mice. TG-1 was positive for Mac1 and/or Gr1 positive but negative for Thy1.2 and B220 and diagnosed with AML, whereas TG-2 was positive for B220 but negative for Thy1.2, Gr1, and Mac1 and accordingly diagnosed with B-lymphocytic leukemia (B-ALL). (D) Survival curves of Tg, Tg/MOL4070LTR (Tg mice infected with MOL4070LTR), control (Ctrl), Ctrl/MOL4070LTR. MOL4070LTR infection induced acute leukemias in all Tg/MOL4070LTR mice and 1 Ctrl/MOL4070LTR mouse. The Tg/MOL4070LTR mice showed significantly accelerated mortality compared with the original Tg mice, whereas WT Ctrl mice did not. The time of MOL4070LTR injection is indicated by an arrow. (E) Survival curves of Tg, Tg/ENU (Tg mice injected with ENU), Ctrl, Ctrl/ENU mice. ENU-induced acute leukemias in all Tg/ENU and 1 Ctrl/ENU mice. The Tg/ENU mice showed significantly accelerated mortality compared with the original Tg mice, whereas Ctrl mice did not. The time of ENU injection is indicated by an arrow. For survival comparisons, the Tg and control cohorts shown in panel A were merged in panels D and E.
Transgenic expression of Fbxl10 in mice develop leukemia. (A) Kaplan-Meier survival plots for the Fbxl10 Tg (Tg) (n = 30) and control WT (Ctrl) (n = 20) mice. (B) Representative peripheral blood smear (left) and macroscopic appearance (right) of a leukemic Tg mouse. Proliferation of blast cells and massive hepatosplenomegaly are shown. (C) Flow cytometric profiles in the leukemic tissues of 2 Tg mice. TG-1 was positive for Mac1 and/or Gr1 positive but negative for Thy1.2 and B220 and diagnosed with AML, whereas TG-2 was positive for B220 but negative for Thy1.2, Gr1, and Mac1 and accordingly diagnosed with B-lymphocytic leukemia (B-ALL). (D) Survival curves of Tg, Tg/MOL4070LTR (Tg mice infected with MOL4070LTR), control (Ctrl), Ctrl/MOL4070LTR. MOL4070LTR infection induced acute leukemias in all Tg/MOL4070LTR mice and 1 Ctrl/MOL4070LTR mouse. The Tg/MOL4070LTR mice showed significantly accelerated mortality compared with the original Tg mice, whereas WT Ctrl mice did not. The time of MOL4070LTR injection is indicated by an arrow. (E) Survival curves of Tg, Tg/ENU (Tg mice injected with ENU), Ctrl, Ctrl/ENU mice. ENU-induced acute leukemias in all Tg/ENU and 1 Ctrl/ENU mice. The Tg/ENU mice showed significantly accelerated mortality compared with the original Tg mice, whereas Ctrl mice did not. The time of ENU injection is indicated by an arrow. For survival comparisons, the Tg and control cohorts shown in panel A were merged in panels D and E.
To further assess the tumor susceptibility caused by the expression of Fbxl10, neonate mice were infected with replication-competent MOL4070LTR mouse leukemia retrovirus, which is capable of inducing both myeloid disease and lymphoid diseases.20 The injection of MOL4070LTR profoundly accelerated the onset of the disease in the Fbxl10 Tg group (median survival = 274 days, P < .0001) (Figure 1D). We also applied a chemically (N-ethyl-N-nitrosourea [ENU]) induced leukemia model to the same cohort, which enhanced leukemogenesis in Tg mice (median survival = 210 days, P < .0001) (Figure 1E). Consistent with the spontaneously developed leukemias, the injection of virus predisposed Tg mice to AML and B-ALL but not T-cell (T-ALL) (supplemental Figure 3). Thus, Tg expression of Fbxl10 in HSCs drives myeloid and B-cell lineage leukemias with enhanced tumor susceptibility.
Transgenic expression of Fbxl10 induces proliferation advantage in HSCs
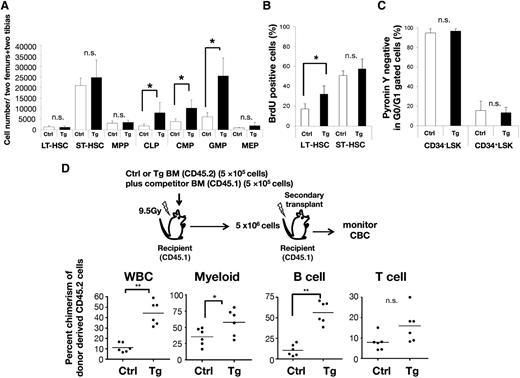
We first analyzed BM cellularity at 8 weeks of age in control and Tg mice, and found similar absolute cell numbers of multiple lineages among these mice (supplemental Figure 4A-B). To characterize more primitive cells, we further compared the HSC compartment and progenitor populations in the BM of control and Tg mice. Flow cytometric analysis revealed that the populations of myeloid progenitor cells (common myeloid progenitor [CMP] and granulocyte/macrophage progenitor [GMP]) and common lymphoid progenitor (CLP) cells were significantly higher in Tg mice than in the controls, whereas the HSC subpopulations (long-term [LT] HSC, short-term [ST] HSC, and multipotent progenitor [MPP]) were apparently not affected by the overexpression of Fbxl10 (Figure 2A).
Ly6A/Sca-1 promoter-driven Fbxl10 overexpression induces cell-autonomous proliferation in HSCs. (A) Absolute numbers of HSCs (LT-HSC, ST-HSC, and MPP) and progenitor (CLP, CMP, GMP, and MEP) cells (n = 4-8, *P < .025). Surface markers to separate mouse stem and progenitor cell subsets are shown in supplemental Table 3. (B) Increased in vivo BrdU incorporation in LT-HSCs of Tg mice. BrdU-positive cells in LT-HSC and ST-HSC were analyzed by flow cytometry (n = 4 per group, *P = .024). (C) Summary of flow cytometric pyronin Y analysis of CD34− LSK or CD34+ LSK fractions in control or Tg BM cells (n = 4 per group). Error bars, SD. (D) Percent chimerism of donor cells in the peripheral blood after serial BM transplantation. Upper panel shows a schematic diagram of the experimental strategies. Each dot shows the percent chimerism of donor cells in the peripheral blood of recipients 12 weeks after the secondary transplant. Bars indicate the average (n = 6 per group, *P < .05, **P < .01). CBC, complete blood cell count; MEP, megakaryocyte/erythroid progenitor; n.s., not significant; SD, standard deviation; WBC, white blood cell count.
Ly6A/Sca-1 promoter-driven Fbxl10 overexpression induces cell-autonomous proliferation in HSCs. (A) Absolute numbers of HSCs (LT-HSC, ST-HSC, and MPP) and progenitor (CLP, CMP, GMP, and MEP) cells (n = 4-8, *P < .025). Surface markers to separate mouse stem and progenitor cell subsets are shown in supplemental Table 3. (B) Increased in vivo BrdU incorporation in LT-HSCs of Tg mice. BrdU-positive cells in LT-HSC and ST-HSC were analyzed by flow cytometry (n = 4 per group, *P = .024). (C) Summary of flow cytometric pyronin Y analysis of CD34− LSK or CD34+ LSK fractions in control or Tg BM cells (n = 4 per group). Error bars, SD. (D) Percent chimerism of donor cells in the peripheral blood after serial BM transplantation. Upper panel shows a schematic diagram of the experimental strategies. Each dot shows the percent chimerism of donor cells in the peripheral blood of recipients 12 weeks after the secondary transplant. Bars indicate the average (n = 6 per group, *P < .05, **P < .01). CBC, complete blood cell count; MEP, megakaryocyte/erythroid progenitor; n.s., not significant; SD, standard deviation; WBC, white blood cell count.
Subsequently, to assess the specific activation of the cell cycle of HSCs, we examined the ratio of 5-bromo 2′-deoxyuridine (BrdU)–positive cells in the BM after the injection of BrdU into mice combined with pyronin Y staining. The ratio of BrdU-incorporated LT-HSCs in Tg mice was higher than that in the controls (Figure 2B), whereas pyronin Y analysis demonstrated no significant difference of G0/G1 ratio between Tg and control mice (Figure 2C). Thus, the transgenic expression of Fbxl10 in HSCs accelerated the progression of cell cycle accompanying a normal G0 to G1 entry, resulting in pleiotropic progenitor expansion. In addition, peripheral blood chimerism in competitive and serial transplantation analyses showed that whereas no difference was observed in the primary transplantation, the secondary transplantation exhibited a significant increase in the long-term repopulating activity of Fbxl10 Tg BM cells as compared with control BM cells (Figure 2D). Tg mouse-derived transplanted cells strongly contributed to myeloid and B-cell lineages but not to T-cell lineage (Figure 2D), which is in good agreement with the lineage specificity of Fbxl10-induced leukemia (Figure 1C; supplemental Table 1). The percentage of donor-derived cells was slightly higher in the BM recipients that received Fbxl10 Tg cells compared with those that received control cells, although the difference was not statistically significant at the time point (supplemental Figure 4C). These results suggest that Fbxl10 enhances self-renewal activity and a long-term BM repopulation capacity with a lasting myeloid and B-lymphoid expansion in the peripheral blood in a cell-autonomous manner.
Nsg2, a neuron-specific gene aberrantly induced by Fbxl10, impairs hematopoietic differentiation in vitro
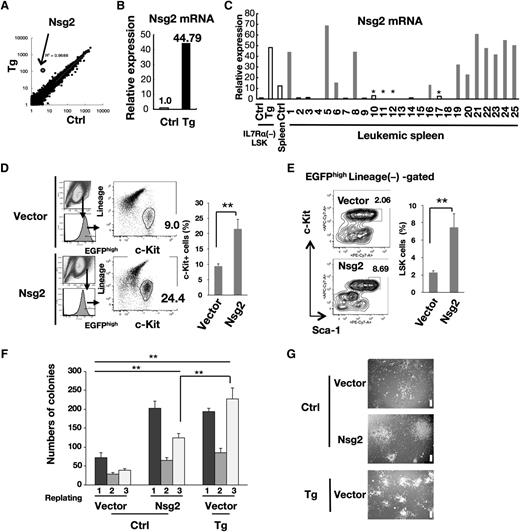
To elucidate the mechanistic link between the overexpression of Fbxl10 and development of leukemia, we analyzed the transcriptional profiles of IL-7Rα− LSK HSCs in nonleukemic young Tg mice (8 weeks of age) together with the corresponding controls (Table 1). A remarkable observation was the markedly increased expression of Nsg2 (also known as HMP19), encoding a 19-kDa protein localized in the Golgi apparatus,21 in Tg cells (Figure 3A; Table 1). We found that the levels of Nsg2 mRNA were low in control HSCs (Table 1), whereas a 40-fold higher expression was observed in the Tg HSCs (Figure 3B). A significantly higher expression of Nsg2 was also detected in most of the Fbxl10 Tg leukemic tissues of myeloid lineage (Figure 3C), suggesting that Nsg2 may be implicated in the pathogenesis of leukemia. To assess this possibility, we performed small interfering RNA (siRNA) knockdown in cells prepared from a Tg leukemic spleen diagnosed as AML exhibiting high Nsg2 expression (mouse no. 8 in Figure 3C and supplemental Table 1) (supplemental Figure 5A). siRNA oligos were successfully introduced into these cultured leukemic cells by electroporation (supplemental Figure 5B), and efficient Nsg2 knockdown was verified by quantitative real-time polymerase chain reaction (PCR) (quantitative PCR [qPCR]) (supplemental Figure 5C). As a result, siRNA-mediated Nsg2 knockdown in the leukemic cells significantly decreased the colony number and cell viability in methylcellulose culture (supplemental Figure 5D-E).
The top 10 differentially regulated genes in Tg HSCs
| Refseq_name . | Gene symbol . | Ctrl . | Tg . | Tg/Ctrl . | Description . |
|---|---|---|---|---|---|
| Genes upregulated in Tg HSCs | |||||
| NM_008741 | Nsg2 | 1.37 | 62.8 | 45.9 | Neuron-specific gene family member 2 |
| NM_013805 | Cldn5 | 0.37 | 2.49 | 6.73 | Claudin 5 |
| NM_001018013 | Izumo1 | 0.80 | 4.93 | 6.16 | Izumo sperm-egg fusion 1 |
| NM_178618 | Fam83g | 0.58 | 2.33 | 4.02 | Family with sequence similarity 83, member G |
| NM_001033227 | Slc5a10 | 1.32 | 5.10 | 3.86 | Solute carrier family 5, member 10 |
| NM_011627 | Tpbg | 0.59 | 2.19 | 3.71 | Trophoblast glycoprotein |
| NM_010738 | Ly6a | 21.9 | 78.5 | 3.58 | Lymphocyte antigen 6 complex, locus A |
| NM_010218 | Fjx1 | 0.70 | 2.38 | 3.40 | Four jointed box 1 |
| NM_001159748 | Serpinb8 | 0.90 | 3.02 | 3.36 | Serine (or cysteine) peptidase inhibitor, clade B, member 8 |
| NM_007388 | Acp5 | 0.75 | 2.41 | 3.21 | Acid phosphatase 5, tartrate resistant |
| Genes downregulated in Tg HSCs | |||||
| NM_181596 | Retnlg | 8.31 | 1.23 | 0.15 | Resistin-like γ |
| NM_008611 | Mmp8 | 1.87 | 0.28 | 0.15 | Matrix metallopeptidase 8 |
| NM_008405 | Itgb2l | 2.97 | 0.55 | 0.19 | Integrin β2-like |
| NM_145143 | Mpp4 | 2.80 | 0.66 | 0.24 | MAGUK p55 subfamily member 4 |
| NM_021407 | Trem3 | 4.36 | 1.07 | 0.25 | Triggering receptor expressed on myeloid cells 3 |
| NM_013599 | Mmp9 | 4.20 | 1.04 | 0.25 | Matrix metallopeptidase 9 |
| NM_009639 | Crisp3 | 2.51 | 0.63 | 0.25 | Cysteine-rich secretory protein 3 |
| NM_011410 | Slfn4 | 1.08 | 0.28 | 0.26 | Schlafen 4 |
| NM_145389 | BC016579 | 4.44 | 1.20 | 0.27 | cDNA sequence, BC016579 |
| NM_178885 | AI182371 | 1.36 | 0.37 | 0.27 | Expressed sequence AI182371 |
| Refseq_name . | Gene symbol . | Ctrl . | Tg . | Tg/Ctrl . | Description . |
|---|---|---|---|---|---|
| Genes upregulated in Tg HSCs | |||||
| NM_008741 | Nsg2 | 1.37 | 62.8 | 45.9 | Neuron-specific gene family member 2 |
| NM_013805 | Cldn5 | 0.37 | 2.49 | 6.73 | Claudin 5 |
| NM_001018013 | Izumo1 | 0.80 | 4.93 | 6.16 | Izumo sperm-egg fusion 1 |
| NM_178618 | Fam83g | 0.58 | 2.33 | 4.02 | Family with sequence similarity 83, member G |
| NM_001033227 | Slc5a10 | 1.32 | 5.10 | 3.86 | Solute carrier family 5, member 10 |
| NM_011627 | Tpbg | 0.59 | 2.19 | 3.71 | Trophoblast glycoprotein |
| NM_010738 | Ly6a | 21.9 | 78.5 | 3.58 | Lymphocyte antigen 6 complex, locus A |
| NM_010218 | Fjx1 | 0.70 | 2.38 | 3.40 | Four jointed box 1 |
| NM_001159748 | Serpinb8 | 0.90 | 3.02 | 3.36 | Serine (or cysteine) peptidase inhibitor, clade B, member 8 |
| NM_007388 | Acp5 | 0.75 | 2.41 | 3.21 | Acid phosphatase 5, tartrate resistant |
| Genes downregulated in Tg HSCs | |||||
| NM_181596 | Retnlg | 8.31 | 1.23 | 0.15 | Resistin-like γ |
| NM_008611 | Mmp8 | 1.87 | 0.28 | 0.15 | Matrix metallopeptidase 8 |
| NM_008405 | Itgb2l | 2.97 | 0.55 | 0.19 | Integrin β2-like |
| NM_145143 | Mpp4 | 2.80 | 0.66 | 0.24 | MAGUK p55 subfamily member 4 |
| NM_021407 | Trem3 | 4.36 | 1.07 | 0.25 | Triggering receptor expressed on myeloid cells 3 |
| NM_013599 | Mmp9 | 4.20 | 1.04 | 0.25 | Matrix metallopeptidase 9 |
| NM_009639 | Crisp3 | 2.51 | 0.63 | 0.25 | Cysteine-rich secretory protein 3 |
| NM_011410 | Slfn4 | 1.08 | 0.28 | 0.26 | Schlafen 4 |
| NM_145389 | BC016579 | 4.44 | 1.20 | 0.27 | cDNA sequence, BC016579 |
| NM_178885 | AI182371 | 1.36 | 0.37 | 0.27 | Expressed sequence AI182371 |
The values indicate reads per kilobase of transcript per million mapped reads and the Tg vs control.
Nsg2 is highly upregulated in HSCs and leukemic tissues of Tg mice and impairs hematopoietic differentiation. (A) Scatter plot comparing the transcriptomes of control and Tg IL-7Rα− LSK HSCs. Transcription levels are shown on a logarithmic scale. A linear regression curve and correlation coefficient are indicated. (B) Nsg2 mRNA in IL-7Rα− LSK HSCs was measured by real-time quantitative RT-PCR, and is presented as the relative expression level in arbitrary units where the mRNA level in the control was defined as 1 unit. (C) Comparison of Nsg2 mRNA levels in IL-7Rα− LSK HSCs from the control and Tg BM, and in the control normal spleen as well as leukemic spleens from Tg mice. Nsg2 mRNA was measured and shown as in panel B. Among leukemic spleens, gray and black bars indicate the leukemic tissues that were diagnosed with AML and B-ALL, respectively. *Unclassified samples. (D) Murine primary c-Kit+ BM cells were retrovirally transduced with pMYs-IRES-EGFP empty vector or the vector expressing Nsg2. Cells were grown in liquid culture with SCF, IL-3, IL-6, GM-CSF, TPO, and Flt3L for 5 days and assessed for the percentage of c-Kit+ population in EGFPhigh cells. (Left) Flow cytometric profiles. (Right) The ratio of c-Kit+ cells (percentage of EGFP+ cells). (E) Cells treated as in panel D were assessed using flow cytometry for the percentage of LSK in total Lin−EGFPhigh cells. (Left) Flow cytometric profiles. (Right) The ratio of LSK cells (percentage of EGFP+ cells). All results are shown as the mean ± SD of independent experiments; **P < .005. (F) c-Kit+ BM cells from the control and Tg littermates were retrovirally transduced with pMYs-IRES-EGFP empty vector. c-Kit+ BM cells from control mice were retrovirally transduced with the vector expressing Nsg2. EGFP+ cells were sorted and then cultured in methylcellulose media as described in “Methods.” Colonies were counted and replated at 2 × 104 cells every 7 days. Error bars, SD. **P < .005. (G) Representative images of colonies from the third plating as shown in panel F. Scale bars, 200 µm. EGFP, enhanced green fluorescent protein; GM-CSF, granulocyte macrophage–colony-stimulating factor; IL, interleukin; IRES, internal ribosome entry site; RT, reverse transcription; SCF, stem cell factor; TPO, thrombopoietin.
Nsg2 is highly upregulated in HSCs and leukemic tissues of Tg mice and impairs hematopoietic differentiation. (A) Scatter plot comparing the transcriptomes of control and Tg IL-7Rα− LSK HSCs. Transcription levels are shown on a logarithmic scale. A linear regression curve and correlation coefficient are indicated. (B) Nsg2 mRNA in IL-7Rα− LSK HSCs was measured by real-time quantitative RT-PCR, and is presented as the relative expression level in arbitrary units where the mRNA level in the control was defined as 1 unit. (C) Comparison of Nsg2 mRNA levels in IL-7Rα− LSK HSCs from the control and Tg BM, and in the control normal spleen as well as leukemic spleens from Tg mice. Nsg2 mRNA was measured and shown as in panel B. Among leukemic spleens, gray and black bars indicate the leukemic tissues that were diagnosed with AML and B-ALL, respectively. *Unclassified samples. (D) Murine primary c-Kit+ BM cells were retrovirally transduced with pMYs-IRES-EGFP empty vector or the vector expressing Nsg2. Cells were grown in liquid culture with SCF, IL-3, IL-6, GM-CSF, TPO, and Flt3L for 5 days and assessed for the percentage of c-Kit+ population in EGFPhigh cells. (Left) Flow cytometric profiles. (Right) The ratio of c-Kit+ cells (percentage of EGFP+ cells). (E) Cells treated as in panel D were assessed using flow cytometry for the percentage of LSK in total Lin−EGFPhigh cells. (Left) Flow cytometric profiles. (Right) The ratio of LSK cells (percentage of EGFP+ cells). All results are shown as the mean ± SD of independent experiments; **P < .005. (F) c-Kit+ BM cells from the control and Tg littermates were retrovirally transduced with pMYs-IRES-EGFP empty vector. c-Kit+ BM cells from control mice were retrovirally transduced with the vector expressing Nsg2. EGFP+ cells were sorted and then cultured in methylcellulose media as described in “Methods.” Colonies were counted and replated at 2 × 104 cells every 7 days. Error bars, SD. **P < .005. (G) Representative images of colonies from the third plating as shown in panel F. Scale bars, 200 µm. EGFP, enhanced green fluorescent protein; GM-CSF, granulocyte macrophage–colony-stimulating factor; IL, interleukin; IRES, internal ribosome entry site; RT, reverse transcription; SCF, stem cell factor; TPO, thrombopoietin.
Next, to evaluate the impact of the aberrant expression of Nsg2 on normal hematopoiesis, WT BM cells were infected with retroviruses expressing Nsg2. The overexpression of Nsg2 in murine primary WT c-Kit+ BM cells increases the proportion of c-Kit+ hematopoietic stem/progenitor cells and LSK cell fraction compared with the cells expressing empty vector in short-term ex vivo liquid culture (Figure 3D-E). In addition, colony numbers of Nsg2-transduced control cells were significantly higher than those of vector-transduced control cells through 3 rounds of plating, and were almost comparable to those of vector-transduced Tg cells after the first and second platings (Figure 3F). The Nsg2-transduced cells formed fewer colonies than vector-transduced Tg cells at the third round, but the colonies retained their concentric and compact morphology (Figure 3F-G). These results suggest that Nsg2 potentially possesses undifferentiated properties and may contribute to leukemogenesis in a manner similar to canonical class II mutations. We further transplanted the WT c-Kit+ BM progenitor cells overexpressing Nsg2 into irradiated mice. These recipients, however, did not show leukemia at least during the observation period of 8 months.
Fbxl10-overexpressing HSCs are metabolically active and characterized by an increased oxidative phosphorylation
Previous studies have reported that Fbxl10 suppresses senescence-associated cyclin-dependent kinase inhibitors (CDKIs) such as p15ink4b, p16ink4a, p18ink4c, and p57kip2 in murine blood cells.12,13 Therefore, we investigated the possibility that suppression of these CDKI genes might account for the survival/proliferation advantage in Tg HSCs, which is reminiscent of the canonical class I mutations. The former 2 genes, p15ink4b and p16ink4a, were very weakly expressed in primary control HSCs, as reported in a previous transcriptome analysis22 ; no expressional change of these genes was observed in Tg HSCs. The latter 2 genes, p18ink4c and p57kip2, were expressed in both genotypes and slightly suppressed by 20% to 45% in Tg HSCs as compared with wild-type cells (supplemental Table 2). Although the suppression of p57kip2 was most remarkable, this would not account for the enhanced self-renewal activity of Fbxl10 Tg HSCs because a previous report showed that p57kip2 was required for HSC quiescence and its deficiency abrogated the self-renewal capacity of HSCs in mice.23
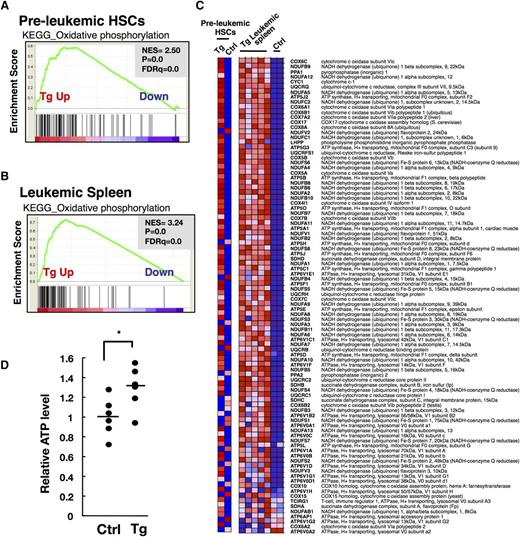
To identify the differentially regulated functional networks accounting for growth advantage of HSCs in Tg mice, we applied gene set enrichment analysis (GSEA) to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, a database that represents molecular interaction networks, including metabolic pathways, regulatory pathways, and molecular complexes. This analysis revealed that in nonleukemic Tg HSCs, several gene sets were significantly upregulated with “oxidative phosphorylation (OXPHOS)” being the highest (Figure 4A; Table 2), whereas no gene sets were significantly downregulated, as compared with the control HSCs. The genes involved in OXPHOS were also most markedly enriched in the leukemic tissues compared with the corresponding controls (Figure 4B-C; Table 3). Adenosine 5′-triphosphate (ATP) is the central cellular metabolic marker and bioenergetic source.24,25 We observed a significant increase in basal intracellular ATP content in IL-7Rα− LSK HSCs purified from nonleukemic Fbxl10 Tg mice compared with the corresponding controls (Figure 4D). These results strongly suggest that Fbxl10 metabolically activates HSCs by increasing OXPHOS, which is linked to the progression of cell cycle and promotes the development of leukemia.
Fbxl10 overexpression leads to mitochondrial metabolic activation characterized by increased oxidative phosphorylation. (A) GSEA enrichment plot of KEGG OXPHOS gene set in nonleukemic (preleukemic) BM HSCs (HSCs from Tg mice vs HSCs from controls). Genes involved in OXPHOS were significantly enriched in Tg HSCs. Pooled RNAs from IL-7Rα− LSK HSCs in Tg and control individuals were used for the analysis. (B) GSEA enrichment plot of KEGG OXPHOS gene set in leukemic spleens (Tg leukemic spleens vs control spleens). Genes involved in OXPHOS were significantly enriched in Tg leukemic spleens. (C) Heat map of OXPHOS genes. Genes involved in OXPHOS are profoundly highly expressed in both pooled Tg HSCs and individual Tg leukemic spleens compared with the corresponding WT control HSCs and spleens. (D) Relative ATP levels in IL-7Rα− LSK HSCs of control and Tg mice at 20 weeks of age (n = 6 per group). Horizontal line, mean of ATP levels. Tg cells produced significantly more ATP than control cells (*P = .032). FDR, false discovery rate; NES, normalized enrichment score.
Fbxl10 overexpression leads to mitochondrial metabolic activation characterized by increased oxidative phosphorylation. (A) GSEA enrichment plot of KEGG OXPHOS gene set in nonleukemic (preleukemic) BM HSCs (HSCs from Tg mice vs HSCs from controls). Genes involved in OXPHOS were significantly enriched in Tg HSCs. Pooled RNAs from IL-7Rα− LSK HSCs in Tg and control individuals were used for the analysis. (B) GSEA enrichment plot of KEGG OXPHOS gene set in leukemic spleens (Tg leukemic spleens vs control spleens). Genes involved in OXPHOS were significantly enriched in Tg leukemic spleens. (C) Heat map of OXPHOS genes. Genes involved in OXPHOS are profoundly highly expressed in both pooled Tg HSCs and individual Tg leukemic spleens compared with the corresponding WT control HSCs and spleens. (D) Relative ATP levels in IL-7Rα− LSK HSCs of control and Tg mice at 20 weeks of age (n = 6 per group). Horizontal line, mean of ATP levels. Tg cells produced significantly more ATP than control cells (*P = .032). FDR, false discovery rate; NES, normalized enrichment score.
Gene sets enriched by Fbxl10 in HSCs on GSEA (NES > 2.0)
| Pathway . | Size* . | NES . | Nominal P . | FDR q value . |
|---|---|---|---|---|
| Upregulated gene set in Tg HSCs | ||||
| Oxidative phosphorylation | 98 | 2.50 | .000 | 0.000 |
| Ribosome | 56 | 2.39 | .000 | 0.000 |
| Proteasome | 22 | 2.13 | .000 | 0.001 |
| Downregulated gene set in Tg HSCs | ||||
| Not applicable |
| Pathway . | Size* . | NES . | Nominal P . | FDR q value . |
|---|---|---|---|---|
| Upregulated gene set in Tg HSCs | ||||
| Oxidative phosphorylation | 98 | 2.50 | .000 | 0.000 |
| Ribosome | 56 | 2.39 | .000 | 0.000 |
| Proteasome | 22 | 2.13 | .000 | 0.001 |
| Downregulated gene set in Tg HSCs | ||||
| Not applicable |
FDR, false discovery rate; NES, normalized enrichment score.
Number of genes in each set.
Gene sets enriched by Fbxl10 in leukemic tissues on GSEA (NES > 2.5)
| Pathway . | Size . | NES . | Nominal P . | FDR q value . |
|---|---|---|---|---|
| Upregulated gene set in Tg leukemic tissues | ||||
| Oxidative phosphorylation | 94 | 3.56 | .000 | 0.000 |
| Ribosome | 56 | 2.56 | .000 | 0.000 |
| Proteasome | 22 | 2.50 | .000 | 0.000 |
| Downregulated gene set in Tg leukemic tissues | ||||
| Not applicable | ||||
| Pathway . | Size . | NES . | Nominal P . | FDR q value . |
|---|---|---|---|---|
| Upregulated gene set in Tg leukemic tissues | ||||
| Oxidative phosphorylation | 94 | 3.56 | .000 | 0.000 |
| Ribosome | 56 | 2.56 | .000 | 0.000 |
| Proteasome | 22 | 2.50 | .000 | 0.000 |
| Downregulated gene set in Tg leukemic tissues | ||||
| Not applicable | ||||
Size, number of genes in each set.
FDR, false discovery rate; NES, normalized enrichment score.
Fbxl10 has direct and indirect effects in proleukemogenic gene expression profiles
To investigate the regulatory mechanism of the proleukemogenic transcription signature caused by the overexpression of Fbxl10, we performed chromatin immunoprecipitation sequencing (ChIP-seq) in Tg and control HSCs (LSKs) using a ChIP-grade polyclonal antibody for Fbxl10.9 Recent studies demonstrated that Fbxl10 preferentially binds to the regions flanking transcription start sites (TSSs) and containing unmethylated CpG sequences at actively transcribed genes but not only repressive genes in ES cells.9 Tag density of ChIP-seq data revealed 3 distinct regions flanking the TSS of the Nsg2 gene, in which additional bindings of Fbxl10 were detected in Tg HSCs (boxed in Figure 5A). CpG sequences associated with an Fbxl10 binding region (+1991 to +2190 of Nsg2 gene locus) were detected using the sequence manipulation suite program26 (Figure 5B). We also identified increased or additional peaks on OXPHOS-related Ndufs6, Ndufb2, and Uqcrfs1 gene regions flanking the TSS in Tg HSCs (Figure 5C). However, the bindings of Fbxl10 were not pronouncedly increased at the TSS and promoter regions of most other OXPHOS genes that were upregulated in Tg HSCs. The activation of OXPHOS genes in Tg HSCs may involve overall effects triggered by the transgenic expression of Fbxl10. These results indicate direct and indirect regulation of proleukemic gene expression signature mediated by Fbxl10. Because Fbxl10 is an H3K36me2 demethylase, we also performed the ChIP-seq for HSCs using an anti-histone H3K36me2 antibody shown to be applicable for the ChIP-seq.27 The result revealed that Fbxl10 overexpression in HSCs reduced H3K36me2 levels more around TSSs (supplemental Figure 6A-B). The H3K36me2 ChIP-seq reads were, however, detected at low levels on Fbxl10-targeted regions of Figure 5, panels A and C, in control HSCs, and therefore, it was unable to clarify the corresponding decrease in reads in Tg HSCs.
Nsg2 and mitochondrial genes are directly targeted by Fbxl10. (A) Fbxl10 ChIP-seq signal at Nsg2 gene regions flanking promoter and TSS in HSCs. Rectangles indicate regions where Fbxl10 bindings are higher in Tg HSCs than in control HSCs (left) and the bar graph demonstrates relative ChIP/Input ratio [ChIP DNA (ppm) divided by Input DNA (ppm)] in the indicated regions (right). (B) CpG sequences detected in Nsg2 gene regions flanking promoter and TSS, which was sought using sequence manipulation suite program.26 Fbxl10 was highly recruited to the region +1991 to +2190 of Nsg2 gene in Tg HSCs, as shown in panel A. (C) Fbxl10 ChIP-seq signal at OXPHOS genes (Ndufs6, Ndufb2, and Uqcrfs1) regions flanking promoter and TSS. Rectangles indicate regions where Fbxl10 bindings are higher in Tg HSCs than in control HSCs (left) and the bar graphs demonstrate relative ChIP/Input ratio in the indicated regions (right).
Nsg2 and mitochondrial genes are directly targeted by Fbxl10. (A) Fbxl10 ChIP-seq signal at Nsg2 gene regions flanking promoter and TSS in HSCs. Rectangles indicate regions where Fbxl10 bindings are higher in Tg HSCs than in control HSCs (left) and the bar graph demonstrates relative ChIP/Input ratio [ChIP DNA (ppm) divided by Input DNA (ppm)] in the indicated regions (right). (B) CpG sequences detected in Nsg2 gene regions flanking promoter and TSS, which was sought using sequence manipulation suite program.26 Fbxl10 was highly recruited to the region +1991 to +2190 of Nsg2 gene in Tg HSCs, as shown in panel A. (C) Fbxl10 ChIP-seq signal at OXPHOS genes (Ndufs6, Ndufb2, and Uqcrfs1) regions flanking promoter and TSS. Rectangles indicate regions where Fbxl10 bindings are higher in Tg HSCs than in control HSCs (left) and the bar graphs demonstrate relative ChIP/Input ratio in the indicated regions (right).
Next, to assess the regulatory status in transformed cells, we performed ChIP-PCR to analyze the recruitment of Fbxl10 to the Nsg2 gene regions in proliferating myeloid leukemic cells in culture from a Tg spleen, using anti-Fbxl10 and anti-Flag Abs (primer sets, P1-P3 in supplemental Figure 7A). A primer set P3 detected a significant binding of Fbxl10 on the Nsg2 gene region, corresponding to regions that were identified in Tg LSK by the ChIP-seq with Fbxl10 Ab (supplemental Figure 7B). In addition, the primer set was able to detect reduced DNA binding for H3K36me2 in this tumor line (supplemental Figure 7C). These results suggest that the ectopic expression of Nsg2 in Tg HSCs is targeted by the overexpression of Fbxl10 and that the recruitment to Nsg2 gene locus is partly sustained in the resulting leukemic cells.
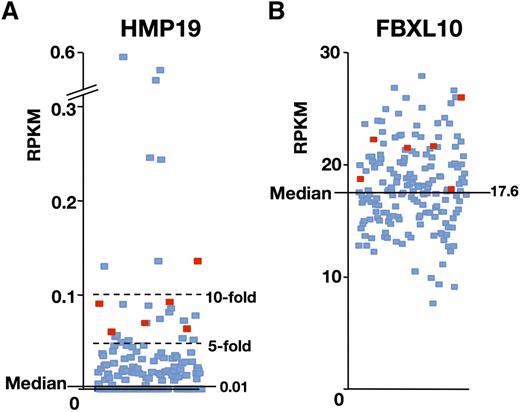
A subpopulation of AML patients exhibits high expression of both HMP19 and FBXL10
To investigate the potential relevance of the findings to human disease, we investigated the expression levels of HMP19 (human homolog of Nsg2) and FBXL10 on The Cancer Genome Atlas (TCGA) database.28 We found that HMP19 mRNA was strongly elevated in 21 cases (12.8%) by more than fivefold (RPKM > 0.05) of the whole AML median value (0.01), whereas 82 of 179 cases (45.8%) showed no expressions of the gene (Figure 6A). Eight cases (4.5%) exhibited even more than 10-fold upregulation (RPKM > 0.1) of the HMP19 gene expression. Of note, 6 cases (3.4%) that exhibited a higher expression of FBXL10 over the median value were coupled with more than fivefold upregulation of the HMP19 gene expression (Figure 6). These data may support that our mouse model is clinically relevant with a specific population of leukemia patients.
A subpopulation of AML patients exhibits high expression of both HMP19 and FBXL10. RNA-Seq data of 179 AML cases available in the TCGA database were analyzed. Expression levels of HMP19 (A) and FBXL10 (B) mRNAs are shown as RPKM. Six that expressed HMP19 mRNA more than fivefold over the average and FBXL10 mRNA over the median value are indicated by red squares (TCGA-AB-2807, 2832, 2892, 2943, 2982, and 3009). RPKM, reads per kilobase per million mapped reads.
A subpopulation of AML patients exhibits high expression of both HMP19 and FBXL10. RNA-Seq data of 179 AML cases available in the TCGA database were analyzed. Expression levels of HMP19 (A) and FBXL10 (B) mRNAs are shown as RPKM. Six that expressed HMP19 mRNA more than fivefold over the average and FBXL10 mRNA over the median value are indicated by red squares (TCGA-AB-2807, 2832, 2892, 2943, 2982, and 3009). RPKM, reads per kilobase per million mapped reads.
Discussion
Genetic and functional studies have so far suggested that there are at least 2 classes of myeloid leukemia disease alleles that contribute to hematopoietic transformation: class I alleles, which confer a growth advantage, and class II alleles, which impair the differentiation.29 However, recent studies have demonstrated that mutations of single genes in epigenetic modifiers can contribute to myeloid malignancies, suggesting another class of somatic functional mutations or altered gene expression besides the canonically categorized class I/class II mutations (TET2, ASXL1, DNMT3A, EZH2, MLL, etc).29 In this study, we used a genetically engineered mouse system to show that the Tg expression of epigenetic regulator Fbxl10 induced acute leukemias (Figure 1).
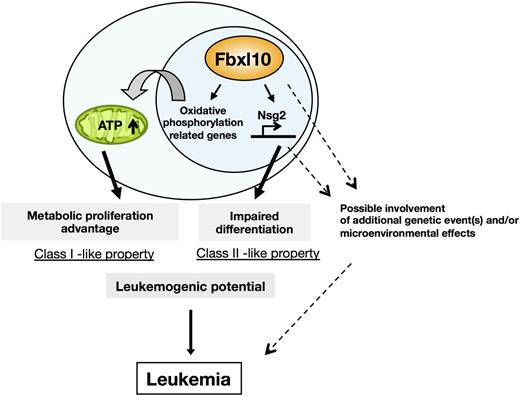
Mechanistically, Fbxl10 confers HSCs on the progression of cell cycle through the metabolic activation, whereas Fbxl10 activated the expression of Nsg2, which predisposes hematopoietic stem/early progenitor cells to an undifferentiated state. Thus, elevated levels of Fbxl10 involve both class I– and class II–like properties (Figure 7). However, the mice did not show an evident HSC differentiation block at a steady state, and the development of leukemia in Tg mice required long latency. Although these properties may underlie tumor susceptibility, additional genetic event(s) and/or microenvironmental effects may be required for full transformation (Figure 7). This idea was validated by the results that leukemogenesis in Tg mice is drastically accelerated by retrovirus insertional mutagenesis or a carcinogenic mutagen (Figure 1D-E).
Schematic illustration of Fbxl10-mediated leukemogenic processes. Fbxl10 induces leukemia involving metabolic proliferation advantage and impaired differentiation mediated by Nsg2. Fbxl10 confers HSCs on cell cycle progression through metabolic activation whereas Fbxl10 upregulates the expression of the Nsg2, impairing the differentiation of HSCs. Thus, Fbxl10 profoundly enhances leukemogenic potential/tumor susceptibility and leads to acute leukemia in mice after a long latency. Additional gene alterations and/or microenvironmental effects possibly intensify the proliferative potential and accelerate leukemic transformation mediated by Fbxl10 overexpression.
Schematic illustration of Fbxl10-mediated leukemogenic processes. Fbxl10 induces leukemia involving metabolic proliferation advantage and impaired differentiation mediated by Nsg2. Fbxl10 confers HSCs on cell cycle progression through metabolic activation whereas Fbxl10 upregulates the expression of the Nsg2, impairing the differentiation of HSCs. Thus, Fbxl10 profoundly enhances leukemogenic potential/tumor susceptibility and leads to acute leukemia in mice after a long latency. Additional gene alterations and/or microenvironmental effects possibly intensify the proliferative potential and accelerate leukemic transformation mediated by Fbxl10 overexpression.
Although the biological function of Nsg2 has not been fully understood so far, structurally similar transmembrane molecules NEEP21 (also known as Nsg1) and Caly (also known as Nsg3) are crucial for glutamate and/or dopamine receptor sorting through endosomes and their recruitment to the plasma membrane in neural cells.30-32 Because the expression of Nsg2 is severely repressed in normal HSCs, its ectopic upregulation might deregulate cytokine receptor recycling/intracellular sorting, thereby resulting in impaired differentiation of HSCs (Figure 3D-F). Wu et al described Fbxl10 binding loci in WT and Fbxl10-overexpressed ES cells.9 Although the binding loci of Fbxl10 were entirely shared between these 2 cell types, Fbxl10 was bound to the Nsg2 locus specifically in the overexpressed condition.9 Likewise, our ChIP-seq for HSCs and ChIP-PCR for the Tg leukemic cells demonstrated that Nsg2 was targeted by the overexpression of Fbxl10 (Figure 5A; supplemental Figure 7). To examine the oncogenic activity of Nsg2, we transplanted BM c-Kit+ cells retrovirally overexpressing Nsg2 into irradiated mice. However, these recipients did not develop leukemia during the limited period of the 8 months, suggesting that overexpression of Nsg2 alone is not leukemogenic.
Another remarkable characteristic of Tg HSCs is metabolic proliferation advantage coupled with mitochondrial energy activation (Figures 2B and 4A,C-D). This feature is also retained in leukemic tissues (Figure 4B), indicating that this change plays a critical role in the leukemogenic process. Accumulating evidence has shown that tumor cells increase their metabolic requirements for cell survival and proliferation.33 In addition, clinical studies also have provided evidence of a close relationship between altered cellular metabolism and the cancer outcome.34 Reactive oxygen species (ROS), which are generated by mitochondrial oxidative phosphorylation, was reported to limit the life span of HSCs.35 However, mitochondrial ROS levels in HSCs were not increased before the development of disease by the overexpression of Fbxl10, as examined by flow cytometry using a fluorogenic molecular probe (supplemental Figure 8). Consistent with these findings, cell cycle analysis demonstrated that Tg HSCs had a growth advantage without exhausting the HSC pool (Figure 2B-D). In mouse embryonic fibroblasts, the overexpression of Fbxl10 was reported to detoxify ROS by directly upregulating antioxidative genes and contributes to the inhibition of cellular senescence.36 Our results suggest that Fbxl10 has a substantially similar effect on HSCs, although the gene expression of individual canonical ROS scavenger was not significantly altered.37 Therefore, we cannot unequivocally state the mechanism underlying the lack of ROS increase in Tg HSCs at this point, and the possible involvement of Fbxl10 to maintain redox homeostasis in the HSC unique context remains to be defined. A recent study demonstrated that leukemic stem cells show relatively low levels of ROS and depend on oxidative respiration rather than glycolysis for energy generation, which may support our results.38 In addition, GSEA revealed that genes involved in ribosomal biogenesis were also enriched in both Tg HSCs and leukemic tissues (Tables 2-3), which would provide a direct connection between metabolic activation and the development of tumor because increased protein synthesis is a highly energy demanding process.
Fbxl10 was recently reported to promote K-RAS–mutated pancreatic cancer through upregulating a module of metabolic genes involved in OXPHOS in a mouse allograft model.5 In addition, IMP2-positive glioma and PPARGC1A (PGC1α)–positive melanoma showed tumor progression associated with mitochondrial energy activation in the absence and presence of enhanced ROS levels, respectively.39,40 Thus, despite the general enhancement of glycolytic bioenergetic requirements in many cancers (so called as “Warburg effect”), several recent studies have revealed unanticipated tumor metabolism by Warburg, that is, the bioenergetic type of tumors can vary widely from glycolysis to OXPHOS.41-44 Our findings suggest that OXPHOS-mediated metabolic activation provides a bioenergetic source to promote leukemogenesis in Fbxl10-upregulated HSCs. However, a number of questions remain to be answered, including the lineage specificity of leukemia and the possible involvement of other factors in Fbxl10-induced leukemogenesis. The search for these effectors will no doubt contribute to elucidating the mechanism by which overexpressed Fbxl10 leads to leukemia.
In summary, we established Fbxl10 as a bona fide oncogene in vivo and provided a novel mechanistic insight into leukemogenesis. Taken together with the upregulation of FBXL10 in human leukemia and solid tumors,3-6 selective inhibition of FBXLl10 may be a therapeutic strategy to achieve improvement in these diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Yuki Sakai for animal care, Yu Iwai and Rika Tai for mouse genotyping and molecular experiments, Dr Tatsuo Miyamoto for technical assistance, and Drs Kazuo Yamamoto and Masato Sasaki for helpful discussions. The authors also thank Dr Elaine Dzierzak (Erasmus University) for providing them with the Ly6A/Sca-1 transgenic cassette.
This work was in part supported by a Grant in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan.
Authorship
Contribution: T.U., H.H., Z.-i.H., T.S., T.I., and A.I. designed the research; T.U., A.N., K.T., N.Y., Y.N., K.I., H.M., A.K., T.K., H.O., and H.H. performed the research; L.W., X.W., and K.H. contributed new reagents or analytic tools; T.U., K.T., H.H., H.M., A.K., T.K., and A.I. analyzed the data; and T.U. and H.H wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Takeshi Ueda, Department of Disease Model, Research Institute for Radiation Biology and Medicine, Hiroshima University, 1-2-3 Kasumi Minami-ku Hiroshima 734-8553, Japan; e-mail: takeshiueda@hiroshima-u.ac.jp; and Hiroaki Honda, Department of Disease Model, Research Institute for Radiation Biology and Medicine, Hiroshima University, 1-2-3 Kasumi Minami-ku Hiroshima 734-8553, Japan; e-mail: hhonda@hiroshima-u.ac.jp.

![Figure 5. Nsg2 and mitochondrial genes are directly targeted by Fbxl10. (A) Fbxl10 ChIP-seq signal at Nsg2 gene regions flanking promoter and TSS in HSCs. Rectangles indicate regions where Fbxl10 bindings are higher in Tg HSCs than in control HSCs (left) and the bar graph demonstrates relative ChIP/Input ratio [ChIP DNA (ppm) divided by Input DNA (ppm)] in the indicated regions (right). (B) CpG sequences detected in Nsg2 gene regions flanking promoter and TSS, which was sought using sequence manipulation suite program.26 Fbxl10 was highly recruited to the region +1991 to +2190 of Nsg2 gene in Tg HSCs, as shown in panel A. (C) Fbxl10 ChIP-seq signal at OXPHOS genes (Ndufs6, Ndufb2, and Uqcrfs1) regions flanking promoter and TSS. Rectangles indicate regions where Fbxl10 bindings are higher in Tg HSCs than in control HSCs (left) and the bar graphs demonstrate relative ChIP/Input ratio in the indicated regions (right).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/22/10.1182_blood-2014-03-562694/4/m_3437f5.jpeg?Expires=1769091858&Signature=Z6zTvSaDiBhsBlxntQrHesWJadOeFp2wnjfZIuNZZcGoBeg2rz8tld7gIUAP3YSkGNzaJIlBVmNDNGs2ayCURx5Qrjsm0cJy4eQOxwreyzCxxa-289dyKso4qG9CNYOXns58TblLPZ50VLqBiHaVx0-8-QAdUPV2X48rf8Ri5O2VRVlcLqeRmd3BDbhoI9yWSezqH6x9d3aexT06Rlv-ofMpEZCMF53~Ln4sntsKKObTB8dVZjUHH443R-C5KQQ8Dt34H6SkH7SIRpTeFy8dbgIzavFFgAw75SPtNnTYsCkpkITkH1rbPWdh5A~SlZN6kQxvarwXjCNIcWem6AqlKw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal