Abstract
Macrophages have been implicated in erythropoiesis historically as a mediator of iron recycling and a key component of the erythroblastic island-consisting of a central macrophage surrounded by erythroid cells in different stages of differentiation. Recently we and others have shown that macrophages contribute to stress erythropoiesis, and such contributions extend beyond the known macrophage function of iron recycling. This finding necessitates the investigation of processes within a macrophage itself that might facilitate stress erythropoiesis and the characterization of macrophage transcriptome signatures associated with the same.
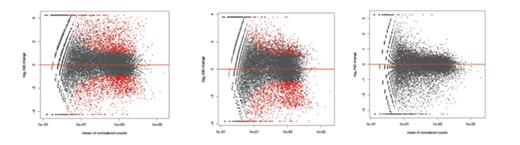
Macrophages in the bone marrow have been identified previously as CD115LowGR1LowF4/80High cells. Extending this to the spleen, we sorted and sequenced singlet macrophages in the bone marrow and spleen of two mice models of stress erythropoiesis: acute stress induced by retro orbital phlebotomy and chronic stress by bone marrow transplanted Polycythemia Vera. We found that splenic macrophages show more significant changes upon induction of stress (both acute and chronic) in comparison of macrophages in the bone marrow. The mean of normalized count of each gene when plotted against fold change (Figure 1) show that the acute and chronically stressed splenic macrophages differ significantly over non stressed macrophages but not over each other.
Acute over non stressed Chronic over non stressed Acute over non stressed
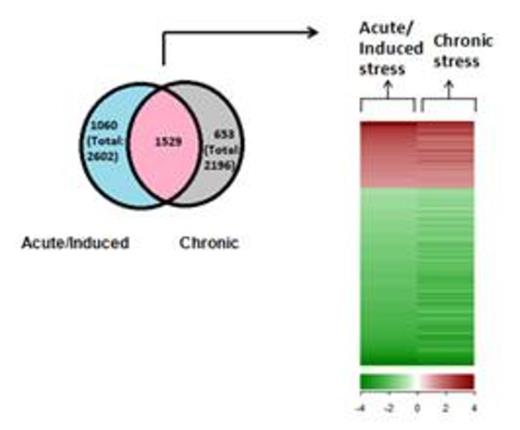
We further found that stressed splenic macrophages have a characteristic gene expression profile associated with erythropoietic stress, common to both models of induced and chronic stress. There were a total of 2602 genes showing significant changes in expression upon induction of acute stress and a total of 2196 genes showing significant changes in expression upon chronic stress. Out of these, 1529 genes common to both acute and chronically stressed splenic macrophages showed an expression profile common to both acute and induced stress. (Fig 2)
We used the open bioinformatics resource DAVID and Ingenuity Pathway Analysis (IPA) to look at biological processes encompassing our significantly up regulated candidates (FDR 10%), upstream analysis of our candidates and further network analysis. Some biological processes of highest significance were cellular growth and proliferation, hematological system development and function, inflammatory response, response to infection, cellular adhesion, cellular movement and immune cell trafficking. We validated some of our key candidates under these biological processes by flow cytometry, such as the iron exporter ferroportin, the receptor for the anti inflammatory cytokine, IL-10 and the cellular adhesion molecule ICAM-1. Some transcription factors significantly upregulated were Hes1, Nfkb1A, RelB, Bcl3 to mention a few. Upstream analysis using IPA predicted activation of transcription factors such as Notch1, Jun, RelA etc.
Along with further validation of our key candidates, we are currently investigating macrophage gene expression in another model of chronic stress, beta thalassemia and how this profile associated with the physiological condition of erythropoietic stress compares to those under in vivo models of classical and alternative macrophage activation.
Casu:Merganser Biotech: Research Funding; Isis Pharmaceuticals Inc.: Research Funding. Rivella:Merganser Biotech: Consultancy, Research Funding, Stock options, Stock options Other; Bayer: Consultancy, Research Funding; Isis Pharmaceuticals. Inc.: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal