Key Points
In cases with intermediate-risk NPM1MUT AML, there are only minor differences in relapse risk according to FLT3ITD level.
When considering allogeneic transplantation in first remission, NPM1MUT cases with low-level FLT3ITD should not be considered as good risk.
Abstract
Some studies have suggested that cases of acute myeloid leukemia (AML) with low levels of FLT3 internal tandem duplications (FLT3ITD) do not have a worse prognosis if there is a concomitant NPM1 mutation, although this is controversial. To clarify this therapeutically important issue, we have analyzed FLT3ITD and NPM1MUT levels in 1609 younger adult cases of cytogenetically intermediate-risk AML. The cumulative incidence of relapse was increased in NPM1MUT cases by the presence of a FLT3ITD, but did not differ markedly according to FLT3ITD level. This remained true when allowance was made for poor leukemic cell purity by adjustment of the FLT3ITD level to the measured NPM1MUT level. If consolidation therapies are to be determined by relapse risk, then NPM1MUT cases with low-level FLT3ITD should not be considered as good risk without further studies. AML 12 and AML 15 are registered at http://www.controlled-trials.com under ISRCTN17833622 and ISRCTN17161961, respectively.
Introduction
In acute myeloid leukemia (AML), cytogenetic and mutational analysis provides valuable prognostic information that is widely used as the basis for risk-adapted therapy. In cytogenetic intermediate-risk (IR) patients, 2 molecular markers are commonly evaluated at diagnosis, NPM1 mutations (NPM1MUT) and FLT3 internal tandem duplications (FLT3ITD).1,2 FLT3ITD cases are associated with a poor prognosis and in the absence of a FLT3ITD, NPM1MUT cases are considered to have a sufficiently good prognosis to be spared allogeneic transplantation in first complete remission (CR1).3,4 However, the FLT3ITD mutant allele level is highly variable, and although it is generally agreed that very high-level mutants (indicative of biallelic disease in at least some cells) are associated with a poor prognosis, there is controversy about the impact of lower FLT3ITD levels.5-7 Furthermore, recent studies have suggested that the impact of FLT3ITD mutant level is modulated by the NPM1 mutant status, in particular for cases with lower FLT3ITD levels. Two studies have reported that in NPM1MUT cases, only a higher-level FLT3ITD with mutant/wild-type (WT) allele ratio >0.5 (equivalent to a mutant allele level ≥33%) was predictive of a significantly worse outcome.8,9 The implication from these studies is that NPM1MUT cases with lower-level FLT3ITD should also be considered in the good risk category together with NPM1MUTFLT3WT cases. Conversely, another study reported that a lower level FLT3ITD (<50%) did significantly impact the outcome of NPM1MUT cases.10 Controversy also exists for NPM1 wild-type (NPM1WT) cases, with either no difference or worse outcome reported for lower-level FLT3ITD cases.9,10
Appreciation of the interaction of NPM1 status and FLT3ITD mutant level is important because this information is relevant to therapeutic stratification, particularly the decision to offer allogeneic transplantation in CR1. The reported discrepancies may arise because of the small size of the minor subgroups in some of the studies, the use of different thresholds for FLT3ITD levels, and because FLT3ITD levels might be underestimated in samples with low leukemic cell purity. We have, therefore, analyzed a larger cohort of IR patients than previously studied, and we also made allowances for samples with apparently low leukemic cell purity as determined by the NPM1MUT allele level.
Study design
DNA was available from 1609 younger adult patients at entry into the United Kingdom Medical Research Council AML 10, AML 12 or AML 15 trials with IR cytogenetics, according to the Medical Research Council classification.11 Median age was 46 years; 1499 (93%) patients were 15 to 59 years of age. The median length of follow-up is 116 months (range, 7-263 months). NPM1 and FLT3ITD were analyzed as previously described,7 and relative mutant level was expressed as a percentage of total alleles. Apparent mutants quantified as <5% were scored as wild-type (WT). Levels of 5% to 25% are referred to as FLT3 internal tandem duplications low (FLT3ITD-LOW), which are indicative of a mutant-positive subclone; 25% to 50% are referred to as FLT3 internal tandem duplications intermediate (FLT3ITD-INT), compatible with a heterozygous mutation in most cells; and >50% are referred to as FLT3 internal tandem duplications high (FLT3ITD-HIGH), indicative of biallelic disease.7 Details of the trials, response end points, statistical methods used, and patient demographics are given in the supplemental Material, available on the Blood Web site (supplemental Tables 1 and 2). Ethical approval for the trials and tissue collection for research was obtained from the Multi-Centre Research Committee of Wales. The study was conducted in accordance with the Declaration of Helsinki.
Results and discussion
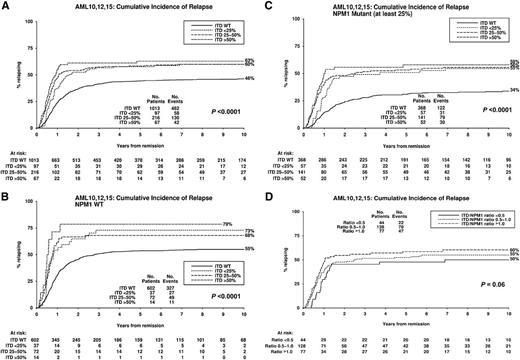
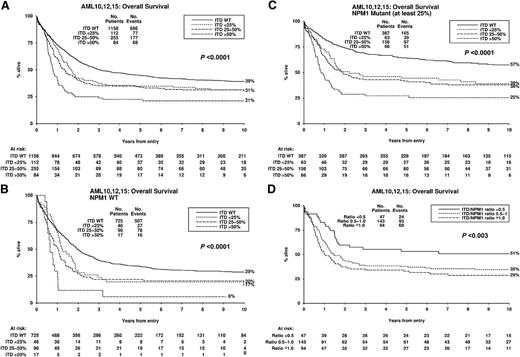
Overall, 45% of cases were NPM1MUT. In accord with previous reports, they had a higher complete remission (CR) rate than NPM1WT cases (92% vs 82%; P < .0001), lower cumulative incidence of relapse (CIR) (42% vs 58%; P < .0001) and higher overall survival (OS) (48% vs 27%; P < .0001) (supplemental Table 3). The frequency of FLT3ITD genotypes was 72% FLT3WT, 7% FLT3ITD-LOW, 16% FLT3ITD-INT, and 5% FLT3ITD-HIGH. There was a lower CR rate with higher FLT3ITD levels (88%, 87%, 85%, and 80% for the 4 groups, respectively; P = .03), although this did not remain significant in the adjusted analysis (supplemental Table 3). The trend in the univariate analysis was not significantly different between NPM1MUT and NPM1WT cases (P = .4 for interaction). CIR was higher in FLT3ITD cases (P < .0001), but only marginally higher in FLT3ITD-HIGH (63%) than FLT3ITD-INT and FLT3ITD-LOW cases (both 60%; Figure 1A). The CIR of 63% in the FLT3ITD-HIGH patients was lower than we reported previously (82%) in a smaller number of patients,7 but that study included all cytogenetic risk groups. OS was lower in FLT3ITD cases (P < .0001) and the reduction was related to FLT3ITD level (FLT3WT 39%, FLT3ITD-LOW 31%, FLT3ITD-INT 30%, and FLT3ITD-HIGH 21%; P < .0001) (Figure 2A) (supplemental Table 3). The same pattern was observed for relapse-free survival (supplemental Figure 1). There was no significant heterogeneity between the impact of FLT3ITD-LOW and FLT3ITD-INT mutants on CIR or OS when they were each compared with FLT3WT (P = .6 for both). The reduced OS in FLT3ITD-HIGH cases, despite only a marginally increased CIR, is due in part to the lower CR rate of such cases and also to a trend to a lower salvage rate in patients with relapsed or resistant disease. Within the whole cohort, the second CR rate was 53% for FLT3WT, 40%, for FLT3ITD-LOW, 37% for FLT3ITD-INT, and only 24% for FLT3ITD-HIGH cases (P < .0001 for trend). The effect was consistent in NPM1WT and NPM1MUT patients (P = .6 for interaction).
Kaplan-Meier curves for cumulative incidence of relapse stratified according to FLT3ITD mutant level. (A) All patients, (B) NPM1WT patients, (C) NPM1MUT patients with ≥25% NPM1MUT, (D) NPM1MUTFLT3ITD patients with ≥25% NPM1MUT where FLT3ITD mutant level has been adjusted to NPM1MUT level. P value is for trend. ITD, FLT3ITD; No., number; WT, wild-type.
Kaplan-Meier curves for cumulative incidence of relapse stratified according to FLT3ITD mutant level. (A) All patients, (B) NPM1WT patients, (C) NPM1MUT patients with ≥25% NPM1MUT, (D) NPM1MUTFLT3ITD patients with ≥25% NPM1MUT where FLT3ITD mutant level has been adjusted to NPM1MUT level. P value is for trend. ITD, FLT3ITD; No., number; WT, wild-type.
Kaplan-Meier curves for overall stratified according to FLT3ITD mutant level. (A) All patients, (B) NPM1WT patients, (C) NPM1MUT patients with ≥25% NPM1MUT, (D) NPM1MUTFLT3ITD patients with ≥25% NPM1MUT where FLT3ITD mutant level has been adjusted to NPM1MUT level. P value is for trend. ITD, FLT3ITD; No., number; WT, wild-type.
Kaplan-Meier curves for overall stratified according to FLT3ITD mutant level. (A) All patients, (B) NPM1WT patients, (C) NPM1MUT patients with ≥25% NPM1MUT, (D) NPM1MUTFLT3ITD patients with ≥25% NPM1MUT where FLT3ITD mutant level has been adjusted to NPM1MUT level. P value is for trend. ITD, FLT3ITD; No., number; WT, wild-type.
To investigate the impact of FLT3ITD level according to NPM1 status, 7% of cases with NPM1MUT level <25% were excluded on the assumption that the NPM1MUT allele would be present in every leukemic cell and a low level must indicate poor leukemic cell purity. The impact of FLT3ITD level on CIR was similar in both NPM1WT and NPM1MUT cases (Figure 1B-C), with no apparent difference between the FLT3ITD-LOW and FLT3ITD-INT categories (P = .8 for heterogeneity for NPM1WT and P = 0.5 for NPM1MUT cases). The impact of the FLT3ITD level on OS was also similar in NPM1WT and NPM1MUT cases, with almost identical outcomes in FLT3ITD-LOW and FLT3ITD-INT cases (P = .4 for heterogeneity for NPM1WT and P = .7 for NPM1MUT cases), although, as expected, the OS was lower in the NPM1WT cases (Figure 2B-C). Some studies have used a single cutoff of 33% FLT3ITD allele burden based on thresholds that show maximal clinical differences. When our data were subdivided this way, there was still no significant difference in CIR or OS between the lower and higher levels of FLT3ITD (supplemental Figures 2A-D).
To further correct for leukemic cell purity, in cases with ≥25% NPM1MUT, we calculated the ratio of FLT3ITD/NPM1MUT mutant allele levels and investigated 3 adjusted groups. The FLT3ITD-HIGH group (FLT3ITD/NPM1MUT >1.0) increased from 66 to 94 patients. Sixteen patients in the unadjusted FLT3ITD-LOW group moved into a higher-level FLT3ITD category. CIR in the adjusted FLT3ITD-LOW group (FLT3ITD/NPM1MUT <0.5) was correspondingly reduced from 55% to 50% (Figure 1D), but this was still higher than in NPM1MUTFLT3WT cases (34%). OS was significantly lower in patients in the adjusted FLT3ITD-HIGH and FLT3ITD-INT groups compared with NPM1MUTFLT3WT cases (29%, 35%, and 57%, respectively; P < .0001 for trend) (Figure 2D), but was only marginally reduced in the adjusted FLT3ITD-LOW cases (51%) compared with the NPM1MUTFLT3WT cases.
Overall, 278 patients (16% of this series) received an allogeneic transplant in CR1, including 34 of 292 NPM1MUTFLT3ITD patients (12%) (supplemental Table 2). Censoring at the time of transplantation did not impact the results.
Thus, this study of a large number of cytogenetically IR patients shows that in our cohort there were only minor differences in CIR according to the FLT3ITD levels in both NPM1MUT and NPM1WT cases, and this remained true after adjustments for lower leukemic cell purity using the NPM1MUT level. The similar CIR rates at all FLT3ITD levels is not surprising in that, if a FLT3ITD is associated with chemoresistance, there may be enrichment of any subclone of FLT3ITD bearing cells during chemotherapy. By contrast, OS was similar in the adjusted FLT3ITD-LOW and the FLT3WT cases, and as the relapse rate in the adjusted FLT3ITD-LOW cases was not different from the other FLT3ITD adjusted groups, this presumably reflects the higher initial CR rate and better outcome of salvage therapy in this group. Decisions about the applicability of allogeneic transplantation in CR1 are usually based on the predicted relapse rate in a particular patient,4 and this study shows that NPM1MUT cases with low-level FLT3ITD should not yet be considered different from patients with higher FLT3ITD levels.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the clinical investigators who entered and managed patients in these trials.
This work was supported by Leukaemia & Lymphoma Research, United Kingdom and the United Kingdom Medical Research Council. The work was undertaken at University College London Hospitals and University College London Cancer Institute, who received a proportion of funding from the Department of Health’s National Institute for Health Research Biomedical Research Centres funding scheme.
Authorship
Contribution: R.E.G., A.K., and D.C.L. designed the study; R.E.G. and R.K.H. analyzed the data; A.K.B. was principal trial coordinator; R.E.G., R.K.H., and D.C.L. wrote the manuscript; and all the authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David C. Linch, Department of Haematology, UCL Cancer Institute, Paul O’Gorman Building, 72 Huntley St, London, WC1E 6DD, United Kingdom; e-mail: d.linch@ucl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal