Key Points
Tet2 loss of function confers a strong functional competitive advantage to Jak2V617F-mutant hematopoietic stem cells.
Jak2V617F expression and Tet2 loss generate distinct and nonoverlapping transcriptional programs in hematopoietic stem cells.
Abstract
Signaling mutations (eg, JAK2V617F) and mutations in genes involved in epigenetic regulation (eg, TET2) are the most common cooccurring classes of mutations in myeloproliferative neoplasms (MPNs). Clinical correlative studies have demonstrated that TET2 mutations are enriched in more advanced phases of MPNs such as myelofibrosis and leukemic transformation, suggesting that they may cooperate with JAK2V617F to promote disease progression. To dissect the effects of concomitant Jak2V617F expression and Tet2 loss within distinct hematopoietic compartments in vivo, we generated Jak2V617F/Tet2 compound mutant genetic mice. We found that the combination of Jak2V617F expression and Tet2 loss resulted in a more florid MPN phenotype than that seen with either allele alone. Concordant with this, we found that Tet2 deletion conferred a strong functional competitive advantage to Jak2V617F-mutant hematopoietic stem cells (HSCs). Transcriptional profiling revealed that both Jak2V617F expression and Tet2 loss were associated with distinct and nonoverlapping gene expression signatures within the HSC compartment. In aggregate, our findings indicate that Tet2 loss drives clonal dominance in HSCs, and Jak2V617F expression causes expansion of downstream precursor cell populations, resulting in disease progression through combinatorial effects. This work provides insight into the functional consequences of JAK2V617F-TET2 comutation in MPNs, particularly as it pertains to HSCs.
Introduction
Whole-genome and whole-exome sequencing studies have provided important insight into the somatic genetic lesions that drive myeloid neoplasms.1-3 Although much can be inferred from the patterns of genetic alterations identified in such studies, we still have an incomplete understanding of the functional significance of these relationships, particularly in how different driver mutations collaborate in the transformation of the hematopoietic stem cell (HSC).
In myeloproliferative neoplasms (MPNs), the majority of driver mutations can be broadly classified within two categories.4 First, virtually all MPN patients are now known to harbor mutations that confer hyperactive JAK-STAT signaling. By far, the JAK2V617F mutation is the most frequent of these mutations,5-8 with a minority of patients also harboring mutations in exon 12 of JAK2,9 MPL,10 LNK,11 or c-CBL.12 Recently, mutations in the gene encoding the endoplasmic reticulum chaperone, calreticulin (CALR) have been identified in the majority of JAK2-unmutated MPN patients,3,13 with early evidence suggesting that mutant CALR also causes constitutive JAK-STAT signaling and cytokine-independent growth.13
The second major class of somatic alterations in the MPN cancer genome is in genes encoding epigenetic regulators.14 In particular, deletions or loss-of-function mutations of the TET2 methylcytosine dioxygenase occur in approximately 7.5% to 17% of MPNs and are enriched in myelofibrosis compared to essential thrombocythemia15,16 and more aggressive forms of mastocytosis.17 Other than JAK2V617F and mutations in CALR, TET2 is the most common somatically altered gene in MPNs and the most commonly comutated gene with JAK2V617F.18 Although JAK2V617F and CALR mutations are mutually exclusive, TET2 mutations cooccur with both,19 suggesting that TET2 impacts distinct downstream oncogenic pathways from those affected by JAK2V617F or mutant CALR.
MPN animal models accurately recapitulate human disease in mice and have been an important tool for the study of MPN biology and therapy.20,21 Genetically engineered Jak2V617F and Tet2 animal models generated by ourselves and others20,21 have permitted a detailed examination of the functional effects of these genetic alterations in different hematopoietic compartments. In this study, we sought to model the co-occurrence of JAK2V617F and TET2 mutations in MPN patients by investigating the consequences of concomitant Jak2V617F expression and Tet2 loss in vivo. We provide new insight into the impact of Tet2 loss on (1) disease progression in Jak2V617F-mediated MPNs; (2) Jak2V617F-mutant hematopoietic stem and progenitor cell (HSPC) function; and (3) the transcriptional program of Jak2V617F-mutant MPN stem cells.
Material and methods
Experimental mice
We have previously described Jak2V617F (Jak2VF) conditional knockin and Tet2 conditional knockout mice.22,23 In this study, we used VavCre transgenic mice to target Cre recombinase expression to the hematopoietic lineage24 and to delete Tet2 in the hematopoietic compartment of Jak2V617F mice (supplemental Figure 1). We generated Jak2VF mice that were wild-type (WT) or nullizygous for Tet2 (Jak2VF or Jak2VF/Tet2null, respectively). We also generated mice that were WT for Jak2 and nullizygous for Tet2 (Tet2null). For controls, we used VavCre-positive mice that were WT for both Jak2 and Tet2. We maintained all mice in pathogen-free facilities at Children’s Hospital Boston. The institutional ethics committee of Children’s Hospital Boston approved all mouse experiments on protocol 13-04-2393R.
Blood analysis
Blood was collected into EDTA-coated containers and analyzed on a Hemavet 950 analyzer (Drew Scientific).
Flow cytometry
Bone marrow, spleen, or peripheral blood was collected and prepared for staining by red blood cell lysis (BD Pharm Lyse; BD Biosciences) and homogenization through a 70-μm filter. Erythroid precursor cell stainings were not pretreated with red cell lysis. All samples were analyzed by flow cytometry using the fluorescence-activated cell sorter BD FACSCanto or BD LSR II (BD Biosciences). All staining steps were performed in ice-cold phosphate-buffered saline containing 2% fetal bovine serum. Postacquisition analysis of data was performed with FlowJo software V9.2.3 (Tree Star, Ashland, OR). For peripheral blood chimerism studies, the following antibodies were used (clone in parentheses): CD45.1 (A20) and CD45.2 (104). For erythroid precursor cells, the following antibodies were used: CD71 (RI7217) and Ter119 (Ter-119). For stem cell and progenitor analysis, the following antibodies were used: a lineage cocktail containing CD3ε (145-2C11), CD5 (53-7.3), Ter-119 (TER-119), Gr-1 (RB6-8C5), Mac-1 (M1/70), and B220 (30-F11); c-Kit (2B8), Sca-1 (D7), CD150 (TC15-12F12.2), CD48 (HM48-1), CD135 (A2F10), CD34 (Ram34), and CD16/32 (93), in addition to CD45.1 (A20) and CD45.2 (104). For dead cell discrimination, SYTOX Blue (Invitrogen) was used. For cell cycle analysis, cells were first stained for lineage, stem, and progenitor markers, then fixed and permeabilized and stained with Ki67, followed by staining with Hoechst 33342. For cell sorting experiments, lineage-positive cells were first depleted with Dynabeads (Invitrogen); c-Kit positive cells were first enriched using CD117 mouse microbeads using magnetic-activated cell sorting (Miltenyi Biotec). The remaining cells were then stained with biotinylated lineage cocktail antibodies (clones listed above), followed by Streptavidin-ApcCy7, cKit (2B8), Sca-1 (D7), CD150 (TC15-12F12.2), and CD48 (HM48-1) where appropriate, and sorted on a BD FACSAria cell sorter (BD Biosciences).
Histopathology
Tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, and stained with hematoxylin- eosin (H&E), or with reticulin when assessing for fibrosis. For megakaryocyte analysis, megakaryocytes were counted in 10 high-power fields (×1000 with oil) per tibia from 4 biological replicates. Images of histologic slides were obtained on a Nikon Eclipse E400 microscope (Nikon, Tokyo, Japan) equipped with a SPOT RT color digital camera, model 2.1.1 (Diagnostic Instruments, Sterling Heights, MI).
Colony-forming unit assays
Myeloid colony–plating assays were performed in methylcellulose-based medium supplemented with complete cytokine mix (MethoCult GF M3434; Stem Cell Technologies). We plated unfractionated bone marrow cells in methylcellulose-based medium, seeded 1 × 104 cells per plate in triplicate, and scored for colony formation 7 to 10 days later. For serial replating assays, we resuspended, pooled, and washed the remaining cells of the same genotype in phosphate-buffered saline (Gibco). We then counted the cells and replated 1 × 104 cells in triplicate under the same culture conditions as previously and scored colonies 7 to 10 days later. We performed serial replating a total of 4 times.
Bone marrow transplantation
Bone marrow cells were resuspended in Hanks balanced salt solution (Gibco) and injected into lethally irradiated (1 × 10 Gy [1000 rads]) WT recipient mice by either lateral tail vein or retroorbital injection.
Competitive transplantation experiments were performed using lineagelowKithighSca1+ (LSK) cells purified from Jak2VF, Tet2null, or Jak2VF/Tet2null mice (n = 2 pooled for each genotype) and WT LSK cells isolated from 45.1 mice. LSK cells were mixed and then injected into lethally irradiated 45.1 SJL recipients (n = 5 in each recipient group). On a single-mouse basis, 2.5 × 104 Jak2VF LSK cells were competed against 1.43 × 104 WT 45.1 LSK cells (ratio 1.7:1; 64% Jak2VF, 36% WT); 1.65 × 104 LSK Tet2null LSK cells were competed against 2.3 × 104 WT 45.1 LSK cells (ratio 0.7:1; 42% Tet2null, 58% WT); 2.0 × 104 Jak2VF/Tet2null LSK cells were competed against 2.0 × 104 WT 45.1 LSK cells (ratio 1:1; 50% Jak2VF/Tet2null, 50% WT). Bone marrow derived from Jak2V617F and Tet2 mice expressed the CD45.2 antigen, and WT competitor bone marrow cells expressed 45.1. Of note, because recipient mice also expressed 45.1, residual recipient hematopoietic cells also contributed to hematopoiesis posttransplantation (at an irradiation dose of 10 Gy, we expect approximately 10%-20% residual recipient hematopoiesis).
Purified bone marrow subpopulation transplants were performed using 2.2 × 103 short-term (ST)-HSCs (CD150− CD48− LSK) or 5.0 × 103 multipotent progenitor (MPP) (CD48+ LSK) donor cells from Jak2VF or Jak2VF/Tet2null mice (n = 2 pooled for each genotype) plus 4 × 105 supportive WT bone marrow cells injected into lethally irradiated 45.1 SJL recipients (n = 5 in each recipient group). Bone marrow derived from Jak2V617F and Tet2 mice expressed the CD45.2 antigen; recipient mice and supportive WT bone marrow cells expressed 45.1.
For bone marrow transplantation experiments, percentage chimerism was defined as the proportion of Jak2V617F or Tet2 or Jak2V617F/Tet2 cells as a percentage of total cells. That is, (%CD45.2)/(%CD45.2 + %45.1 WT) × 100%.
Gene expression profiling
LSK cells (3 × 104 to 5 × 104 per mouse) were isolated from WT (n = 4), Tet2null (n = 3), Jak2VF (n = 3), or Jak2VF/Tet2null (n = 4) mice. RNA was extracted using a PicoPure RNA isolation kit (Invitrogen) according to the manufacturer’s instructions. The samples were amplified with the Illumina TotalPrep RNA Amplification Kit (Invitrogen) and hybridized on Illumina MouseRef-8 v2.0 gene expression arrays. The data were analyzed with GenePattern online analysis software, including quantile normalization.25 Gene set enrichment analysis (GSEA) was performed across the complete list of genes ranked by signal-to-noise ratio according to their differential expression.26 STAT5A and HSC self-renewal signatures were extracted from previously published gene expression data.27,28 The microarray data set reported in this article has been deposited in the ArrayExpress repository at European Molecular Biology Laboratory–European Bioinformatics Institute (http://www.ebi.ac.uk/arrayexpress/) and is accessible through the ArrayExpress accession number E-MTAB-2986.
Statistical analysis
GraphPad Prism (GraphPad Software, La Jolla, CA) was used to analyze results and create graphs. All comparisons represent 2-tailed unpaired Student t test analysis (using the Welch correction where appropriate) unless otherwise specified.
Results
Tet2 loss accelerates the MPN phenotype of Jak2V617F mice
We have previously described a conditional knockin, which expresses Jak2V617F under the control of the endogenous Jak2 promoter (Jak2VF mice).22 The phenotype of the model closely recapitulates many of the clinical features of human MPNs, including prominent splenomegaly as a result of extramedullary hematopoiesis. To evaluate the effect of concomitant Jak2V617F expression and Tet2 loss on MPN phenotype, we generated Jak2VF/Tet2null mice and analyzed the splenic phenotype. As previously reported, Jak2VF mice exhibited marked splenomegaly (Figure 1A). Strikingly, the spleens of Jak2VF/Tet2null mice were significantly larger than those of Jak2VF mice (Figure 1A-B). We next performed histopathologic examination of the spleen and found expansion of the white pulp and enlarged partially confluent lymphoid follicles in Tet2null mice compared with WT mice (Figure 1C). In contrast, the spleens of Jak2VF and Jak2VF/Tet2null mice showed marked effacement of the white pulp and trilineage hyperplasia consistent with MPNs (Figure 1C). Similar to Jak2VF mice,22 the phenotype of Jak2VF/Tet2null mice was apparent at the time of genotyping (1 month), and mice were followed until 6 months of age.
Tet2 loss accelerates the MPN phenotype of Jak2V617F mice. (A) Spleen weights of age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice 24 to 30 weeks old (mean ± standard error of the mean [SEM]; n = 4 in each group). (B) Photograph of spleens from Jak2VF and Jak2VF/Tet2null mice. (C) Histopathologic sections of spleen from representative WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (original magnification ×4; H&E stain). (D) Frequency of CD71+ Ter119+ erythroid precursor cells in spleen from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). (E) Frequency of Mac1+ Gr1+ myeloid precursor cells in spleen from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). (F) Frequency of CD150+ CD48− LSK cells (LT-HSC), CD150− CD48− LSK cells (ST-HSC), and CD48+ LSK cells (MPP) in spleen from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). *P < .05; **P < .005; ***P < .001.
Tet2 loss accelerates the MPN phenotype of Jak2V617F mice. (A) Spleen weights of age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice 24 to 30 weeks old (mean ± standard error of the mean [SEM]; n = 4 in each group). (B) Photograph of spleens from Jak2VF and Jak2VF/Tet2null mice. (C) Histopathologic sections of spleen from representative WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (original magnification ×4; H&E stain). (D) Frequency of CD71+ Ter119+ erythroid precursor cells in spleen from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). (E) Frequency of Mac1+ Gr1+ myeloid precursor cells in spleen from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). (F) Frequency of CD150+ CD48− LSK cells (LT-HSC), CD150− CD48− LSK cells (ST-HSC), and CD48+ LSK cells (MPP) in spleen from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). *P < .05; **P < .005; ***P < .001.
We next assessed the cellular composition of the spleen using flow cytometry and found marked expansion of CD71+ Ter119+ erythroid precursor cells in Jak2VF and Jak2VF/Tet2null mice (Figure 1D) and additional expansion of Mac1+ Gr1+ myeloid precursor cells in the spleens of Jak2VF/Tet2null mice compared with Jak2VF animals (Figure 1E). Given the marked trilineage hyperplasia that we observed in the spleens of Jak2VF and Jak2VF/Tet2null mice, we performed a quantitative assessment of the splenic HSC compartment. Compared with WT or Tet2null mice, we found expansion of long-term (LT)-HSCs, (ST)-HSCs, and MPP cells in the spleens of Jak2-mutant mice irrespective of Tet2 genotype (Figure 1F). The spleens of Jak2VF/Tet2null mice were significantly larger than those of Jak2VF animals (Figure 1A), indicating that the splenic LSK compartment of Jak2VF/Tet2null mice was expanded absolutely compared with that of Jak2VF animals. In aggregate, these data indicate that Tet2 loss augments the extramedullary hematopoiesis phenotype of Jak2V617F mice through additional LSK and myeloid precursor cell expansion, resulting in enhanced splenomegaly.
Tet2 loss influences hematopoietic differentiation within the myeloid progenitor compartment of Jak2V617F mice but is insufficient to induce leukemic transformation
To evaluate the effect of Tet2 loss on the hematopoietic differentiation of Jak2V617F mice, we began by analyzing peripheral blood counts. As previously reported, Jak2V617F mice demonstrate leukocytosis (Figure 2A), erythrocytosis (Figure 2B and supplemental Figure 2B), and thrombocytosis (Figure 2C and supplemental Figure 2C-D) compared with WT animals. Homozygous loss of Tet2 led to a trend toward a further elevation in white cell count in mutant Jak2 mice (Figure 2A), whereas hematocrit and platelet numbers were similarly elevated in both Jak2VF and Jak2VF/Tet2null animals (Figure 2B-C). More marked leukocytosis developed in Jak2VF/Tet2null mice that were older than 6 months (supplemental Figure 2A). Finally, we reviewed peripheral blood smears and found that platelets from Jak2VF/Tet2null mice were large and showed signs of dysplasia, including increased basophilia of the cytoplasm, a tendency to aggregate, abnormal shape, and pseudopod formation (supplemental Figure 2D).
Tet2 loss influences hematopoietic differentiation within the myeloid progenitor compartment of Jak2V617F mice but is insufficient to induce leukemic transformation. (A-C) White blood cell (WBC) count, hematocrit, and platelet (PLT) count of age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice 24 to 30 weeks old (mean ± SEM; n = 4 in each group). (D-F) Relative frequency of common myeloid progenitor (CMP), granulocyte macrophage progenitor (GMP), and megakaryocyte erythroid progenitor (MEP) cells in bone marrow from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). (G-I) Histopathologic sections of bone marrow from representative WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (original magnifications ×10 for G1-G4, ×40 for H1-H4, and ×100 for I1-I4; H&E stain).
Tet2 loss influences hematopoietic differentiation within the myeloid progenitor compartment of Jak2V617F mice but is insufficient to induce leukemic transformation. (A-C) White blood cell (WBC) count, hematocrit, and platelet (PLT) count of age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice 24 to 30 weeks old (mean ± SEM; n = 4 in each group). (D-F) Relative frequency of common myeloid progenitor (CMP), granulocyte macrophage progenitor (GMP), and megakaryocyte erythroid progenitor (MEP) cells in bone marrow from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). (G-I) Histopathologic sections of bone marrow from representative WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (original magnifications ×10 for G1-G4, ×40 for H1-H4, and ×100 for I1-I4; H&E stain).
Next, we focused on the bone marrow. We have previously reported that Jak2V617F induces erythroid skewing in the myeloid progenitor compartment.22 Consistent with these findings, we found a relative decrease in common myeloid progenitor cells and a relative increase in megakaryocyte-erythroid progenitor cells in the myeloid progenitor bone marrow compartment of Jak2VF mice compared with WT mice, which was not seen in Jak2VF/Tet2null mice (Figure 2D-F). These findings indicate that Tet2 loss reduces Jak2V617F-induced erythroid skewing in the myeloid progenitor compartment. We also performed a quantitative analysis of HSCs in the bone marrow and found a significant expansion of LT-HSCs in Tet2null, Jak2VF, and Jak2VF/Tet2null mice compared with WT animals, and a significant expansion of MPP cells in Jak2VF/Tet2null mice compared with WT animals (supplemental Figure 3A).
TET2 loss-of-function mutations have been associated with leukemic transformation in MPNs.18,29-31 Therefore, we compared the propensity for leukemic transformation in Jak2VF/Tet2null mice relative to control animals. In the bone marrow of both Jak2VF and Jak2VF/Tet2null mice, we found trilineage hyperplasia consistent with MPNs, with more prominent myeloid expansion and less prominent eythroid hyperplasia observed in Jak2VF/Tet2null marrow relative to Jak2VF marrow (Figure 2G-H). We did not see, however, histopathologic evidence of acute myeloid leukemia (AML) in the bone marrow of Jak2VF/Tet2null mice up to 6 months old (n = 12). Because Jak2VF mice die prematurely at approximately 6 months22 and Jak2VF/Tet2null mice have a comparable survival, it was not possible to follow Jak2VF/Tet2null animals for an extended period beyond 6 months. To further evaluate for leukemic transformation, we measured the percentage of circulating c-Kit+ cells using flow cytometry and found no difference between Jak2VF and Jak2VF/Tet2null animals (supplemental Figure 3B).
Finally, given that TET2 mutations are enriched in myelofibrosis compared to essential thrombocythemia16 and that megakaryocytes are a key cellular driver of fibrotic transformation in MPNs,32 we focused on megakaryocytes within the bone marrow of Jak2VF/Tet2null mice. We found that the number of megakaryocytes was increased in Jak2VF/Tet2null compared with Jak2VF mice and that there was greater heterogeneity in megakaryocyte size in Jak2VF/Tet2null compared with Jak2VF animals (supplemental Figure 4A-B). We next performed a detailed histopathologic analysis and noted emperipolesis and abnormal clustering of megakaryocytes consistent with MPNs in the bone marrow of both Jak2VF and Jak2VF/Tet2null mice (Figure 2I). With more detailed morphologic analysis, we further noted that Jak2VF/Tet2null megakaryocytes appeared apoptotic and showed additional atypical features including pyknotic nuclei and atypical mitotic figures. In aggregate, these findings suggest higher megakaryocyte turnover in Jak2VF/Tet2null compared with Jak2VF animals (Figure 2I and supplemental Figure 4C). Finally, we performed reticulin staining but did not see evidence of reticulin fibrosis in the bone marrow of Jak2VF/Tet2null mice up to 6 months old (n = 12) (supplemental Figure 4D).
Tet2 loss confers enhanced self-renewal to Jak2V617F bone marrow cells in vitro
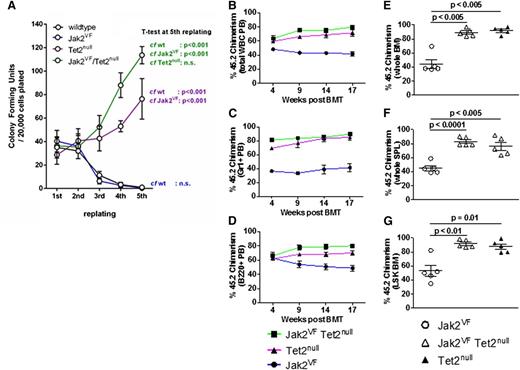
Next, we turned to the functional effects of concomitant Jak2V617F expression and Tet2 loss on HSPCs by performing serial replating colony-forming unit assays. We compared colony-forming unit assay formation using unfractionated bone marrow cells derived from Jak2VF, Tet2null, or Jak2VF/Tet2null mice. We found that from the third replating onward, Tet2null and Jak2VF/Tet2null cells had markedly enhanced replating activity compared to Jak2VF or WT cells (Figure 3A). These findings indicate that homozygous Tet2 loss confers increased self-renewal potential in vitro to Jak2V617F-expressing hematopoietic cells.
Tet2 loss confers enhanced self-renewal to Jak2V617F HSPCs in vitro and in vivo. (A) Colony-forming unit assays from unfractionated bone marrow derived from WT, Jak2VF, Tet2null, and Jak2VF/Tet2null mice. Results represent the average of triplicate assays (mean ± SEM). The percentage of 45.2 donor chimerism assessed in peripheral blood (PB) total WBCs (B), PB Gr1+ cells (C), and PB B220+ cells (D) from lethally irradiated secondary recipients of Jak2VF, Tet2null, or Jak2VF/Tet2null LSK cells competed against an approximately equal number of 45.1 WT LSK cells, measured 4 to 17 weeks posttransplantation (mean ± SEM; n = 5 in each group). The percentage of 45.2 donor chimerism assessed in whole bone marrow (BM) cells (E), whole spleen (SPL) (F), and LSK BM cells (G) from lethally irradiated secondary recipients of Jak2VF, Tet2null, or Jak2VF/Tet2null LSK cells competed against an approximately equal number of 45.1 WT LSK cells, measured 18 weeks posttransplantation (mean ± SEM; n = 5 in each group). P values for each of the comparisons are indicated in the figure.
Tet2 loss confers enhanced self-renewal to Jak2V617F HSPCs in vitro and in vivo. (A) Colony-forming unit assays from unfractionated bone marrow derived from WT, Jak2VF, Tet2null, and Jak2VF/Tet2null mice. Results represent the average of triplicate assays (mean ± SEM). The percentage of 45.2 donor chimerism assessed in peripheral blood (PB) total WBCs (B), PB Gr1+ cells (C), and PB B220+ cells (D) from lethally irradiated secondary recipients of Jak2VF, Tet2null, or Jak2VF/Tet2null LSK cells competed against an approximately equal number of 45.1 WT LSK cells, measured 4 to 17 weeks posttransplantation (mean ± SEM; n = 5 in each group). The percentage of 45.2 donor chimerism assessed in whole bone marrow (BM) cells (E), whole spleen (SPL) (F), and LSK BM cells (G) from lethally irradiated secondary recipients of Jak2VF, Tet2null, or Jak2VF/Tet2null LSK cells competed against an approximately equal number of 45.1 WT LSK cells, measured 18 weeks posttransplantation (mean ± SEM; n = 5 in each group). P values for each of the comparisons are indicated in the figure.
Tet2 loss confers a competitive repopulating advantage to Jak2V617F HSCs
To further evaluate the functional impact of Tet2 loss on Jak2V617F HSCs in vivo, we performed competitive bone marrow transplantation experiments. We transplanted LSK cells derived from Jak2VF, Tet2null, or Jak2VF/Tet2null mice into lethally irradiated congenic recipients, together with an approximately equal number of 45.1 WT competitor LSK cells (see “Materials and methods” for precise ratios). The blood counts were broadly similar in all recipient groups (supplemental Figure 5A-C). We assessed peripheral blood chimerism in total white blood cells, myeloid cells (Gr1+), B-lymphoid cells (B220+), or T-lymphoid cells (CD3+) and found a competitive advantage over WT cells for the Jak2VF/Tet2null and Tet2null cells in each of these compartments (Figure 3B-D and supplemental Figure 5D). This competitive advantage was present at 4 weeks posttransplantation and was sustained at 17 weeks. We assessed chimerism in the bone marrow and spleen at 18 weeks post bone marrow transplantation and found a strong competitive advantage for Jak2VF/Tet2null and Tet2null cells (Figure 3E-F). Chimerism in the LSK compartment at 18 weeks showed that >90% of the cells were derived from Jak2VF/Tet2null or Tet2null donors in these groups (Figure 3G). We next determined the cell cycle status of LSK cells from primary mice from each of the genotypes. Consistent with our previously published findings,33 we found a more activated cell cycle in Jak2VF LSK cells compared to WT LSK cells (supplemental Figure 5G). We also found a more activated cell cycle in both Jak2VF/Tet2null LSK and linage c-Kitlo/c-Kithi cells compared to WT LSK and linage c-Kitlo/c-Kithi cells (supplemental Figure 5H). We have previously demonstrated that MPN disease-propagating cells are contained exclusively in the LT-HSC (CD150+ CD48− LSK) compartment of Jak2V617F mice and that Jak2V617F expression in ST-HSCs and MPP cells does not confer long-term self-renewal capability to these cell populations.34 To determine whether Tet2 loss enhanced the self-renewal of Jak2V617F-mutant ST-HSCs or MPP cells, we compared the competitive repopulation of ST-HSCs (CD150− CD48− LSK) or MPP cells (CD48+ LSK) purified from Jak2VF or Jak2VF/Tet2null mice using bone marrow transplantation and found no difference in self-renewal capacity between the groups at 19 weeks (supplemental Figure 3E-F). In aggregate, these data demonstrate that (1) homozygous Tet2 loss confers a competitive advantage to Jak2V617F-mutant LT-HSCs similar to that seen in Tet2-deficient LT-HSCs that are WT for Jak2 and (2) deleting Tet2 in Jak2V617F-mutant ST-HSCs or MPP cells does not confer long-term self-renewal capability to these cell populations.
Jak2V617F expression and Tet2 loss cause distinct and nonoverlapping gene expression changes
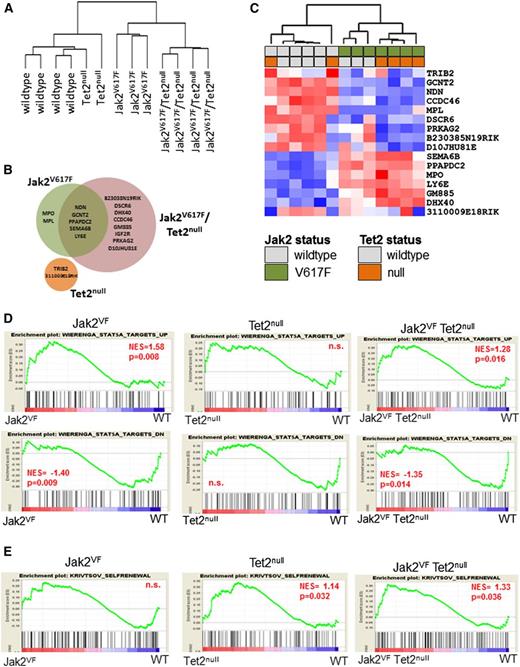
To interrogate the molecular pathways responsible for the different HSC phenotypes of Jak2VF and Jak2VF/Tet2null mice, we performed gene expression profiling of LSK cells derived from WT, Jak2VF, Tet2null, or Jak2VF/Tet2null animals (n = 2-4 mice per group). Unsupervised hierarchical clustering of the global gene expression signatures revealed two main branches in the gene expression hierarchy, with WT and Tet2null samples clustering closely along one branch, and Jak2VF and Jak2VF/Tet2null samples composing the other (Figure 4A). This finding indicates that Jak2V617F expression imparts a larger effect on global gene expression patterns than Tet2 loss. We successfully identified 17 unique transcripts that were differentially regulated by Jak2V617F expression, Tet2 loss, or both (false discovery rate <10%; minimum fold change relative to WT samples = 1.3) (Figure 4B; supplemental Table 1). These transcripts included those unique to LSK cells harboring the Jak2VF allele (n = 2), Tet2null allele (n = 2), or the Jak2VF/Tet2null alleles (n = 8), as well as transcripts that were common to Jak2VF and Jak2VF/Tet2null LSK cells (n = 5) (Figure 4B-C).
Jak2V617F expression and Tet2 loss cause distinct and nonoverlapping gene expression changes. (A) Dendrogram constructed from unsupervised hierarchical clustering of all 13 data sets from WT (n = 4), Tet2null (n = 2), Jak2VF (n = 3), and Jak2VF/Tet2null (n = 4) LSK cells using Pearson correlation. (B) Venn diagram depicting differentially expressed genes in LSK cells from Jak2VF, Tet2null, and Jak2VF/Tet2null mice (false discovery rate = 10%; minimum fold change relative to WT samples = 1.3). (C) Hierarchical clustering of expression profiles of all 12 data sets according to the 17 genes differentially expressed in either Jak2VF, Tet2null, or Jak2VF/Tet2null mice relative to WT controls. A red/blue color scale depicts normalized gene expression levels (red: high; blue: low). Dendrograms were constructed using Pearson correlation. (D) GSEA demonstrating enrichment for STAT5A target genes in Jak2VF and Jak2VF/Tet2null LSK cells but not in Tet2null LSK cells (top row: STAT5A targets UP; bottom row: STAT5A targets DOWN). (E) GSEA demonstrating enrichment of an HSC self-renewal signature in Tet2null and Jak2VF/Tet2null LSK cells but not in Jak2VF LSK cells. P values for each of the comparisons are indicated in the figure. NES, net enrichment score; n.s., not significant.
Jak2V617F expression and Tet2 loss cause distinct and nonoverlapping gene expression changes. (A) Dendrogram constructed from unsupervised hierarchical clustering of all 13 data sets from WT (n = 4), Tet2null (n = 2), Jak2VF (n = 3), and Jak2VF/Tet2null (n = 4) LSK cells using Pearson correlation. (B) Venn diagram depicting differentially expressed genes in LSK cells from Jak2VF, Tet2null, and Jak2VF/Tet2null mice (false discovery rate = 10%; minimum fold change relative to WT samples = 1.3). (C) Hierarchical clustering of expression profiles of all 12 data sets according to the 17 genes differentially expressed in either Jak2VF, Tet2null, or Jak2VF/Tet2null mice relative to WT controls. A red/blue color scale depicts normalized gene expression levels (red: high; blue: low). Dendrograms were constructed using Pearson correlation. (D) GSEA demonstrating enrichment for STAT5A target genes in Jak2VF and Jak2VF/Tet2null LSK cells but not in Tet2null LSK cells (top row: STAT5A targets UP; bottom row: STAT5A targets DOWN). (E) GSEA demonstrating enrichment of an HSC self-renewal signature in Tet2null and Jak2VF/Tet2null LSK cells but not in Jak2VF LSK cells. P values for each of the comparisons are indicated in the figure. NES, net enrichment score; n.s., not significant.
The paucity of differentially expressed genes in multiple comparisons and the modest magnitudes of the fold change suggested that individually, these genes were unlikely to be the sole drivers of the phenotypic differences in the Jak2VF, Tet2null, or Jak2VF/Tet2null animals. Therefore, we used GSEA to identify more subtle perturbations downstream of Jak2V617F expression or Tet2 loss in LSK cells by leveraging the data set in its entirety for the presence of specific gene signatures (supplemental Tables 2 and 3). We found that targets of STAT5A signaling27 were enriched among genes that were differentially expressed in Jak2VF and Jak2VF/Tet2null LSK cells, but not in genes differentially expressed in Tet2null LSK cells (Figure 4D). In addition, we found enrichment of genes that harbor putative STAT5 binding sites (defined by the motif NAWTTCYN within +/−2kb of transcriptional start site) only in mice harboring a mutant Jak2 allele but not in Tet2-deleted LSK cells (supplemental Figure 6A). Concomitantly, increased STAT5A signatures were correlated with enrichment of gene signatures from erythroid-, megakaryocytic-, and granulocyte macrophage–committed precursors,35 a feature of Jak2VF and Jak2VF/Tet2null (but not Tet2null) LSK cells (supplemental Figure 6B). Taken together, these findings demonstrate that the LSK compartment of mice harboring mutant Jak2 exhibits genetic signatures indicative of robust activation of the Jak2-Stat5 signaling axis.
Next, we performed GSEA using a 363-gene signature that has previously been shown to be associated with a murine leukemic stem cell self-renewal signature.28 We found significant enrichment of an HSC self-renewal signature in LSK cells from Jak2VF/Tet2null and Tet2null mice but not in LSK cells from Jak2VF animals (Figure 4E). These findings are consistent with our in vitro and in vivo data demonstrating both increased serial replating capacity and enhanced competitive repopulation activity for Tet2-deficient LSK cells, irrespective of Jak2 status.
Discussion
MPNs are primarily disorders of activated intracellular signaling, with the majority of patients harboring mutations in genes that encode proteins that regulate cytokine signaling.4 The recent identification of CALR mutations provides further evidence that MPNs are diseases driven by aberrant signal transduction.3,13 Mutations in genes involved in epigenetic regulation (eg, TET2, ASXL1, DNMT3A, and EZH2) are the most frequently comutated genes with signaling mutations in MPNs,18,36 suggesting that genetic cooperation may occur between these two classes of genes in MPNs. TET2 mutations are known to occur within the HSC compartment of MPN patients,37 and although an initial clonal analysis of patient samples demonstrated that TET2 mutations can precede or follow the JAK2V617F mutation,38 a more recent larger study has shown that TET2 mutations are predominantly acquired prior to JAK2V617F.18 Initial functional studies in immunodeficient mice demonstrated that TET2-JAK2V617F comutated CD34+ HSCs have increased repopulating capacity over JAK2V617F-mutated CD34+ HSCs.37 In this study, using syngeneic genetic murine models that we have developed, we determined the individual and combinatorial effects of Jak2V617F expression and Tet2 loss on (1) disease phenotype; (2) HSPC function; and (3) HSC signaling and self-renewal transcriptional signatures.
We found that homozygous Tet2 loss accelerated the MPN phenotype of Jak2V617F mice, as evidenced by expansion of the splenic HSC compartment, by enhanced extramedullary hematopoiesis, and by splenomegaly. Although mutations in epigenetic regulators are enriched in more advanced phases of MPN such as myelofibrosis and secondary AML,14,16,18,29-31 in our study, combining a mutant Jak2 allele with a Tet2null allele was insufficient to induce fibrotic or leukemic transformation at 6 months of age. The fact that AML which arises out of JAK2V617F-mutant MPNs retains the JAK2V617F allele only approximately 50% of the time39,40 suggests that only a subset of genetic lesions seen in secondary AML cooperate with JAK2V617F and that cell nonautonomous mechanisms of transformation may also occur. One additional note is that in our study we used VavCre to target Cre recombinase to the hematopoietic lineage,24 and in so doing, Jak2V617F expression and Tet2 loss occurred simultaneously in VavCre-positive cells. In MPN patients, JAK2V617F and TET2 mutations typically occur sequentially,18 and it is possible that the order in which these mutations are acquired influences the disease phenotype. Because VavCre is noninducible, we were unable to address the impact of the temporal order of mutation acquisition on MPN phenotype.
In functional studies, we found a strong competitive advantage for Jak2VF/Tet2null compound mutant HSPCs, together with marked expansion of Jak2VF/Tet2null myeloid and erythroid precursor cell populations, resulting in an enhanced MPN phenotype in Jak2VF/Tet2null mice compared with Jak2VF or Tet2null animals. These findings are consistent with a model in which Tet2 loss drives clonal dominance in HSCs, and Jak2V617F expression causes expansion of downstream progenitor and precursor cell populations, resulting in disease progression through combinatorial effects. Furthermore, these findings are consistent with the facts that the JAK2V617F allele burden in the HSC compartment of polycythemia vera and essential thrombocythemia patients is low and that JAK2V617F-mutant HSCs predominate in more advanced phases of MPNs such as myelofibrosis in which epigenetic mutations are enriched.41
The results of the functional studies outlined above are supported by our findings from the gene expression profiling of LSK cells. In this study, we found that both Jak2V617F expression and Tet2 loss were associated with distinct and nonoverlapping gene expression signatures in the HSC compartment. Using GSEA, we found that a STAT5A signature was enriched only in Jak2VF LSK cells, that an HSC self-renewal signature was enriched only in Tet2null LSK cells, but that both signatures were enriched in Jak2VF/Tet2null LSK cells. These data demonstrate that Jak2V617F expression and Tet2 loss each exert a specific effect on the transcriptional program of LSK cells. Specifically, Jak2V617F facilitates increased STAT5 signaling, which is known to potentiate erythroid differentiation, whereas Tet2 loss gives rise to a leukemic stem cell transcriptional program that is associated with increased HSC self-renewal. The combined effects of these transcriptional changes in the HSC compartment drive the development of a more florid MPN phenotype (Jak2VF/Tet2null mice) compared to the phenotype that arises when either transcriptional program manifests alone (Jak2VF or Tet2null mice).
In conclusion, we report the effects of homozygous Tet2 loss on Jak2V617F-mediated MPNs. Overall, we find accelerated myeloproliferation but no overt fibrotic or leukemic transformation. In aggregate, this work elucidates the functional effects of combined Jak2V617F expression and Tet2 loss in distinct hematopoietic compartments in vivo and provides insight into the mechanisms of clonal dominance and disease progression in MPNs.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (K08 HL109734) (A.M.), the MPN Foundation (A.M.), and the Leukemia Research Foundation (A.M.). A.M. has received support from the Jeanne D. Housman Fund for Research on Myeloproliferative Disorders and is a recipient of a Damon Runyon clinical investigator award. E.C. is a recipient of a Lady Tata Memorial Trust Award, and E.A.R. is a recipient of an American Society of Hematology HONORS Award.
Authorship
Contribution: A.M., E.C., R.L., and B.L.E. designed experiments and interpreted data; A.M., L.J.B., E.A.R., L.P., S.E., A.K., and K.B. performed experiments; E.C. performed experiments and analyzed gene expression data; R.K.S. performed experiments and reviewed, interpreted, and photographed histopathology; and A.M. and E.C. wrote the manuscript. All authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ann Mullally, Brigham and Women’s Hospital, Karp Building 5.215, 1 Blackfan Circle, Boston, MA 02115; e-mail: amullally@partners.org.

![Figure 1. Tet2 loss accelerates the MPN phenotype of Jak2V617F mice. (A) Spleen weights of age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice 24 to 30 weeks old (mean ± standard error of the mean [SEM]; n = 4 in each group). (B) Photograph of spleens from Jak2VF and Jak2VF/Tet2null mice. (C) Histopathologic sections of spleen from representative WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (original magnification ×4; H&E stain). (D) Frequency of CD71+ Ter119+ erythroid precursor cells in spleen from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). (E) Frequency of Mac1+ Gr1+ myeloid precursor cells in spleen from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). (F) Frequency of CD150+ CD48− LSK cells (LT-HSC), CD150− CD48− LSK cells (ST-HSC), and CD48+ LSK cells (MPP) in spleen from age-matched WT, Tet2null, Jak2VF, and Jak2VF/Tet2null mice (mean ± SEM; n = 4 in each group). *P < .05; **P < .005; ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/2/10.1182_blood-2014-04-567024/4/m_327f1.jpeg?Expires=1769805521&Signature=X9ovvhk80gZ22qCDEvldtSz4GN6eSAIu4vWlzq0~VBuQaWWSM-Dbbuaxe5Szvd9QpC4Lax~2H4HSp35fXkCutmVcvlwaVSBIr5KLOg9Zvs385kmlyjzHrUbwCMbIILo9VUMgIKNH2-XFmGZRtsjDGGe4ZPN~ksuxW0YOO4nrDlmGjO1pqnQQLNolWeStEXNy9MApcLTspOkHlF3Lb1dx-sQOU7-aKe09nM7vFQ8K-feNcNtt0a6Fkiy~7N1hb8JyGYmxhY3oAptwWrRgnXWdGym9PvTtDRP--CElKyzGITlKqELZO4vOL7Uxm1gzyow~Tuywz8VZG3~wPsu7POaR8w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal