Abstract
Background. Emerging evidence indicates that microRNAs (miRNAs) may regulate immunity. Their role in acquired aplastic anemia (AA), a T cell-mediated autoimmune disease, has not been investigated. We characterized miRNA signatures in patient T cells to identify novel genes involved in the regulation of immune responses of CD4+ and CD8+ T cells in AA. Cells from age-matched healthy donors and patients with myelodysplastic syndrome (MDS) and transfusion-dependent sickle cell disease (SCD) were used for comparison.
Method. MiRNAs and mRNAs from flow cytometrically sorted T cells from patients and controls were extracted and subjected to profiling using miRNA PCR array platform (miScript miRNA PCR array, MIHS-111Z, Qiagen Inc). Real-time PCR was employed for validation of specific miRNA expression changes. To define identified miRNA functions, their target genes were predicted using bioinformatic software (TargetScan, miRBase, DIANA-miRPath). MiRNA target gene profilings were performed using custom PCR array platform (Custom PCR array, Qiagen Inc) in CD4+ and CD8+ T cells in AA. MiRNA knockdown (miR-126-3p and/or miR-145-5p) on healthy CD4+ and CD8+ T cells was performed using the Human T Cell Nucleofector kit (Lonza AG). Cell proliferation was estimated by CFSE assays. Effects of miRNA knockdown on the target genes were evaluated by qPCR and immunoblot. MiRNA expression profilings were examined in CD4+ and CD8+ T cells in a mouse model of lymph node cell infusion-induced bone marrow failure (BMF).
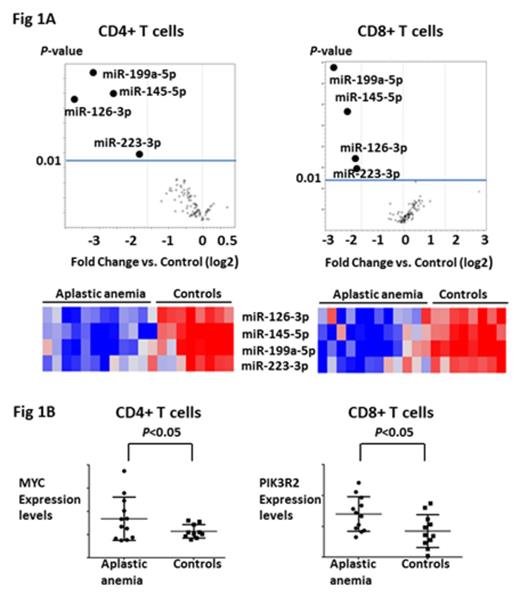
Results and Discussion. As compared to healthy controls, miRNA expression profiling revealed significant down-regulation of miRNAs in a disease-specific pattern. MiR-199a-5p expression was reduced in CD4+ and CD8+ T cells from patients with all diseases that were tested (AA, MDS, SCD). Reduced expression of miR-223-3p (more than 3 fold change (FC), P <0.05) in T cells was seen in low-risk MDS. Concurrent downregulation of miR-126-3p, miR-145-5p, miR-223-3p and miR-199a-5p (more than 3 FC, P <0.05) was uniquely observed only in CD4+ and CD8+ T cells from AA patients (Figure 1A). Analysis of these four miRNAs in T cells from AA patients after successful immunosuppressive therapy (IST) at 3 or 6 months (N=6) revealed restoration of the expression levels of miR-126-3p and miR-145-5p (more than 2 FC, P <0.05) as compared to those of patients before IST. Bioinformatic analysis predicted MYC, PIK3R2, ETS1, STAT1, CD28, and HIF1A as potential target transcriptomes for these four miRNAs. Gene expression profiling revealed significant up-regulation of MYC in CD4+ T cells (more than 1.5 FC, P <0.05) and PIK3R2 in CD8+ T cells (more than 1.5 FC, P <0.05) in AA patients compared to healthy controls (Figure 1B). qPCR and immunoblot confirmed up-regulation of MYC (~2.0 FC) in CD4+ T cells and PIK3R2 (~2.0 FC) in CD8+ T cells after transfection with anti-miR-145-5p and anti-miR-126-3p, respectively. Knock-down of the miR-126-3p and miR-145-5p promoted proliferation of both CD4+ and CD8+ T cells and increased IFN-γ production in CD8+ T cells, suggesting enhanced effector differentiation and functionality of this subset as a result of reduced activity of both miRNAs. Expression levels of miR-126-3p and miR-145-5p were not decreased in CD4+ and CD8+ T cells in a BMF mouse model. However, expression levels of miR-142-5p and miR-29a-3p, which have been reported as miRNAs associated with allo-reactivity, were decreased (more than 2 FC, P <0.05) in this model, concordant with the mouse models basis in H2 mismatched reactivity.
Conclusion. This work describes previously unknown potentially regulatory role of miRNAs 145-5p and 126-3p in T cell activation in AA. Further investigation is warranted to determine the role of these miRNAs in the more general pathogenesis of autoimmune diseases. Understanding these novel pathways might offer new therapeutic strategies and a novel mechanistic insight into the epigenetic control of human T cell effector responses.
Townsley:GSK: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal