Key Points
Intestinal diversity is predictive of mortality in allo-HSCT.
Abstract
Highly diverse bacterial populations inhabit the gastrointestinal tract and modulate host inflammation and promote immune tolerance. In allogeneic hematopoietic stem cell transplantation (allo-HSCT), the gastrointestinal mucosa is damaged, and colonizing bacteria are impacted, leading to an impaired intestinal microbiota with reduced diversity. We examined the impact of intestinal diversity on subsequent mortality outcomes following transplantation. Fecal specimens were collected from 80 recipients of allo-HSCT at the time of stem cell engraftment. Bacterial 16S rRNA gene sequences were characterized, and microbial diversity was estimated using the inverse Simpson index. Subjects were classified into high, intermediate, and low diversity groups and assessed for differences in outcomes. Mortality outcomes were significantly worse in patients with lower intestinal diversity; overall survival at 3 years was 36%, 60%, and 67% for low, intermediate, and high diversity groups, respectively (P = .019, log-rank test). Low diversity showed a strong effect on mortality after multivariate adjustment for other clinical predictors (transplant related mortality: adjusted hazard ratio, 5.25; P = .014). In conclusion, the diversity of the intestinal microbiota at engraftment is an independent predictor of mortality in allo-HSCT recipients. These results indicate that the intestinal microbiota may be an important factor in the success or failure in allo-HSCT.
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT), a potentially curative treatment of a range of hematologic malignancies and primary immune disorders, is associated with a variety of complications including infections and graft-versus-host disease (GVHD). The gastrointestinal tract is particularly impacted by allo-HSCT, serving as both the origin and target of many posttransplant complications.1
Under normal circumstances, the human gastrointestinal tract harbors a highly diverse population of microbes that, to a large degree, consists of obligate anaerobic bacteria.2 During allo-HSCT, many patients undergo dramatic alterations of their intestinal microbiota, with marked decreases in overall bacterial diversity. In many instances, a single bacterial taxon can predominate and replace a previously rich and complex distribution of organisms.3 Some allo-HCST patients, however, maintain intestinal bacterial diversity throughout the course of transplantation. The clinical consequences of these differences are still unclear.
We sought to explore the implications of microbiota diversity loss by correlating them with long-term clinical outcomes. In this study, we examined the intestinal microbiota of patients following allo-HSCT by analyzing fecal specimens collected at the time of stem cell engraftment and correlating those changes with mortality over the subsequent 3 years.
Methods
Patients and specimen collection
Subjects consisted of adult patients undergoing allo-HSCT at Memorial Sloan-Kettering Cancer Center (MSKCC). Patients were enrolled in a fecal collection protocol, where feces were collected during the initial transplant hospitalization and stored in a biospecimen bank. This fecal biospecimen cohort has been described in a previous study.3 For each subject, we analyzed fecal biospecimens collected within 7 days following stem cell engraftment (absolute neutrophil count >500 cells/μL for 3 consecutive days), which we referred to as the engraftment sample. Subjects were included for study if stem cell engraftment was successfully achieved and if a fecal biospecimen was collected within 7 days after engraftment. Clinical characteristics of the study patients were compared with those of patients who were excluded, as well as nonenrolled patients during the same time period. The study was approved by the institutional review board at MSKCC. All study patients provided written informed consent for biospecimen collection and analysis. The study was conducted in accordance with the Declaration of Helsinki.
Transplantation practices
At our institution, antimicrobial prophylaxis is given routinely to patients undergoing allo-HSCT. Subjects receiving myeloablative or reduced intensity conditioning regimens were given ciprofloxacin and intravenous vancomycin for prophylaxis of gram-negative infections and infections with viridans streptococci, respectively. Routine ciprofloxacin prophylaxis was discontinued January 2011 but was later resumed during July 2011. Antibiotic prophylaxis against Pneumocystis carinii was generally administered, but this practice was not officially standardized at our institution. Medications typically used included trimethoprim-sulfamethoxazole, aerosolized pentamadine, and atovaquone; the time at which prophylaxis was initiated (during conditioning or after engraftment) varied. Our institution does not administer antibiotics such as metronidazole for purposes of gut decontamination and/or prevention of GVHD. Piperacillin-tazobactam was the standard default antibiotic given for fever during neutropenia in adult patients.
Administration of ursodeoxycholic acid for prophylaxis of cholestasis was given according to the discretion of the transplant physician.
Analysis of specimens
For each fecal specimen, DNA was extracted and purified, and the V1 to V3 region of the 16S rRNA gene was polymerase chain reaction (PCR)-amplified using modified universal bacterial primers. Purified PCR products were sequenced on a 454 GS FLX Titanium platform. Sequence data were compiled and processed using mothur version 1.33.4 Sequence data were screened and filtered for quality5 and then aligned to the full-length 16S rRNA gene, using as a template the SILVA reference alignment.6 Sequences were grouped into operational taxonomic units of 97% similarity. Additional details are provided in supplemental Methods, available on the Blood Web site.
Microbial diversity was estimated by calculating the inverse Simpson index,7 an ecological estimate of α diversity, calculated to represent the reciprocal of the expected probability of 2 randomly selected bacterial sequences as belonging to the same operational taxonomic unit. Subjects were grouped into 3 levels of postengraftment microbial diversity, based on the inverse Simpson index: high (>4), intermediate (2-4), and low (<2). Phylogenetic classification to genus level was performed using the naïve Bayesian classification scheme described and Wang et al8 using the Greengenes reference database.9
Statistical analysis
We examined the ability of microbial diversity at stem cell engraftment to predict overall survival, our primary outcome of interest. Analysis time began at the time of postengraftment biospecimen collection and continued for up to 3 years until death from any cause or last follow-up. Kaplan-Meier estimates were calculated, and differences in survival were assessed using log-rank test. A similar analysis was performed for transplant-related mortality, defined as death not caused by relapse or progression of malignancy. Other cause-specific mortality end points were also assessed: death due to relapse or progression and death due to GVHD or infection. For these end points, cause of death was determined using criteria from the American Society for Blood and Marrow Transplantation (ASBMT), and defined by Copelan et al.10
Cox proportional hazards modeling was used to examine microbial diversity as a predictor of overall survival. Other clinical variables were also examined as predictors: age (at transplant), sex, underlying disease, pretransplant comorbidity assessed by the hematopoietic cell transplantation comorbidity index (HCT-CI),11 disease risk based on the 2014 ASBMT Request for Information (RFI) Disease Classifications schema (http://www.asbmt.org), conditioning regimen intensity,12 stem cell source, and ex-vivo T-cell depletion of stem cells. In addition, the following transplant events were also included in the analysis: time to stem cell engraftment, administration of ursodeoxycholic acid, acute kidney injury (serum creatinine concentration 2 times as high as baseline value), liver dysfunction (total serum bilirubin 2 times as high as baseline value), whether total parenteral nutrition (TPN) was given, whether antibiotics were administered (intravenous vancomycin, fluoroquinolones, metronidazole, and β-lactams), and diagnosed infections (Clostridium difficile infection, gram-negative bloodstream infection, and vancomycin-resistant Enterococcus [VRE] bloodstream infection). These event variables were assessed during the transplant hospitalization period occurring prior to the start of analysis time (ie, prior to postengraftment specimen collection). These variables were first examined as univariate predictors of mortality; variables with univariate P < .20 were included in a multivariate analysis. These analyses were performed using R version 3.0 (R Development Core Team, Vienna, Austria).
To further examine differences in postengraftment microbiota, we compared the relative abundances of phylogenetic taxa between subjects who died within the 3-year follow-up period compared with those who did not. Phylogenetic abundance comparisons were performed to identify biomarkers of overall survival using linear discriminant analysis (LDA) effect size (LEfSe) analysis,13 where conditioning regimen intensity was used for subclass comparisons. We used a logarithmic LDA cutoff of 2.0, and α of 0.10 for Kruskal-Wallis testing among classes and for Wilcoxon testing between subclasses. Data from this study are stored in the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra).
Results
A total of 80 patients from the biospecimen cohort met study criteria and were selected for analysis. From the original cohort of 94 patients,3 14 patients were excluded either due to primary graft failure or because a fecal specimen was not collected within 7 days after engraftment. The study subjects underwent allo-HSCT from September 4, 2009 to August 4, 2011; pretransplant characteristics were similar to the excluded subjects, as well as the overall allo-HSCT case mix at MSKCC during the same period (supplemental Table 1). The fecal specimens collected from each subject after engraftment yielded an average of 4000 high-quality 16S rRNA bacterial sequences per specimen (ranging from 1330 to 8259).
Subjects were classified into 1 of 3 groups based on overall bacterial diversity using the inverse Simpson index as described above. Table 1 shows the clinical characteristics of subjects within the 3 microbial diversity groups. At the time of stem cell engraftment, high microbial diversity was maintained in 26 (32.5%) patients, intermediate diversity was observed in 20 (25.0%) patients, and low diversity was observed in 34 (42.5%) patients. Characteristics of patients within each of the diversity groups differed in terms of relative distribution. Patients with lower bacterial diversity were likely to have received conditioning regimens of greater intensity compared with those with higher bacterial diversity. In examining each individual element comprising the conditioning regimens, we found no evidence that this association was due to any 1 particular drug or to total body irradiation (supplemental Table 2); rituximab was associated with higher diversity but was exclusively given in nonmyeloablative regimens. Patients who developed acute kidney injury, received any of several antibiotics (intravenous vancomycin, metronidazole, β-lactams), or were diagnosed with C difficile infection more frequently fell into the low diversity group. Underlying disease, pretransplant morbidity, and time to engraftment were not different by Fisher’s exact test, but still demonstrated distributional differences suggestive of some degree of association with microbial diversity.
Clinical characteristics of study patients
| Variables . | Intestinal bacterial diversity,* postengraftment . | P value† . | ||
|---|---|---|---|---|
| High diversity (N = 26) . | Intermediate diversity (N = 20) . | Low diversity (N = 34) . | ||
| Pretransplant characteristics | ||||
| Age (years) | .671 | |||
| <35 | 4 (15.4%) | 3 (15.0%) | 6 (17.6%) | |
| 35-49 | 6 (23.1%) | 5 (25.0%) | 13 (38.2%) | |
| ≥50 | 16 (61.5%) | 12 (60.0%) | 15 (44.1%) | |
| Gender (female) | 13 (50.0%) | 6 (30.0%) | 16 (47.1%) | .346 |
| Underlying disease | .077 | |||
| Leukemia | 8 (30.8%) | 8 (40.0%) | 22 (64.7%) | |
| Lymphoma | 11 (42.3%) | 6 (30.0%) | 4 (11.8%) | |
| Multiple myeloma | 2 (7.7%) | 3 (15.0%) | 3 (8.8%) | |
| Myelodysplastic syndrome | 5 (19.2%) | 2 (10.0%) | 3 (8.8%) | |
| Other | 0 (0.0%) | 1 (5.0%) | 2 (5.9%) | |
| Pretransplant comorbidity (HCT-CI) | .121 | |||
| 0-1 | 15 (57.7%) | 7 (35.0%) | 8 (23.5%) | |
| 2-3 | 6 (23.1%) | 8 (40.0%) | 15 (44.1%) | |
| 4+ | 5 (19.2%) | 5 (25.0%) | 11 (32.4%) | |
| Disease risk (ASBMT RFI) | .388 | |||
| Low | 9 (34.6%) | 3 (15.0%) | 13 (38.2%) | |
| Intermediate | 6 (23.1%) | 4 (20.0%) | 7 (20.6%) | |
| High | 11 (42.3%) | 13 (65.0%) | 14 (41.2%) | |
| Transplant characteristics | ||||
| Conditioning intensity | .019 | |||
| Nonmyeloablative | 8 (30.8%) | 6 (30.0%) | 1 (2.9%) | |
| Reduced intensity | 7 (26.9%) | 7 (35.0%) | 12 (35.3%) | |
| Myeloablative | 11 (42.3%) | 7 (35.0%) | 21 (61.8%) | |
| Stem cell source | .168 | |||
| Related identical sibling | 11 (42.3%) | 9 (45.0%) | 9 (26.5%) | |
| Unrelated identical | 11 (42.3%) | 3 (15.0%) | 11 (32.4%) | |
| Unrelated nonidentical | 2 (7.7%) | 2 (10.0%) | 4 (11.8%) | |
| Umbilical cord | 2 (7.7%) | 6 (30.0%) | 10 (29.4%) | |
| T-cell depletion (ex-vivo) | 11 (42.3%) | 8 (40.0%) | 17 (50.0%) | .744 |
| Transplant course | ||||
| Time to engraftment (≥14 days posttransplant) | 3 (11.5%) | 6 (30.0%) | 10 (29.4%) | .197 |
| Ursodeoxycholic acid‡ | 3 (11.5%) | 5 (25.0%) | 9 (26.5%) | .328 |
| Liver dysfunction (total bilirubin > twice baseline) | 3 (11.5%) | 2 (10.0%) | 5 (14.7%) | 1.000 |
| Acute kidney injury (creatinine > twice baseline) | 2 (7.7%) | 5 (25.0%) | 15 (44.1%) | .006 |
| TPN‡ | 17 (65.4%) | 13 (65.0%) | 29 (85.3%) | .133 |
| Antibiotic‡ | ||||
| Vancomycin (intravenous) | 20 (76.9%) | 18 (90.0%) | 34 (100.0%) | .007 |
| Fluoroquinolone§ | 7 (26.9%) | 5 (25.0%) | 10 (29.4%) | 1.000 |
| Metronidazole | 4 (15.4%) | 4 (20.0%) | 15 (44.1%) | .031 |
| β-lactam¶ | 17 (65.4%) | 18 (90.0%) | 33 (97.1%) | .002 |
| Infection‡ | ||||
| C difficile infection | 1 (3.8%) | 1 (5.0%) | 12 (35.3%) | .002 |
| Gram-negative bloodstream infection | 1 (3.8%) | 3 (15.0%) | 6 (17.6%) | .262 |
| VRE bloodstream infection | 1 (3.8%) | 0 (0.0%) | 3 (8.8%) | .454 |
| Variables . | Intestinal bacterial diversity,* postengraftment . | P value† . | ||
|---|---|---|---|---|
| High diversity (N = 26) . | Intermediate diversity (N = 20) . | Low diversity (N = 34) . | ||
| Pretransplant characteristics | ||||
| Age (years) | .671 | |||
| <35 | 4 (15.4%) | 3 (15.0%) | 6 (17.6%) | |
| 35-49 | 6 (23.1%) | 5 (25.0%) | 13 (38.2%) | |
| ≥50 | 16 (61.5%) | 12 (60.0%) | 15 (44.1%) | |
| Gender (female) | 13 (50.0%) | 6 (30.0%) | 16 (47.1%) | .346 |
| Underlying disease | .077 | |||
| Leukemia | 8 (30.8%) | 8 (40.0%) | 22 (64.7%) | |
| Lymphoma | 11 (42.3%) | 6 (30.0%) | 4 (11.8%) | |
| Multiple myeloma | 2 (7.7%) | 3 (15.0%) | 3 (8.8%) | |
| Myelodysplastic syndrome | 5 (19.2%) | 2 (10.0%) | 3 (8.8%) | |
| Other | 0 (0.0%) | 1 (5.0%) | 2 (5.9%) | |
| Pretransplant comorbidity (HCT-CI) | .121 | |||
| 0-1 | 15 (57.7%) | 7 (35.0%) | 8 (23.5%) | |
| 2-3 | 6 (23.1%) | 8 (40.0%) | 15 (44.1%) | |
| 4+ | 5 (19.2%) | 5 (25.0%) | 11 (32.4%) | |
| Disease risk (ASBMT RFI) | .388 | |||
| Low | 9 (34.6%) | 3 (15.0%) | 13 (38.2%) | |
| Intermediate | 6 (23.1%) | 4 (20.0%) | 7 (20.6%) | |
| High | 11 (42.3%) | 13 (65.0%) | 14 (41.2%) | |
| Transplant characteristics | ||||
| Conditioning intensity | .019 | |||
| Nonmyeloablative | 8 (30.8%) | 6 (30.0%) | 1 (2.9%) | |
| Reduced intensity | 7 (26.9%) | 7 (35.0%) | 12 (35.3%) | |
| Myeloablative | 11 (42.3%) | 7 (35.0%) | 21 (61.8%) | |
| Stem cell source | .168 | |||
| Related identical sibling | 11 (42.3%) | 9 (45.0%) | 9 (26.5%) | |
| Unrelated identical | 11 (42.3%) | 3 (15.0%) | 11 (32.4%) | |
| Unrelated nonidentical | 2 (7.7%) | 2 (10.0%) | 4 (11.8%) | |
| Umbilical cord | 2 (7.7%) | 6 (30.0%) | 10 (29.4%) | |
| T-cell depletion (ex-vivo) | 11 (42.3%) | 8 (40.0%) | 17 (50.0%) | .744 |
| Transplant course | ||||
| Time to engraftment (≥14 days posttransplant) | 3 (11.5%) | 6 (30.0%) | 10 (29.4%) | .197 |
| Ursodeoxycholic acid‡ | 3 (11.5%) | 5 (25.0%) | 9 (26.5%) | .328 |
| Liver dysfunction (total bilirubin > twice baseline) | 3 (11.5%) | 2 (10.0%) | 5 (14.7%) | 1.000 |
| Acute kidney injury (creatinine > twice baseline) | 2 (7.7%) | 5 (25.0%) | 15 (44.1%) | .006 |
| TPN‡ | 17 (65.4%) | 13 (65.0%) | 29 (85.3%) | .133 |
| Antibiotic‡ | ||||
| Vancomycin (intravenous) | 20 (76.9%) | 18 (90.0%) | 34 (100.0%) | .007 |
| Fluoroquinolone§ | 7 (26.9%) | 5 (25.0%) | 10 (29.4%) | 1.000 |
| Metronidazole | 4 (15.4%) | 4 (20.0%) | 15 (44.1%) | .031 |
| β-lactam¶ | 17 (65.4%) | 18 (90.0%) | 33 (97.1%) | .002 |
| Infection‡ | ||||
| C difficile infection | 1 (3.8%) | 1 (5.0%) | 12 (35.3%) | .002 |
| Gram-negative bloodstream infection | 1 (3.8%) | 3 (15.0%) | 6 (17.6%) | .262 |
| VRE bloodstream infection | 1 (3.8%) | 0 (0.0%) | 3 (8.8%) | .454 |
Measured by inverse Simpson index: high diversity, >4; intermediate diversity, 2 to 4; low diversity, <2.
P values are 2-sided and based on Fisher’s exact test.
Evaluated during pre-engraftment period prior to specimen collection.
Fluoroquinolone antibiotics consist of ciprofloxacin and levofloxacin.
β-lactams include cephalosporins, β-lactam-β-lactamase combinations, and carbapenems.
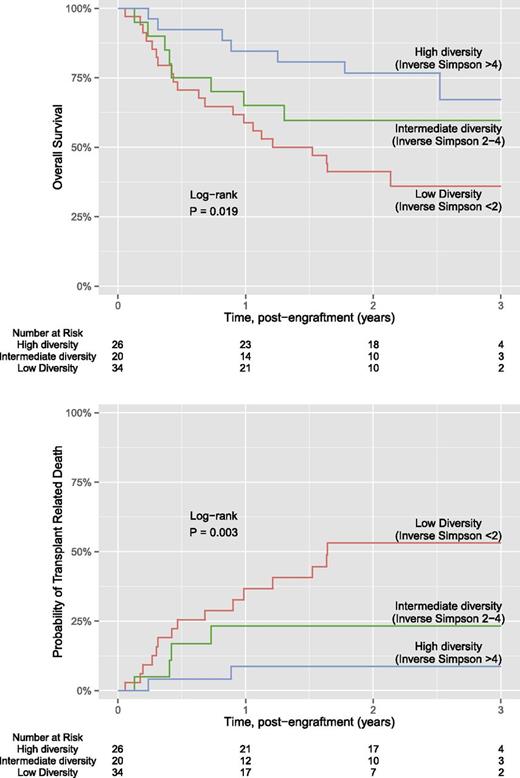
Kaplan-Meier estimates for overall survival and transplant-related mortality for each of the 3 diversity groups are shown in Figure 1. Of the 80 patients, a total of 36 (45%) patients died. The median length of follow-up was 2.3 years for overall survival and 2.2 years for transplant-related mortality. Overall survival was significantly different between the 3 diversity groups (P = .019), where subjects with lower intestinal diversity were more likely to die. Estimated overall survival at 3 years was 67% for the high diversity group, 60% for the intermediate diversity group, and 36% for the low diversity group. Similar differences between diversity groups were found for transplant-related mortality (P = .003), where estimated transplant mortality at 3 years was 9% for the high diversity group, 23% for the intermediate diversity group, and 53% for the low diversity group. Findings were similar if microbial diversity was estimated using Shannon index, an alternative measure to the inverse Simpson (supplemental Figure 1). Further analysis using the end point of death due to GVHD or infection also showed significant differences between the diversity groups; in contrast, we did not find significant associations between diversity and death due to relapse or progression (supplemental Figure 2).
Kaplan-Meier plot of diversity and overall survival and transplant related mortality.
Kaplan-Meier plot of diversity and overall survival and transplant related mortality.
Cox proportional hazards analysis for overall survival is shown in Table 2. Microbial diversity at engraftment was significantly associated with increased risk of death from any cause. In particular, patients with low diversity were approximately 3 times more likely to die within the follow-up period compared with those with higher microbial diversity. This effect remained significant in the multivariate analysis, after adjusting for pretransplant comorbidity, disease risk, and antibiotic administration (intravenous vancomycin, fluoroquinolones, and β-lactams). Other variables that were found to be independently associated with overall survival were disease risk and fluoroquinolone administration. Further analysis showed microbial diversity to be an independent predictor of transplant-related mortality as well, with a higher estimated magnitude of association compared with that of overall survival (Table 3). Not surprisingly, 1 difference between models for these 2 outcomes was that disease risk was predictive of overall survival but not transplant-related mortality. The follow-up and outcomes are depicted individually for each subject in Figure 2. Of the 36 deaths, 21 (58%) were transplant related and 15 (42%) were due to relapse or progression of disease. Transplant-related deaths were frequently due to GVHD or infection and were more frequent in patients with low microbiota diversity. We did not observe any evidence of censoring bias. As inverse Simpson diversity decreased, the corresponding taxonomic composition of the fecal microbiota appeared less complex, with fewer distinct members. In subjects with lower diversity, the microbiota was generally dominated by a single bacterial genus. Dominating genera included Enterococcus, Streptococcus, Enterobacteriaceae (Escherichia and Kluyvera), and Lactobacillus. These bacterial taxa were also observed in a prior report characterizing the microbiota of this allo-HSCT patient cohort.3
Predictors of overall survival
| Predictor . | Univariate . | Multivariate . | ||
|---|---|---|---|---|
| Hazard ratio . | P value . | Hazard ratio . | P value . | |
| Age (years) | 1.01 (0.98-1.04) | .511 | ||
| Gender (female) | 1.05 (0.54-2.03) | .874 | ||
| Underlying disease (leukemia vs other) | 1.11 (0.57-2.14) | .763 | ||
| Pretransplant comorbidity (HCT-CI >3) | 2.18 (1.08-4.26) | .030 | 1.42 (0.68-2.88) | .339 |
| Disease risk* | ||||
| Low | 1.0 | — | 1.0 | — |
| Intermediate | 2.39 (0.97-6.19) | .059 | 4.17 (1.55-11.68) | .005 |
| High | 1.47 (0.65-3.62) | .357 | 2.04 (0.87-5.18) | .103 |
| Conditioning intensity | ||||
| Nonmyeloablative | 1.0 | — | ||
| Reduced intensity | 1.89 (0.71-5.89) | .209 | ||
| Myeloablative | 1.62 (0.64-4.91) | .324 | ||
| Stem cell source | ||||
| Related identical sibling | 1.0 | — | ||
| Unrelated identical | 1.32 (0.58-2.98) | .500 | ||
| Unrelated nonidentical | 1.52 (0.42-4.39) | .486 | ||
| Cord blood | 1.40 (0.55-3.38) | .472 | ||
| T-cell depletion (ex vivo) | 0.92 (0.47-1.77) | .803 | ||
| Time to engraftment (≥14 d post-HSCT) | 1.10 (0.47-2.30) | .819 | ||
| Ursodeoxycholic acid† | 0.94 (0.38-2.04) | .885 | ||
| Liver dysfunction (total bilirubin > twice baseline)† | 1.55 (0.58-3.48) | .353 | ||
| Acute kidney injury (creatinine > twice baseline)† | 1.40 (0.66-2.79) | .360 | ||
| TPN† | 1.41 (0.67-3.32) | .377 | ||
| Antibiotics† | ||||
| Vancomycin (intravenous) | 2.60 (0.79-16.07) | .131 | 1.28 (0.28-9.02) | .764 |
| Fluoroquinolone‡ | 0.52 (0.21-1.12) | .100 | 0.34 (0.13-0.78) | .010 |
| Metronidazole | 1.47 (0.71-2.90) | .286 | ||
| β-lactam§ | 3.97 (1.21-24.48) | .019 | 3.22 (0.84-21.14) | .094 |
| Infections† | ||||
| C difficile infection | 0.76 (0.26-1.80) | .564 | ||
| Gram-negative bloodstream infection | 1.58 (0.59-3.55) | .331 | ||
| VRE bloodstream infection | 1.10 (0.18-3.60) | .901 | ||
| Microbial diversity (engraftment) | ||||
| High diversity (inverse Simpson >4) | 1.0 | — | 1.0 | — |
| Intermediate diversity (inverse Simpson 2-4) | 1.83 (0.65-5.22) | .245 | 1.26 (0.42-3.86) | .673 |
| Low diversity (inverse Simpson <2) | 3.13 (1.39-7.98) | .005 | 2.56 (1.03-7.23) | .042 |
| Predictor . | Univariate . | Multivariate . | ||
|---|---|---|---|---|
| Hazard ratio . | P value . | Hazard ratio . | P value . | |
| Age (years) | 1.01 (0.98-1.04) | .511 | ||
| Gender (female) | 1.05 (0.54-2.03) | .874 | ||
| Underlying disease (leukemia vs other) | 1.11 (0.57-2.14) | .763 | ||
| Pretransplant comorbidity (HCT-CI >3) | 2.18 (1.08-4.26) | .030 | 1.42 (0.68-2.88) | .339 |
| Disease risk* | ||||
| Low | 1.0 | — | 1.0 | — |
| Intermediate | 2.39 (0.97-6.19) | .059 | 4.17 (1.55-11.68) | .005 |
| High | 1.47 (0.65-3.62) | .357 | 2.04 (0.87-5.18) | .103 |
| Conditioning intensity | ||||
| Nonmyeloablative | 1.0 | — | ||
| Reduced intensity | 1.89 (0.71-5.89) | .209 | ||
| Myeloablative | 1.62 (0.64-4.91) | .324 | ||
| Stem cell source | ||||
| Related identical sibling | 1.0 | — | ||
| Unrelated identical | 1.32 (0.58-2.98) | .500 | ||
| Unrelated nonidentical | 1.52 (0.42-4.39) | .486 | ||
| Cord blood | 1.40 (0.55-3.38) | .472 | ||
| T-cell depletion (ex vivo) | 0.92 (0.47-1.77) | .803 | ||
| Time to engraftment (≥14 d post-HSCT) | 1.10 (0.47-2.30) | .819 | ||
| Ursodeoxycholic acid† | 0.94 (0.38-2.04) | .885 | ||
| Liver dysfunction (total bilirubin > twice baseline)† | 1.55 (0.58-3.48) | .353 | ||
| Acute kidney injury (creatinine > twice baseline)† | 1.40 (0.66-2.79) | .360 | ||
| TPN† | 1.41 (0.67-3.32) | .377 | ||
| Antibiotics† | ||||
| Vancomycin (intravenous) | 2.60 (0.79-16.07) | .131 | 1.28 (0.28-9.02) | .764 |
| Fluoroquinolone‡ | 0.52 (0.21-1.12) | .100 | 0.34 (0.13-0.78) | .010 |
| Metronidazole | 1.47 (0.71-2.90) | .286 | ||
| β-lactam§ | 3.97 (1.21-24.48) | .019 | 3.22 (0.84-21.14) | .094 |
| Infections† | ||||
| C difficile infection | 0.76 (0.26-1.80) | .564 | ||
| Gram-negative bloodstream infection | 1.58 (0.59-3.55) | .331 | ||
| VRE bloodstream infection | 1.10 (0.18-3.60) | .901 | ||
| Microbial diversity (engraftment) | ||||
| High diversity (inverse Simpson >4) | 1.0 | — | 1.0 | — |
| Intermediate diversity (inverse Simpson 2-4) | 1.83 (0.65-5.22) | .245 | 1.26 (0.42-3.86) | .673 |
| Low diversity (inverse Simpson <2) | 3.13 (1.39-7.98) | .005 | 2.56 (1.03-7.23) | .042 |
ASBMT RFI Classification.
Evaluated during pre-engraftment period prior to specimen collection.
Fluoroquinolone antibiotics consist of ciprofloxacin and levofloxacin.
β-lactams include cephalosporins, β-lactam-β-lactamase combinations, and carbapenems.
Predictors of transplant-related mortality
| Predictor . | Univariate . | Multivariate . | ||
|---|---|---|---|---|
| Hazard ratio . | P value . | Hazard ratio . | P value . | |
| Age (years) | 0.99 (0.96-1.03) | .753 | ||
| Gender (female) | 1.18 (0.49-2.79) | .708 | ||
| Underlying disease (leukemia vs other) | 0.65 (0.26-1.55) | .338 | ||
| Pretransplant comorbidity (HCT-CI >3) | 2.58 (1.05-6.14) | .040 | 1.85 (0.72-4.65) | .195 |
| Disease risk* | ||||
| Low | 1.0 | — | ||
| Intermediate | 2.02 (0.63-6.49) | .229 | ||
| High | 1.14 (0.41-3.40) | .803 | ||
| Conditioning intensity | ||||
| Nonmyeloablative | 1.0 | — | ||
| Reduced intensity | 1.78 (0.51-8.12) | .378 | ||
| Myeloablative | 1.36 (0.41-6.05) | .635 | ||
| Stem cell source | ||||
| Related identical sibling | 1.0 | — | ||
| Unrelated identical | 0.71 (0.19-2.35) | .581 | ||
| Unrelated nonidentical | 1.66 (0.36-5.97) | .480 | ||
| Cord blood | 1.91 (0.65-5.59) | .230 | ||
| T-cell depletion (ex-vivo) | 0.98 (0.41-2.33) | .969 | ||
| Time to engraftment (≥4 d post-HSCT) | 1.61 (0.57-3.97) | .344 | ||
| Ursodeoxycholic acid† | 0.85 (0.25-2.31) | .774 | ||
| Liver dysfunction (total bilirubin > twice baseline)† | 1.32 (0.31-3.90) | .668 | ||
| Acute kidney injury (creatinine > twice baseline)† | 1.88 (0.74-4.46) | .175 | 1.32 (0.48-3.42) | .572 |
| TPN† | 2.15 (0.73-9.20) | .179 | 1.68 (0.50-7.76) | .426 |
| Antibiotics† | ||||
| Vancomycin (intravenous) | 2.65 (0.55-47.46) | .269 | ||
| Fluoroquinolone‡ | 0.50 (0.14-1.36) | .186 | 0.43 (0.12-1.24) | .124 |
| Metronidazole | 1.43 (0.54-3.45) | .446 | ||
| β-lactam§ | 4.89 (1.02-87.84) | .047 | 1.65 (0.28-31.89) | .630 |
| Infections† | ||||
| C difficile infection | 1.00 (0.29-2.71) | .998 | ||
| Gram-negative bloodstream infection | 1.35 (0.32-4.00) | .643 | ||
| VRE bloodstream infection | 1.79 (0.29-6.19) | .468 | ||
| Microbial diversity (engraftment) | ||||
| High diversity (inverse Simpson >4) | 1.0 | — | 1.0 | — |
| Intermediate diversity (inverse Simpson 2-4) | 3.07 (0.60-22.14) | .179 | 3.02 (0.56-22.92) | .202 |
| Low diversity (inverse Simpson <2) | 7.54 (2.12-47.88) | .001 | 5.25 (1.36-35.07) | .014 |
| Predictor . | Univariate . | Multivariate . | ||
|---|---|---|---|---|
| Hazard ratio . | P value . | Hazard ratio . | P value . | |
| Age (years) | 0.99 (0.96-1.03) | .753 | ||
| Gender (female) | 1.18 (0.49-2.79) | .708 | ||
| Underlying disease (leukemia vs other) | 0.65 (0.26-1.55) | .338 | ||
| Pretransplant comorbidity (HCT-CI >3) | 2.58 (1.05-6.14) | .040 | 1.85 (0.72-4.65) | .195 |
| Disease risk* | ||||
| Low | 1.0 | — | ||
| Intermediate | 2.02 (0.63-6.49) | .229 | ||
| High | 1.14 (0.41-3.40) | .803 | ||
| Conditioning intensity | ||||
| Nonmyeloablative | 1.0 | — | ||
| Reduced intensity | 1.78 (0.51-8.12) | .378 | ||
| Myeloablative | 1.36 (0.41-6.05) | .635 | ||
| Stem cell source | ||||
| Related identical sibling | 1.0 | — | ||
| Unrelated identical | 0.71 (0.19-2.35) | .581 | ||
| Unrelated nonidentical | 1.66 (0.36-5.97) | .480 | ||
| Cord blood | 1.91 (0.65-5.59) | .230 | ||
| T-cell depletion (ex-vivo) | 0.98 (0.41-2.33) | .969 | ||
| Time to engraftment (≥4 d post-HSCT) | 1.61 (0.57-3.97) | .344 | ||
| Ursodeoxycholic acid† | 0.85 (0.25-2.31) | .774 | ||
| Liver dysfunction (total bilirubin > twice baseline)† | 1.32 (0.31-3.90) | .668 | ||
| Acute kidney injury (creatinine > twice baseline)† | 1.88 (0.74-4.46) | .175 | 1.32 (0.48-3.42) | .572 |
| TPN† | 2.15 (0.73-9.20) | .179 | 1.68 (0.50-7.76) | .426 |
| Antibiotics† | ||||
| Vancomycin (intravenous) | 2.65 (0.55-47.46) | .269 | ||
| Fluoroquinolone‡ | 0.50 (0.14-1.36) | .186 | 0.43 (0.12-1.24) | .124 |
| Metronidazole | 1.43 (0.54-3.45) | .446 | ||
| β-lactam§ | 4.89 (1.02-87.84) | .047 | 1.65 (0.28-31.89) | .630 |
| Infections† | ||||
| C difficile infection | 1.00 (0.29-2.71) | .998 | ||
| Gram-negative bloodstream infection | 1.35 (0.32-4.00) | .643 | ||
| VRE bloodstream infection | 1.79 (0.29-6.19) | .468 | ||
| Microbial diversity (engraftment) | ||||
| High diversity (inverse Simpson >4) | 1.0 | — | 1.0 | — |
| Intermediate diversity (inverse Simpson 2-4) | 3.07 (0.60-22.14) | .179 | 3.02 (0.56-22.92) | .202 |
| Low diversity (inverse Simpson <2) | 7.54 (2.12-47.88) | .001 | 5.25 (1.36-35.07) | .014 |
ASBMT RFI classification.
Evaluated during pre-engraftment period prior to specimen collection.
Fluoroquinolone antibiotics consist of ciprofloxacin and levofloxacin.
β-lactams include cephalosporins, β-lactam-β-lactamase combinations, and carbapenems.
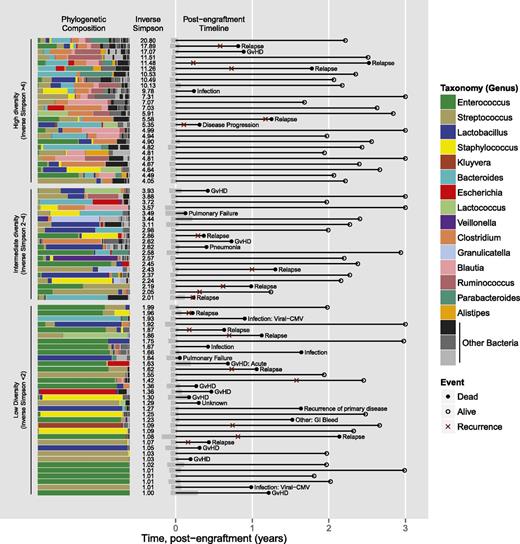
Intestinal microbiota at engraftment and subsequent transplant course by subject (N = 80). Each row represents a study subject. Horizontal stacked bars on the left side represent the phylogenetic composition of each subject at the time of stem cell engraftment. Microbial diversity, measured by inverse Simpson index, is listed in the next column. Timeline plots are shown in the next column; black lines represent survival time, closed circles represent death, and open circles represent censoring. Red x denotes relapse or progression of disease. For death events, cause of death is listed.
Intestinal microbiota at engraftment and subsequent transplant course by subject (N = 80). Each row represents a study subject. Horizontal stacked bars on the left side represent the phylogenetic composition of each subject at the time of stem cell engraftment. Microbial diversity, measured by inverse Simpson index, is listed in the next column. Timeline plots are shown in the next column; black lines represent survival time, closed circles represent death, and open circles represent censoring. Red x denotes relapse or progression of disease. For death events, cause of death is listed.
To determine whether the presence or absence of specific bacterial taxa correlated with mortality, we compared the microbiota composition of patients who died or survived by the LEfSe method (Table 4). The postengraftment microbiota of subjects who died differed significantly from the microbiota of patients who survived, harboring a greater abundance of γ-Proteobacteria, including Enterobacteriaceae. Conversely, Lachnospiraceae and Actinomycetaceae were observed in greater relative abundance in subjects who remained alive during the follow-up period. We did not observe significant associations with mortality for any of the remaining taxa.
Phylogenetic biomarkers of overall survival, using LEfSe analysis
| Biomarker prediction . | Taxonomic rank* . | Bacterial taxon† . | Logarithmic LDA score . | Kruskal-Wallis (among classes) P value . | Median relative abundance‡ . | |
|---|---|---|---|---|---|---|
| Dead (N = 36) . | Alive (N = 44) . | |||||
| Dead | 3 | Bacteria.Proteobacteria.Gammaproteobacteria | 4.15 | .042 | 0.0043 | 0.00039 |
| Dead | 4 | Bacteria.Proteobacteria.Gammaproteobacteria.Enterobacteriales | 4.13 | .083 | 0.002 | 0.00025 |
| Dead | 5 | Bacteria.Proteobacteria.Gammaproteobacteria.Enterobacteriales.Enterobacteriaceae | 4.13 | .083 | 0.002 | 0.00025 |
| Alive | 5 | Bacteria.Firmicutes.Clostridia.Clostridiales.Lachnospiraceae | 4.44 | .098 | 0.00035 | 0.0022 |
| Alive | 5 | Bacteria.Actinobacteria.Actinobacteria.Actinomycetales.Actinomycetaceae | 3.80 | .071 | 0.00017 | 0.00086 |
| Alive | 6 | Bacteria.Actinobacteria.Actinobacteria.Actinomycetales.Actinomycetaceae.Actinomyces | 3.81 | .068 | 0.00017 | 0.00083 |
| Biomarker prediction . | Taxonomic rank* . | Bacterial taxon† . | Logarithmic LDA score . | Kruskal-Wallis (among classes) P value . | Median relative abundance‡ . | |
|---|---|---|---|---|---|---|
| Dead (N = 36) . | Alive (N = 44) . | |||||
| Dead | 3 | Bacteria.Proteobacteria.Gammaproteobacteria | 4.15 | .042 | 0.0043 | 0.00039 |
| Dead | 4 | Bacteria.Proteobacteria.Gammaproteobacteria.Enterobacteriales | 4.13 | .083 | 0.002 | 0.00025 |
| Dead | 5 | Bacteria.Proteobacteria.Gammaproteobacteria.Enterobacteriales.Enterobacteriaceae | 4.13 | .083 | 0.002 | 0.00025 |
| Alive | 5 | Bacteria.Firmicutes.Clostridia.Clostridiales.Lachnospiraceae | 4.44 | .098 | 0.00035 | 0.0022 |
| Alive | 5 | Bacteria.Actinobacteria.Actinobacteria.Actinomycetales.Actinomycetaceae | 3.80 | .071 | 0.00017 | 0.00086 |
| Alive | 6 | Bacteria.Actinobacteria.Actinobacteria.Actinomycetales.Actinomycetaceae.Actinomyces | 3.81 | .068 | 0.00017 | 0.00083 |
Statistically significant taxonomic predictors of mortality are reported; all remaining taxa were not significantly associated.
The following LEfSe parameters were used: conditioning regimen intensity was used for subclass comparisons, logarithmic LDA cutoff of 2.0, and α of 0.1 for Kruskal-Wallis testing among classes and for Wilcoxon testing between subclasses.
Taxonomic ranks correspond to the following divisions: 1, kingdom; 2, phylum; 3, class; 4, order; 5, family; 6, genus.
For each bacterial taxon, classification at each preceding level is denoted and separated by periods.
Median proportion of bacterial sequences belonging to the specified taxon.
Discussion
The intestinal microbiota consists of hundreds of distinct bacterial species that contribute to host health through a variety of functions including stimulation of gut immune system development, inhibition of potentially pathogenic microbes, and breakdown of otherwise unabsorbable nutrients.14,15 Prior studies have shown that the intestinal microbiota of patients undergoing allo-HSCT often loses diversity, resulting in domination by a range of microbes that can enter the bloodstream and cause septicemia, particularly in times of neutropenia and mucosal barrier injury.3,16 In this study, we find that loss of intestinal microbiota diversity is associated with overall mortality after engraftment, independent of known predictors such as pretransplant comorbidity and disease status. Furthermore, a stronger association was observed with death due to transplant-related causes, with no discernible association with mortality arising from relapse or progression of disease, suggesting that lack of microbial diversity is primarily linked to transplant-related deaths such as infection and GVHD. We reanalyzed these data using a series of pairwise models containing diversity and each of the other clinical variables; results remained unchanged, and diversity continued to demonstrate strong independent effects (data not shown).
The membership and structure of different bacterial species in the intestinal microbiota of healthy individuals is remarkably variable; characterization of these microbial populations in a clinically practical and meaningful manner is currently an active area of microbiota research. Although there is some debate, some studies have suggested that the individual microbiota can be categorized into enterotypes based on microbial composition.17,18 The composition of the intestinal microbiota has been implicated in several areas of human disease. A recent study found that patients with new onset rheumatoid arthritis harbor an intestinal microbiota with a higher frequency of Prevotellaceae, a family of anaerobic gram-negative bacteria that defines 1 of the 3 enterotypes.17,19,20 Other studies have demonstrated that microbiota composition influences malnutrition and the development of kwashiorkor,21 development of obesity,22 and metabolism of orally ingested drugs.23 Some members of the microbiota induce development of Th17 cells, which contribute to antimicrobial defense but also contribute to the pathogenesis of inflammatory diseases, whereas other microbes produce short-chain fatty acids and contribute to the development of regulatory T cells and intestinal tolerance.24,25 Older studies have demonstrated that a healthy microbiota enhances resistance to infection by intestinal pathogens through the poorly understood process of colonization resistance.26,27 Given the broad impact of an intact intestinal microbiota on human health, it is perhaps not surprising that the loss of microbiota diversity can result in increased mortality. The relevance and impact of an abnormal intestinal gut flora has been described in a variety of disorders such as inflammatory bowel disease, C difficile infection, obesity, and many others.28-31
In allo-HSCT, pretransplant conditioning regimens consisting of chemotherapy and/or total body irradiation damage the intestinal epithelium and mucosal barrier while simultaneous broad spectrum antibiotic administration dramatically alters the microbiota, enhancing susceptibility to infection.3,32,33 Immune attack by graft-derived T lymphocytes further damages intestinal epithelial cells. The pattern of repeated, self-propagating intestinal insults may explain why many transplant complications involve or originate in the gastrointestinal tract; these include bloodstream infections, C difficile infection, GVHD of the gut, bile salt malabsorption, pancreatic insufficiency, and viral infections such as cytomegalovirus, adenovirus, and norovirus.34 On the basis of data from recent studies, an aberrant microbiota contributes to at least some of these complications.3,32
A wide range of gastrointestinal syndromes can occur in allo-HSCT recipients, such as diarrhea, nausea, vomiting, malabsorption, and anorexia. Not uncommonly, these symptoms can be nonspecific and remain undiagnosed despite extensive workup. The relatively recent characterization of cord colitis syndrome, an antibiotic-responsive diarrheal syndrome following cord blood allo-HSCT,35,36 suggests that there may be other posttransplant disorders related to the intestinal microbiota that are yet to be identified. Moreover, adverse events following allo-HSCT do not occur independently of one another; associations have been observed between separate complications such as C difficile infection and gut GVHD37,38 or GVHD and adenovirus infections,39 which suggests that these overlapping gastrointestinal complications may be symptoms of a more central disorder.
The role played by intestinal bacteria in adverse outcomes in allo-HSCT is by no means a new consideration. Previously, total or selective gut decontamination with antibiotics was performed for allo-HSCT recipients, based on several early studies that suggested that the intervention could lead to lower rates of GVHD and infection.40 Although some centers may administer antibiotics for this purpose,41 the practice of total gut decontamination has largely fallen out of favor and is not standard practice in most allo-HSCT centers. These prior studies were performed at a time when advanced sequencing methods were unavailable, and reliance on culture-based methods provided only a partial picture of the intestinal microbiota. Given the findings of this study and others demonstrating the beneficial effects of a diverse microbiota, total gut decontamination may be ill advised.
In this study, our analysis showed some degree of correlation between administration of certain antibiotics and mortality. Interpretation of these results may be not straightforward, because many antibiotics may have been confounded by the situation they were given in. In this study, patients were given antibiotics for a variety of purposes, such as infectious prophylaxis of a specific subpopulation, or treatment that could either be targeted or empiric. In many cases, antibiotics were not directly associated after multivariate adjustment for other allo-HSCT factors. Interestingly, in the case of β-lactam antibiotics, administration was associated with mortality in the unadjusted analysis but was not in the multivariate model that included microbial diversity, particularly for transplant-related mortality. It is possible that intestinal diversity serves as a causal mediator of β-lactam administration, where the microbial impact of β-lactams is encapsulated by intestinal diversity, which lies on the causal pathway to death. We surmise that the cumulative effects of several preceding adverse factors such as antibiotics and conditioning regimen intensity on the intestinal microbiota may be summarized by microbial diversity.
Along similar lines, pretransplant comorbidity, which we assessed using the HCT-CI, was a significant univariate predictor of outcomes but became nonsignificant in a simultaneous model with microbial diversity. As a known predictor of transplant-related mortality, the HCT-CI is presumably mediated by a number of transplant related complications. In this case, it could be that patients with underlying comorbidities are most likely to suffer derangements of the intestinal microbiota that lead to decreased microbial diversity, and subsequently, death.
In our examination of associations between antibiotics and microbial diversity, we found that β-lactams and metronidazole were associated with decreased diversity, whereas fluoroquinolones were not. Although it may not be entirely certain, we suspect the differences in diversity observed with each antibiotic may be partly explained by its degree of antianaerobic activity. Fluoroquinolones have previously been shown to impact diversity,42 but likely to a lesser degree than antibiotics with broad activity against obligate anaerobic bacteria. In our prior work, we found that fluoroquinolone administration decreased colonization by Proteobacteria, a phylum that includes oxygen-tolerant gram-negative pathogens such as the Enterobactericeae and Pseudmonas, but which exists as a minor population in the absence of antibiotic-mediated microbiota perturbation. Intravenous vancomycin was also significantly associated with decreased diversity; administration of intravenous vancomycin can result in some biliary excretion, although prolonged exposure is usually required, and there is significant interpatient variability in vancomycin levels in the gut.43 We suspect that the vancomycin/diversity association is confounded by the fact that it is frequently given concurrently with β-lactams, which readily access the gut lumen and significantly alter the intestinal microbiota. Although diversity provides one measure of microbiota health, identification of more specific microbial alterations might provide insight into the mechanisms of specific transplant complications. In our explorative analysis of microbial composition, we identified Lachnospiraceae and Actinomycetaceae as commensal members that were associated with survival. Both families of bacteria commonly inhabit the human intestinal tract under normal circumstances. Specifically, other studies have identified Lachnospiraceae bacteria as potential promoters of gut health; for instance, Lachnospiraceae have been shown to have protective effects against C difficile infection in preclinical studies.44,45 In contrast, we found Enterobacteriaceae and other gram-negative bacteria from the Proteobacteria phylum to be positively correlated with death. Although some Proteobacteria are pathogens, it is not clear whether Proteobacteria contribute directly to death or simply emerge as a result of diversity loss or intestinal inflammation. Indeed, recent studies have demonstrated that products of host inflammation can enhance the growth of bacteria belonging to the Enterobacteriaceae family.46
Given the importance of the intestinal microbiota suggested by our findings, interventions to maintain intestinal diversity may lead to improved outcomes in allo-HSCT. Limiting exposure to antibiotics that destroy obligate anaerobic bacterial populations of the intestine may reduce untoward microbial disruptions. Replenishment of commensal bacterial populations that are lost during transplantation, through fecal microbiota transplantation or targeted restoration of specific beneficial bacteria, may also improve outcomes. These approaches should be evaluated carefully; although we have shown the intestinal microbiota to be highly predictive of mortality, independent of other clinical factors, it still remains to be seen if this relationship is directly causal. Ultimately, controlled interventional studies are needed to define the full extent to which the intestinal microbiota contributes to the outcome of allo-HSCT.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by National Institutes of Health, National Institute of Allergy and Infectious Diseases grants 1K23 AI095398-01 (Y.T.) and 1RO1 AI42135 (E.G.P.), the Lucille Castori Center for Microbes, Inflammation, and Cancer, and the Tow Foundation.
Authorship
Contribution: Y.T. and E.G.P. were involved in conception and design of the study; E.R.L., L.L., D.N., A.G., and A.V. were involved in the collection, processing, and sequencing of specimens; Y.T., M.-A.P., E.R.L., S.M., J.N.B, P.B.D., and D.M.P. were involved in clinical data collection; Y.T. performed analysis of microbial sequences and clinical data; and Y.T., R.R.J., M.-A.P., E.R.L., S.G., M.v.d.B., and E.G.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ying Taur, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10065; e-mail: taury@mskcc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal