Key Points
Prognostic tool for CLL patients with high discriminatory power compared with conventional clinical staging systems.
Prognostication on the individual patient level independent of clinical stage.
Abstract
In addition to clinical staging, a number of biomarkers predicting overall survival (OS) have been identified in chronic lymphocytic leukemia (CLL). The multiplicity of markers, limited information on their independent prognostic value, and a lack of understanding of how to interpret discordant markers are major barriers to use in routine clinical practice. We therefore performed an analysis of 23 prognostic markers based on prospectively collected data from 1948 CLL patients participating in phase 3 trials of the German CLL Study Group to develop a comprehensive prognostic index. A multivariable Cox regression model identified 8 independent predictors of OS: sex, age, ECOG status, del(17p), del(11q), IGHV mutation status, serum β2-microglobulin, and serum thymidine kinase. Using a weighted grading system, a prognostic index was derived that separated 4 risk categories with 5-year OS ranging from 18.7% to 95.2% and having a C-statistic of 0.75. The index stratified OS within all analyzed subgroups, including all Rai/Binet stages. The validity of the index was externally confirmed in a series of 676 newly diagnosed CLL patients from Mayo Clinic. Using this multistep process including external validation, we developed a comprehensive prognostic index with high discriminatory power and prognostic significance on the individual patient level. The studies were registered as follows: CLL1 trial (NCT00262782, http://clinicaltrials.gov), CLL4 trial (ISRCTN 75653261, http://www.controlled-trials.com), and CLL8 trial (NCT00281918, http://clinicaltrials.gov).
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia of the Western world, with an incidence of 4.1 per 100 000 persons per year.1 The disease displays a high heterogeneity in its clinical course.2,3 Patients presenting with an indolent form often do not require treatment, whereas others experience a very aggressive course, leading to death within months.
Currently, staging and prognostication of CLL is performed by 2 similar clinical staging systems developed 30 to 35 years ago by Binet et al4 and Rai et al.5 Both systems use inexpensive, simple components such as blood counts and physical examination to identify 3 major prognostic subgroups. Despite these advantages, the clinical staging systems do not fully reflect the high variability of CLL, nor do they account for known biological characteristics of CLL cells predicting survival and response to therapy.3,6,7
Recently an impressive array of novel effective therapies has been developed that hold the potential of increasingly individualized treatments if patient risk could be accurately characterized.8-10 Unfortunately, the large number of novel prognostic markers in CLL, limited information on their independent prognostic value, and a lack of understanding of how to interpret discordant markers are still major barriers to integrate these in routine clinical CLL practice.11 To address this issue, we used the German CLL Study Group (GCLLSG) database to conduct a comprehensive evaluation of 23 clinical, biological, and genetic markers in CLL. The aim was to develop a prognostic index that identifies and combines the prognostic markers of independent importance that are already available. The utility of the developed prognostic index was subsequently validated using an external cohort of newly diagnosed CLL patients from the Mayo Clinic, Rochester, Minnesota.
Methods
Study population
Data from 3 prospective randomized phase 3 trials conducted between 1997 and 2006 by the GCLLSG were used as a training data set. All patients were untreated and had a diagnosis of CLL according to NCI Working Group Criteria.12
The CLL1 trial13 (#NCT00262782) included 876 Binet stage A patients and compared a watch-and-wait (W&W) strategy to early fludarabine (F) monotherapy in patients with a high risk for progression. The CLL4 trial14 (#ISRCTN 75653261) included 375 patients younger than 65 years requiring treatment and compared F to F plus cyclophosphamide (FC). The CLL8 trial15 (#NCT00281918) included 817 patients in need of treatment comparing FC to FC plus rituximab (FCR). Patients who were initially allocated to W&W in CLL1 and received first-line treatment later within CLL4 or CLL8 (N = 61) were only accounted for once. For those, data from first presentation (CLL1) were considered, including the longest observation period and corresponding baseline values.
All trials were approved by the leading ethics committee. Written informed consent was obtained from all patients according to the Declaration of Helsinki.
Data collection
Pretherapeutic features evaluated for potential prognostic relevance were sex, age, time between diagnosis and registration/randomization, Binet/Rai stages, ECOG performance status (PS),16 B-symptoms, blood counts, genetic abnormalities,17,18 expressions of ZAP-70/CD38,19,20 IGHV mutation status (MS),20-23 serum lactate dehydrogenase (LDH) level, serum thymidine kinase (s-TK) level,24-27 and serum β2-microglobulin (s-β2m) level.24,28,29 s-TK and s-β2m were evaluated centrally; s-β2m was analyzed by immunometric chemiluminescence assay and s-TK by either radioimmunoassay or quantitative immunoassay, respectively. Leukemic cells isolated from the peripheral blood were used for the determination of IGHV MS and assessment of ZAP-70/CD38 expression. Detailed descriptions of the diagnostic methods have been published previously.17,19,30-32 Data on ZAP-70/CD38 were not available for CLL1.
Statistical analysis
The main end point of statistical analyses was OS defined as the time between registration/randomization and death. Treatment-free survival (TFS) and progression-free survival (PFS) were calculated from registration/randomization to start of the first CLL treatment or from registration/randomization to disease progression or death, respectively. Subjects without a documented event were censored at time of last follow-up. Survival rates and standard errors were estimated by Kaplan-Meier methods,33 and survival curves were compared using log-rank tests. The prognostic relevance of each factor was evaluated applying the Kaplan-Meier methodology and Cox proportional hazards regression analyses.34 Continuous biological variables were dichotomized using published thresholds, laboratory norms, and quartiles. Threshold analysis including ROC curves35 and Youden Index36 were applied to identify additional thresholds. Dichotomized variables were only considered for further analysis if the continuous analog was of prognostic importance in univariate proportional hazards Cox regression.
All variables that showed significant association with OS on univariate analysis were consequently included in multivariate analysis applying forward and backward stepwise proportional hazards Cox regressions.
The analysis was further controlled for the variables “study” (CLL1/CLL4/CLL8); “type of first-line treatment” (W&W/F/FC/FCR); treatment indication status; B symptoms; time between diagnosis and registration/randomization; lymphadenopathy; splenomegaly/hepatomegaly; and hemoglobin, lymphocyte, and platelet counts to account for possible treatment effects as well as for the heterogeneous data set consisting of patients with and without treatment indication. For testing interactions in the final model, the multivariate analysis was repeated including the independent factors, the variable “type of first-line treatment,” as well as terms for interactions between factors and treatment.
Robustness of the multivariable Cox model was verified by bootstrapping techniques.37-39 A complete case analysis was applied to avoid the problem of missing data.
Factors independently associated with OS in the final model were included in the prognostic index. To account for differences in the magnitude of association between the individual independent factors and OS, we assigned a weighted risk score to each factor based on ranges of their corresponding hazard ratios. The total risk score was then calculated by the sum of the ratings of individual factors. To identify risk groups, the following criteria for the combination of risk categories were defined: (1) statistically significant differences in OS of risk groups, (2) absence of heterogeneities concerning independent factors within each risk group, and (3) adherence of smallest loss of information in terms of log-likelihood change. C-statistics were calculated to further evaluate discriminatory value of the prognostic index (c = 1 indicates perfect discrimination; c = 0.5 is equivalent to chance).37,40 All tests were 2-sided and significance was defined as P < .05. The analyses were performed using SPSS Statistics 21.
External validation
The validation data set was composed of a consecutive series of 676 newly diagnosed, prospectively followed CLL patients cared for at Mayo Clinic who had baseline data on all considered variables except s-TK and/or s-β2m available and who had stored serum collected ≤36 months (median, 1 month) of diagnosis available for s-TK and s-β2m analysis. Stored serum was shipped to the Institute for Clinical Chemistry at the University Hospital of Cologne for subsequent s-TK assessment. Because the s-TK assays in the training data set measured s-TK using a radioimmunoassay and the validation cohort used a non–radio-labeled immunoassay, interassay calibration of both assays was performed (correlation R2 = 0.89) to allow mathematical conversion before applying the s-TK threshold for assigning index point score. For the validation cohort, OS was defined as the time between diagnosis and death. TFS was calculated from the date of diagnosis to the start of the first CLL treatment. Subjects without a documented event were censored at time of last follow-up. The outcome of individuals was prospectively assessed.
Results
Patients’ characteristics of the training data set
After excluding patients with missing baseline data (N = 47) and those with insufficient follow-up (N = 12), 1948 eligible patients were available as a training data set (flowchart, supplemental material I). Median age was 60.0 years (range, 30.0-81.0); 485 deaths from all causes were reported after a median observation time of 63.4 months.
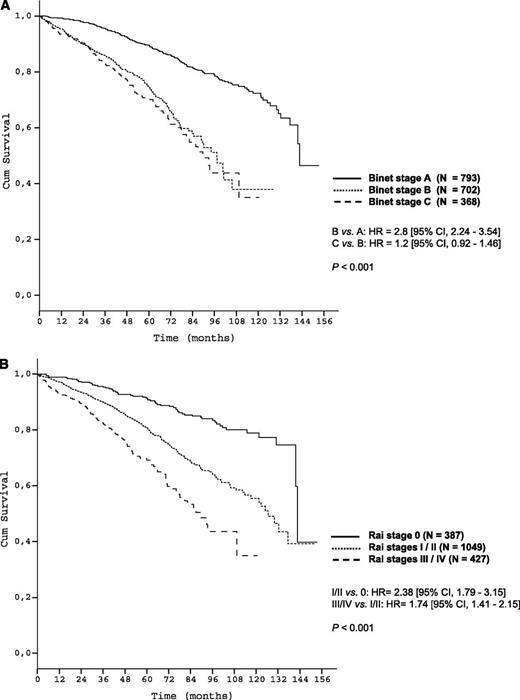
Patient characteristics are shown in Table 1. Survival curves according to Binet/Rai stages are shown in Figure 1A-B.
Patients’ characteristics and results of the univariate analyses of OS after five years
| Training data set . | No. of patients, N (%) . | 5-y survival (%) . | Log rank, P . | Univariable comparison . | HR* (95% CI†) . |
|---|---|---|---|---|---|
| All patients | 1948 | 80.3% | |||
| Sex | 1948 | <.001 | |||
| Female | 615 (31.6) | 84.0 | |||
| Male | 1333 (68.4) | 78.6 | vs female | 1.5 (1.2-1.9) | |
| Time from diagnosis to study entry (y) | 1877 | .004 | |||
| ≤1 | 1187 (63.3) | 84.1 | |||
| >1 | 690 (36.7) | 78.1 | vs ≤1 | 1.3 (1.1-1.6) | |
| Age (y) | 1948 | .003 | |||
| Median (range) | 60.0 (30.0 - 81.0) | ||||
| ≤60 | 1019 (52.3) | 83.5 | |||
| >60 and ≤65 | 511 (26.2) | 76.7 | vs ≤60 | 1.5 (1.2-1.8) | |
| >65 and ≤70 | 275 (14.1) | 78.9 | vs ≤60 | 1.3 (1.0-1.7) | |
| >70 | 143 (7.3) | 73.2 | vs ≤60 | 1.4 (1.0-1.9) | |
| B symptoms | 1856 | <.001 | |||
| No | 1313 (70.7) | 83.5 | |||
| Yes | 543 (29.3) | 72.3 | vs no | 1.8 (1.5-2.2) | |
| Binet stage | 1863 | <.001 | |||
| A | 793 (42.6) | 89.5 | |||
| B | 702 (37.7) | 74.6 | |||
| C | 368 (19.8) | 70.1 | B/C vs A | 3.0 (2.4-3.6) | |
| Rai stage | 1863 | <.001 | |||
| 0 | 387 (20.8) | 91.4 | |||
| I | 342 (18.4) | 82.5 | |||
| II | 707 (37.9) | 80.0 | |||
| III | 161 (8.6) | 64.4 | |||
| IV | 266 (14.3) | 72.0 | 0-I vs II-IV | 2.1 (1.7-2.6) | |
| ECOG performance status | 1826 | <.001 | |||
| 0 | 1277 (69.9) | 84.6 | |||
| 1 | 533 (29.2) | 70.3 | 0 vs >0 | 2.1 (1.7-2.5) | |
| 2 | 16 (0.9) | 62.5 | |||
| IGHV MS | 1430 | <.001 | |||
| Mutated | 661 (46.2) | 89.1 | |||
| Unmutated | 769 (53.8) | 70.9 | vs mutated | 3.4 (2.7-4.3) | |
| Deletion 6q | 779 | .318 | |||
| No | 746 (95.8) | 84.0 | |||
| Yes | 33 (4.2) | 82.2 | vs no | 1.3 (0.7-2.4) | |
| Categories according to hierarchical model17 | 1557 | <.001 | |||
| Del(17p) | 89 (5.7) | 34.4 | vs del(13q)# | 5.9 (4.3-8.2) | |
| Del(11q)‡ | 250 (16.1) | 75.4 | vs del(13q)# | 2.1 (1.6-2.8) | |
| Trisomy 12§ | 163 (10.5) | 83.0 | vs del(13q)# | 1.1 (0.8-1.6) | |
| Normal¶ | 555 (35.6) | 86.9 | vs del(13q)# | 0.8 (0.6-1.0) | |
| Del(13q)# | 500 (32.1) | 85.3 | |||
| Hemoglobin (g/dL) | 1863 | <.001 | |||
| ≤10.0 | 182 (9.8) | 65.0 | vs >10.0 | 2.1 (1.6-2.7) | |
| >10.0 | 1681 (90.2) | 82.1 | |||
| Platelet count (× 103/µL) | 1859 | <.001 | |||
| <100.0 | 262 (14.1) | 70.0 | vs ≥100.0 | 1.9 (1.5-2.4) | |
| ≥100.0 | 1597 (85.9) | 82.3 | |||
| Leukocyte count (× 103/µL) | 1866 | <.001 | |||
| ≤50.0 | 1069 (57.3) | 86.0 | |||
| >50.0 | 797 (42.7) | 75.6 | vs <50.0 | 2.4 (2.0-2.9) | |
| Lymphocyte count (× 103/µL) | 1832 | <.001 | |||
| ≤50.0 | 1149 (62.7) | 85.3 | |||
| >50.0 | 683 (37.3) | 72.7 | vs ≤50.0 | 2.3 (1.9-2.8) | |
| Absolute neutrophil count (× 103/µL) | 1698 | <.001 | |||
| ≤6.5 | 1284 (75.6) | 83.2 | |||
| >6.5 | 414 (24.4) | 75.2 | vs ≤6.5 | 1.6 (1.3-1.9) | |
| LDH (U/L) | 1762 | <.001 | |||
| ≤210.0 | 1149 (65.2) | 84.7 | |||
| >210.0 | 613 (34.8) | 71.6 | vs ≤210.0 | 2.1 (1.7-2.5) | |
| s-TK (U/L) | 1670 | <.001 | |||
| ≤10.0 | 801 (48.0) | 91.7 | |||
| >10.0 and ≤24.0 | 450 (27.0) | 78.6 | vs ≤10.0 | 31 (2.4-4.1) | |
| >24.0 | 419 (25.0) | 66.4 | vs ≤10.0 | 54 (4.2-7.0) | |
| s-β2m (mg/L) | 1676 | <.001 | |||
| ≤1.7 | 403 (24.0) | 94.2 | |||
| >1.7 and ≤3.5 | 861 (51.4) | 82.7 | vs ≤1.7 | 2.8 (2.0-3.9) | |
| >3.5 | 412 (24.6) | 67.9 | vs ≤1.7 | 6.5 (4.6-9.3) | |
| ZAP-70 (%) | 502 | .2 | |||
| ≤20.0 | 310 (61.8) | 73.5 | |||
| >20.0 | 192 (38.2) | 68.6 | vs ≤20.0 | 1.2 (0.9-1.7) | |
| CD38 (%) | 914 | .8 | |||
| ≤30.0 | 619 (67.7) | 75.2 | |||
| >30.0 | 295 (32.3) | 70.9 | vs ≤30.0 | 1.1 (0.9-1.4) | |
| Mayo validation data set | |||||
| All patients | 676 | ||||
| Sex | 676 | .2 | |||
| Female | 223 (33.0) | 92.9 | |||
| Male | 453 (67.0) | 87.1 | vs female | 1.4 (0.9-2.2) | |
| Age (y) | 676 | <.001 | |||
| Median (range) | 61.5 (32.0 - 89.0) | ||||
| ≤60 | 314 (46.4) | 94.3 | |||
| >60 and ≤65 | 116 (17.2) | 91.6 | |||
| >65 and ≤70 | 102 (15.1) | 88.3 | |||
| >70 | 144 (21.3) | 77.0 | >60 vs ≤60 | 2.3 (1.4-3.7) | |
| Rai stage | 676 | <.001 | |||
| 0 | 386 (57.1) | 91.8 | |||
| I | 230 (34.0) | 85.0 | |||
| II | 40 (5.9) | 88.9 | 0 vs I-II | 1.6 (1.01-2.4) | |
| III | 7 (1.0) | 51.4 | |||
| IV | 13 (1.9) | 92.3 | 0 vs III-IV | 4.5 (1.9-10.6) | |
| ECOG performance status | 676 | <.001 | |||
| 0 | 635 (93.9) | 89.8 | |||
| >0 | 41 (6.1) | 70.6 | 0 vs >0 | 1.8 (1.3-2.6) | |
| IGHV MS | 676 | <.001 | |||
| Mutated | 382 (56.5) | 91.4 | |||
| Unmutated | 294 (43.5) | 85.1 | vs mutated | 2.6 (1.7-4.0) | |
| Categories according to hierarchical model17 | 676 | <.001 | |||
| Del(17p) | 30 (4.4) | 39.7 | vs del(13q)# | 8.9 (4.5-17.5) | |
| Del(11q)‡ | 66 (9.8) | 81.0 | vs del(13q)# | 2.6 (1.3-5.1) | |
| Trisomy 12§ | 133 (19.7) | 88.6 | vs del(13q)# | 1.7 (1.0-3.2) | |
| Normal¶ | 160 (23.7) | 90.5 | vs del(13q)# | 1.2 (0.7-2.3) | |
| Del(13q)# | 287 (42.5) | 93.8 | |||
| s-TK (U/L) | 676 | .01 | |||
| ≤10.0 | 493 (72.9) | 91.0 | |||
| >10.0 | 183 (27.1) | 81.7 | vs ≤10.0 | 1.7 (1.1-2.8) | |
| s-β2m (mg/L) | 672 | <.001 | |||
| ≤1.7 | 88 (13.1) | 98.2 | |||
| >1.7 and ≤3.5 | 468 (69.6) | 92.5 | vs ≤1.7 | 2.7 (0.9-8.8) | |
| >3.5 | 116 (17.3) | 65.5 | vs ≤1.7 | 11.6 (3.6-37.6) | |
| Training data set . | No. of patients, N (%) . | 5-y survival (%) . | Log rank, P . | Univariable comparison . | HR* (95% CI†) . |
|---|---|---|---|---|---|
| All patients | 1948 | 80.3% | |||
| Sex | 1948 | <.001 | |||
| Female | 615 (31.6) | 84.0 | |||
| Male | 1333 (68.4) | 78.6 | vs female | 1.5 (1.2-1.9) | |
| Time from diagnosis to study entry (y) | 1877 | .004 | |||
| ≤1 | 1187 (63.3) | 84.1 | |||
| >1 | 690 (36.7) | 78.1 | vs ≤1 | 1.3 (1.1-1.6) | |
| Age (y) | 1948 | .003 | |||
| Median (range) | 60.0 (30.0 - 81.0) | ||||
| ≤60 | 1019 (52.3) | 83.5 | |||
| >60 and ≤65 | 511 (26.2) | 76.7 | vs ≤60 | 1.5 (1.2-1.8) | |
| >65 and ≤70 | 275 (14.1) | 78.9 | vs ≤60 | 1.3 (1.0-1.7) | |
| >70 | 143 (7.3) | 73.2 | vs ≤60 | 1.4 (1.0-1.9) | |
| B symptoms | 1856 | <.001 | |||
| No | 1313 (70.7) | 83.5 | |||
| Yes | 543 (29.3) | 72.3 | vs no | 1.8 (1.5-2.2) | |
| Binet stage | 1863 | <.001 | |||
| A | 793 (42.6) | 89.5 | |||
| B | 702 (37.7) | 74.6 | |||
| C | 368 (19.8) | 70.1 | B/C vs A | 3.0 (2.4-3.6) | |
| Rai stage | 1863 | <.001 | |||
| 0 | 387 (20.8) | 91.4 | |||
| I | 342 (18.4) | 82.5 | |||
| II | 707 (37.9) | 80.0 | |||
| III | 161 (8.6) | 64.4 | |||
| IV | 266 (14.3) | 72.0 | 0-I vs II-IV | 2.1 (1.7-2.6) | |
| ECOG performance status | 1826 | <.001 | |||
| 0 | 1277 (69.9) | 84.6 | |||
| 1 | 533 (29.2) | 70.3 | 0 vs >0 | 2.1 (1.7-2.5) | |
| 2 | 16 (0.9) | 62.5 | |||
| IGHV MS | 1430 | <.001 | |||
| Mutated | 661 (46.2) | 89.1 | |||
| Unmutated | 769 (53.8) | 70.9 | vs mutated | 3.4 (2.7-4.3) | |
| Deletion 6q | 779 | .318 | |||
| No | 746 (95.8) | 84.0 | |||
| Yes | 33 (4.2) | 82.2 | vs no | 1.3 (0.7-2.4) | |
| Categories according to hierarchical model17 | 1557 | <.001 | |||
| Del(17p) | 89 (5.7) | 34.4 | vs del(13q)# | 5.9 (4.3-8.2) | |
| Del(11q)‡ | 250 (16.1) | 75.4 | vs del(13q)# | 2.1 (1.6-2.8) | |
| Trisomy 12§ | 163 (10.5) | 83.0 | vs del(13q)# | 1.1 (0.8-1.6) | |
| Normal¶ | 555 (35.6) | 86.9 | vs del(13q)# | 0.8 (0.6-1.0) | |
| Del(13q)# | 500 (32.1) | 85.3 | |||
| Hemoglobin (g/dL) | 1863 | <.001 | |||
| ≤10.0 | 182 (9.8) | 65.0 | vs >10.0 | 2.1 (1.6-2.7) | |
| >10.0 | 1681 (90.2) | 82.1 | |||
| Platelet count (× 103/µL) | 1859 | <.001 | |||
| <100.0 | 262 (14.1) | 70.0 | vs ≥100.0 | 1.9 (1.5-2.4) | |
| ≥100.0 | 1597 (85.9) | 82.3 | |||
| Leukocyte count (× 103/µL) | 1866 | <.001 | |||
| ≤50.0 | 1069 (57.3) | 86.0 | |||
| >50.0 | 797 (42.7) | 75.6 | vs <50.0 | 2.4 (2.0-2.9) | |
| Lymphocyte count (× 103/µL) | 1832 | <.001 | |||
| ≤50.0 | 1149 (62.7) | 85.3 | |||
| >50.0 | 683 (37.3) | 72.7 | vs ≤50.0 | 2.3 (1.9-2.8) | |
| Absolute neutrophil count (× 103/µL) | 1698 | <.001 | |||
| ≤6.5 | 1284 (75.6) | 83.2 | |||
| >6.5 | 414 (24.4) | 75.2 | vs ≤6.5 | 1.6 (1.3-1.9) | |
| LDH (U/L) | 1762 | <.001 | |||
| ≤210.0 | 1149 (65.2) | 84.7 | |||
| >210.0 | 613 (34.8) | 71.6 | vs ≤210.0 | 2.1 (1.7-2.5) | |
| s-TK (U/L) | 1670 | <.001 | |||
| ≤10.0 | 801 (48.0) | 91.7 | |||
| >10.0 and ≤24.0 | 450 (27.0) | 78.6 | vs ≤10.0 | 31 (2.4-4.1) | |
| >24.0 | 419 (25.0) | 66.4 | vs ≤10.0 | 54 (4.2-7.0) | |
| s-β2m (mg/L) | 1676 | <.001 | |||
| ≤1.7 | 403 (24.0) | 94.2 | |||
| >1.7 and ≤3.5 | 861 (51.4) | 82.7 | vs ≤1.7 | 2.8 (2.0-3.9) | |
| >3.5 | 412 (24.6) | 67.9 | vs ≤1.7 | 6.5 (4.6-9.3) | |
| ZAP-70 (%) | 502 | .2 | |||
| ≤20.0 | 310 (61.8) | 73.5 | |||
| >20.0 | 192 (38.2) | 68.6 | vs ≤20.0 | 1.2 (0.9-1.7) | |
| CD38 (%) | 914 | .8 | |||
| ≤30.0 | 619 (67.7) | 75.2 | |||
| >30.0 | 295 (32.3) | 70.9 | vs ≤30.0 | 1.1 (0.9-1.4) | |
| Mayo validation data set | |||||
| All patients | 676 | ||||
| Sex | 676 | .2 | |||
| Female | 223 (33.0) | 92.9 | |||
| Male | 453 (67.0) | 87.1 | vs female | 1.4 (0.9-2.2) | |
| Age (y) | 676 | <.001 | |||
| Median (range) | 61.5 (32.0 - 89.0) | ||||
| ≤60 | 314 (46.4) | 94.3 | |||
| >60 and ≤65 | 116 (17.2) | 91.6 | |||
| >65 and ≤70 | 102 (15.1) | 88.3 | |||
| >70 | 144 (21.3) | 77.0 | >60 vs ≤60 | 2.3 (1.4-3.7) | |
| Rai stage | 676 | <.001 | |||
| 0 | 386 (57.1) | 91.8 | |||
| I | 230 (34.0) | 85.0 | |||
| II | 40 (5.9) | 88.9 | 0 vs I-II | 1.6 (1.01-2.4) | |
| III | 7 (1.0) | 51.4 | |||
| IV | 13 (1.9) | 92.3 | 0 vs III-IV | 4.5 (1.9-10.6) | |
| ECOG performance status | 676 | <.001 | |||
| 0 | 635 (93.9) | 89.8 | |||
| >0 | 41 (6.1) | 70.6 | 0 vs >0 | 1.8 (1.3-2.6) | |
| IGHV MS | 676 | <.001 | |||
| Mutated | 382 (56.5) | 91.4 | |||
| Unmutated | 294 (43.5) | 85.1 | vs mutated | 2.6 (1.7-4.0) | |
| Categories according to hierarchical model17 | 676 | <.001 | |||
| Del(17p) | 30 (4.4) | 39.7 | vs del(13q)# | 8.9 (4.5-17.5) | |
| Del(11q)‡ | 66 (9.8) | 81.0 | vs del(13q)# | 2.6 (1.3-5.1) | |
| Trisomy 12§ | 133 (19.7) | 88.6 | vs del(13q)# | 1.7 (1.0-3.2) | |
| Normal¶ | 160 (23.7) | 90.5 | vs del(13q)# | 1.2 (0.7-2.3) | |
| Del(13q)# | 287 (42.5) | 93.8 | |||
| s-TK (U/L) | 676 | .01 | |||
| ≤10.0 | 493 (72.9) | 91.0 | |||
| >10.0 | 183 (27.1) | 81.7 | vs ≤10.0 | 1.7 (1.1-2.8) | |
| s-β2m (mg/L) | 672 | <.001 | |||
| ≤1.7 | 88 (13.1) | 98.2 | |||
| >1.7 and ≤3.5 | 468 (69.6) | 92.5 | vs ≤1.7 | 2.7 (0.9-8.8) | |
| >3.5 | 116 (17.3) | 65.5 | vs ≤1.7 | 11.6 (3.6-37.6) | |
Hazard ratio.
Confidence interval.
Not including deletion 17p.
Not including deletion 17p and deletion 11q.
No abnormalities according to the hierarchical model including deletion 17p, deletion 11q, trisomy 12, and deletion13q.
Not including deletion 17p, deletion 11q, and trisomy 12.
The survival curves according to clinical staging in the training data set. (A) OS of Binet stages within the training data set (N = 1863); (B) OS of Rai stages within the training data set (N = 1863).
The survival curves according to clinical staging in the training data set. (A) OS of Binet stages within the training data set (N = 1863); (B) OS of Rai stages within the training data set (N = 1863).
Univariate and multivariate analyses
Except for ZAP-70/CD38 status, and del(6q), all parameters showed a significant correlation with OS using univariate analysis (Table 1), and variables were subsequently considered for multivariate analysis.
Eight parameters were identified as independent predictors of OS in 1223 patients, with all parameters significant on univariate analysis available: sex, age, ECOG PS, genetic aberrations del(17p) and del(11q), IGHV MS, s-TK, and s-β2m (Table 2). These 1223 patients were representative of the entire population training data set. All variables used to control for possible confounding effects—as specified in Methods—were proven to not be independent factors for OS. Internal validation was performed by bootstrapping techniques: Based on 100 generated resamples of the training data set, regressions were repeated, and the robustness of the 8-parameter model was confirmed uniquely (Table 2).
Results of the final Cox regression model and risk scores of independent factors based on 1223 patients with all parameters available (training data set, score population)
| Independent factor . | HR* (95% CI†) . | P . | Risk score . | |
|---|---|---|---|---|
| Category according hierarchical model17 | Del(17p) | 6.0 (4.2 – 8.6) | <.001 | 6 |
| s-TK | >10.0 U/L | 2.1 (1.5-2.9) | <.001 | 2 |
| s-β2m | >3.5 mg/L | 2.3 (1.4-3.6) | .001 | 2 |
| s-β2m | >1.7 and ≤3.5 mg/L | 1.7 (1.1-2.7) | .01 | 1 |
| IGHV MS | Unmutated | 1.9 (1.5-2.5) | <.001 | 1 |
| ECOG PS | >0 | 1.7 (1.3-2.1) | <.001 | 1 |
| Category according hierarchical model17 | Del(11q) | 1.4 (1.03-2.0) | .03 | 1 |
| Sex | Male | 1.3 (1.01-1.6) | .026 | 1 |
| Age | >60 y | 1.3 (1.04-1.7) | .045 | 1 |
| Independent factor . | HR* (95% CI†) . | P . | Risk score . | |
|---|---|---|---|---|
| Category according hierarchical model17 | Del(17p) | 6.0 (4.2 – 8.6) | <.001 | 6 |
| s-TK | >10.0 U/L | 2.1 (1.5-2.9) | <.001 | 2 |
| s-β2m | >3.5 mg/L | 2.3 (1.4-3.6) | .001 | 2 |
| s-β2m | >1.7 and ≤3.5 mg/L | 1.7 (1.1-2.7) | .01 | 1 |
| IGHV MS | Unmutated | 1.9 (1.5-2.5) | <.001 | 1 |
| ECOG PS | >0 | 1.7 (1.3-2.1) | <.001 | 1 |
| Category according hierarchical model17 | Del(11q) | 1.4 (1.03-2.0) | .03 | 1 |
| Sex | Male | 1.3 (1.01-1.6) | .026 | 1 |
| Age | >60 y | 1.3 (1.04-1.7) | .045 | 1 |
Hazard ratio.
Confidence interval.
We also repeated the multivariate analysis analyzing CLL1 (an early intervention trial) and CLL4/8 (first-line treatment trials) separately (supplemental Figures 4 and 5). The key molecular biomarkers/serum factors identified for inclusion in the model (s-TK, s-β2m, IGHV MS, del(17p)) were similar in both models. Notably, no other/unique molecular characteristics were identified for either models, with the exception of deletion 11q23, which was significant in the CLL1 model but not in the CLL4/8 model (potentially because of small sample size and inadequate power). These findings provide support for pooling the data from the CLL1, CLL4, and CLL8 trials to determine whether additional factors enter the model with larger sample size and greater power.
Prognostic index
Next, given a large range of hazard ratios (HR) of the independent factors (eg, HR = 1.3 for sex; HR = 6.0 for del(17p)), a risk score was assigned to each of the independent factors in the final model (Table 2). The weighting was based on a simple algorithm assigning the integer value of the corresponding HR to each factor (ie, 1 point for HR 1.1-1.9; 2 points for HR 2.0-2.9, etc). Finally, we defined the total risk score as the sum of the risk scores of the 8 individual factors (range, 0-14).
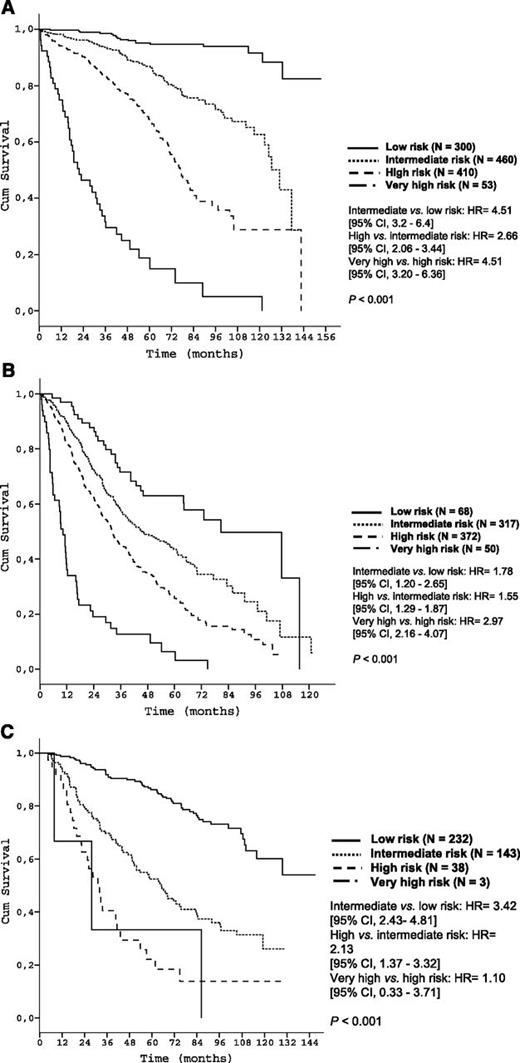
According to the predefined criteria (see Methods), 4 different risk categories for OS were determined: low (score 0-2, N = 300), intermediate (score 3-5, N = 460), high (score 6-10, N = 410), and very high (score 11-14, N = 53) (Table 3). The proposed risk categories segregated 5-year OS rates from 95.2% (low risk) to 18.7% (very high risk) (P < .001) with c = 0.75 (Table 3 and Figure 2A).
Rates of 5-y OS according to the single risk scores and the risk groups of the prognostic index (training and validation data set)
| . | Risk score . | No. of patients, N (%) . | 5-y OS, % . | 6-y OS, % . | HR* (95% CI†) . |
|---|---|---|---|---|---|
| 0 | 40 (3.3) | (all censored) | (all censored) | ||
| 1 | 112 (9.2) | 96.0 | 94.9 | ||
| 2 | 148 (12.1) | 93.2 | 93.2 | ||
| 3 | 122 (10.0) | 91.3 | 85.7 | ||
| 4 | 154 (12.6) | 87.9 | 84.5 | ||
| 5 | 184 (15.0) | 83.0 | 70.5 | ||
| 6 | 192 (15.7) | 72.4 | 62.9 | ||
| 7 | 138 (11.3) | 67.9 | 50.7 | ||
| 8 | 54 (4.4) | 56.2 | 46.7 | ||
| 9 | 16 (1.3) | 56.1 | 42.1 | ||
| 10 | 10 (0.8) | 45.0 | 45.0 | ||
| ≥11 | 53 (4.3) | 18.7 | 15.0 | ||
| Risk group GCLLSG | |||||
| Low | 0-2 | 300 (24.5) | 95.2 | 94.8 | |
| Intermediate | 3-5 | 460 (37.6) | 86.9 | 80.4 | 4.8 (2.9-8.0) |
| High | 6-10 | 410 (33.5) | 67.6 | 55.6 | 12.5 (7.7-20.5) |
| Very high | 11-14 | 53 (4.3) | 18.7 | 15.0 | 57.7 (33.0-101.2) |
| Risk group Mayo Clinic | |||||
| Low | 0-2 | 226 (33.6) | 95.2 | 92.1 | |
| Intermediate | 3-5 | 336 (49.9) | 91.4 | 86.7 | 1.9 (1.03-3.5) |
| High | 6-10 | 95 (14.1) | 71.7 | 69.1 | 5.5 (2.9-10.7) |
| Very high | 11-14 | 16 (2.4) | 13.6 | 13.6 | 28.9 (12.3-68.3) |
| . | Risk score . | No. of patients, N (%) . | 5-y OS, % . | 6-y OS, % . | HR* (95% CI†) . |
|---|---|---|---|---|---|
| 0 | 40 (3.3) | (all censored) | (all censored) | ||
| 1 | 112 (9.2) | 96.0 | 94.9 | ||
| 2 | 148 (12.1) | 93.2 | 93.2 | ||
| 3 | 122 (10.0) | 91.3 | 85.7 | ||
| 4 | 154 (12.6) | 87.9 | 84.5 | ||
| 5 | 184 (15.0) | 83.0 | 70.5 | ||
| 6 | 192 (15.7) | 72.4 | 62.9 | ||
| 7 | 138 (11.3) | 67.9 | 50.7 | ||
| 8 | 54 (4.4) | 56.2 | 46.7 | ||
| 9 | 16 (1.3) | 56.1 | 42.1 | ||
| 10 | 10 (0.8) | 45.0 | 45.0 | ||
| ≥11 | 53 (4.3) | 18.7 | 15.0 | ||
| Risk group GCLLSG | |||||
| Low | 0-2 | 300 (24.5) | 95.2 | 94.8 | |
| Intermediate | 3-5 | 460 (37.6) | 86.9 | 80.4 | 4.8 (2.9-8.0) |
| High | 6-10 | 410 (33.5) | 67.6 | 55.6 | 12.5 (7.7-20.5) |
| Very high | 11-14 | 53 (4.3) | 18.7 | 15.0 | 57.7 (33.0-101.2) |
| Risk group Mayo Clinic | |||||
| Low | 0-2 | 226 (33.6) | 95.2 | 92.1 | |
| Intermediate | 3-5 | 336 (49.9) | 91.4 | 86.7 | 1.9 (1.03-3.5) |
| High | 6-10 | 95 (14.1) | 71.7 | 69.1 | 5.5 (2.9-10.7) |
| Very high | 11-14 | 16 (2.4) | 13.6 | 13.6 | 28.9 (12.3-68.3) |
Hazard ratio.
Confidence interval.
Time-to-event curves according to the prognostic index in the training data set. (A) OS of risk groups of the training data set according to the prognostic index (score population: N = 1223); (B) PFS of patients starting treatment according to the prognostic index (training data set N = 807); (C) TFS of untreated patients according to the prognostic index (training data set N = 416).
Time-to-event curves according to the prognostic index in the training data set. (A) OS of risk groups of the training data set according to the prognostic index (score population: N = 1223); (B) PFS of patients starting treatment according to the prognostic index (training data set N = 807); (C) TFS of untreated patients according to the prognostic index (training data set N = 416).
Analyses for PFS in treated patients (N = 807) and TFS in patients managed with W&W (N = 416) also demonstrated validity of the prognostic index for these end points. Among treated patients, 5-year PFS rates of the 4 risk groups were 62.9%, 43.6%, 25.6%, and 6.4%, respectively (P < .001; Figure 2B). Among patients initially managed with W&W, 5-year TFS rates were 86.2%, 52.4%, 22.1%, and 0.0% respectively (P < .001; Figure 2C).
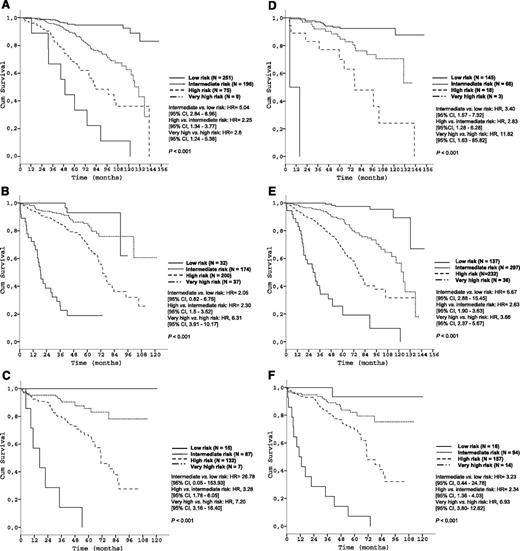
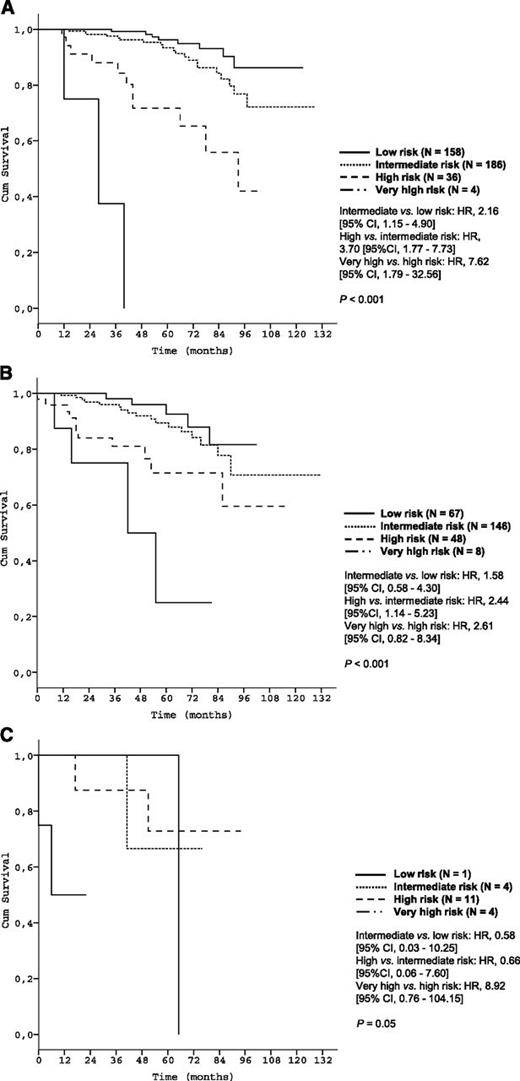
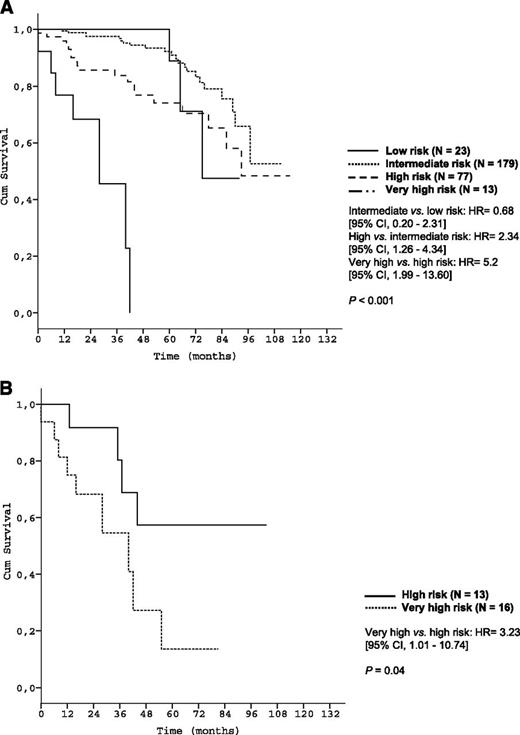
Subgroup analyses corroborated the discriminative strength of the prognostic index. The 4 risk groups were reproduced within each Binet/Rai stage (P < .001, respectively) (Figure 3) and within IGHV-unmutated patients (Figure 4A). Within the group of patients with del(17p), the index was able to distinguish patients with high risk from patients with very high risk (P < .001) (Figure 4B).
Survival curves of the Binet and Rai stages by prognostic index risk categories in the training data set. (A) Patients in Binet stage A (N = 531); (B) Patients in Binet stage B (N = 443); (C) Patients in Binet stage C (N = 241); (D) Patients in Rai stage 0 (N = 232); (E) Patients in Rai stage I/II (N = 702); (F) Patients in Rai stage III/IV (N = 281).
Survival curves of the Binet and Rai stages by prognostic index risk categories in the training data set. (A) Patients in Binet stage A (N = 531); (B) Patients in Binet stage B (N = 443); (C) Patients in Binet stage C (N = 241); (D) Patients in Rai stage 0 (N = 232); (E) Patients in Rai stage I/II (N = 702); (F) Patients in Rai stage III/IV (N = 281).
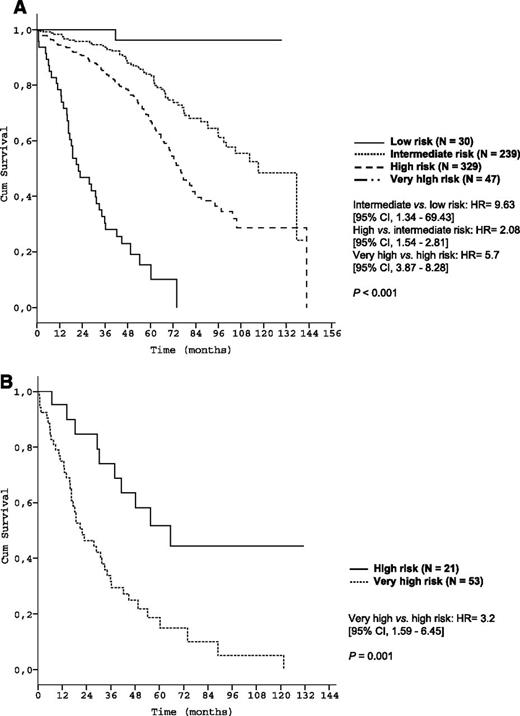
Survival curves of genetic subgroups by prognostic index risk categories in the training data set. (A) OS of patients with unmutated IGHV mutation status within risk groups of the prognostic index (training data set N = 645); (B) OS of patients with deletion 17p13 within risk groups of the prognostic index (training data set N = 74).
Survival curves of genetic subgroups by prognostic index risk categories in the training data set. (A) OS of patients with unmutated IGHV mutation status within risk groups of the prognostic index (training data set N = 645); (B) OS of patients with deletion 17p13 within risk groups of the prognostic index (training data set N = 74).
External validation
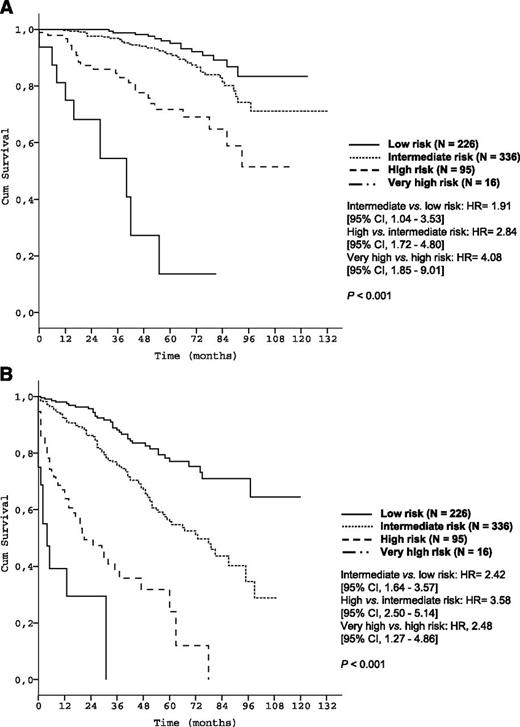
The utility of the prognostic index was subsequently evaluated in a prospectively followed validation cohort of 676 newly diagnosed CLL patients cared for at Mayo Clinic (Table 1). Three patients were excluded because of missing data for s-β2m. The median observation time was 57.0 months, and 85 deaths (12.6%) were observed. The median age was 61.5 years (range, 32.0-89.0). Within this cohort the 5-year OS for the respective risk groups were 95.2%, 91.4%, 71.7%, and 13.6% (P < .001, Table 3 and Figure 5A) (C-statistic: c = 0.83).
Time-to-event curves according to the prognostic index in the validation data set. (A) OS of risk groups of the Mayo validation cohort according to the prognostic index (N = 673); (B) TFS of the Mayo validation cohort according to the prognostic index (N = 673).
Time-to-event curves according to the prognostic index in the validation data set. (A) OS of risk groups of the Mayo validation cohort according to the prognostic index (N = 673); (B) TFS of the Mayo validation cohort according to the prognostic index (N = 673).
At last follow-up, 486 patients (71.9%) were still untreated, and the validity of the prognostic index for predicting TFS was also confirmed: after 5 years, 77.1%, 55.7%, 23.9%, and 0.0%, respectively, were untreated (P < .001; Figure 5B).
The discriminative strength of the prognostic index within clinical stages (Figure 6) and biological risk groups (Figure 7) was confirmed in the validation data set as well.
Survival curves of Rai stages by prognostic index risk categories in the validation data set. (A) Patients in Rai stage 0 (N = 384); (B) Patients in rai stage I/II (N = 269); (C) Patients in Rai stage III/IV (N = 20).
Survival curves of Rai stages by prognostic index risk categories in the validation data set. (A) Patients in Rai stage 0 (N = 384); (B) Patients in rai stage I/II (N = 269); (C) Patients in Rai stage III/IV (N = 20).
Survival curves of genetic subgroups by prognostic index risk categories in the validation data set. (A) OS of patients with unmutated IGHV mutation status within risk groups of the prognostic index (validation data set N = 292); (B) OS of patients with deletion 17p13 withihn risk groups of the prognostic index (validation data set N = 29).
Survival curves of genetic subgroups by prognostic index risk categories in the validation data set. (A) OS of patients with unmutated IGHV mutation status within risk groups of the prognostic index (validation data set N = 292); (B) OS of patients with deletion 17p13 withihn risk groups of the prognostic index (validation data set N = 29).
Comparisons with the prognostic index
Wierda and colleagues29 have proposed a prognostic model for OS including 3 factors contained in our model (age, s-β2m, sex) but without genomic aberrations, IGHV MS, and s-TK. To explore how our index improved on this model, we identified 1144 patients in our training data set with the necessary variables to classify patients according to both systems. The C-statistic of the previous model was c = 0.61, a level below that of the full index (c = 0.75) and below the accepted 0.7 threshold necessary to have value at the individual patient level.40 When the incremental prognostic value of genomic aberrations, IGHV MS, and s-TK was assessed, each of these parameters improved prediction of OS compared with the previous model29 without these factors: del(17p) (HR = 5.6 [95% confidence interval (CI), 4.0-7.8], P < .001), del(11q) (HR = 1.5 [95% CI, 1.1-2.0], P = .005), IGHV MS (HR = 2.1 [95% CI, 1.6-2.7], P < .001), s-TK >10.0 U/L (HR = 2.5 [95% CI, 1.8-3.4], P < .001). In addition, classifying the patients of each risk strata (low, intermediate, high) of the previous model29 according to our prognostic index provided substantial improvement in prognostication (supplemental Material II).
Discussion
For almost 40 years, the Rai/Binet staging classifications have formed the backbone of CLL management. However, it has become apparent that both systems lack precision in discriminating prognostic subgroups of CLL patients and that the ability to predict outcomes for individual patients is limited, as demonstrated by a C-statistic of only c = 0.56 and c = 0.58 for Rai and Binet staging, respectively, within our training data set.11,41 Furthermore, a multitude of new prognostic markers have been identified in the past few decades. The aim of this analysis was to identify markers that have independent prognostic value among assays in routine clinical practice in the US and/or Europe. We also sought to determine how these factors can be combined into an integrated prognostic model that allows clinicians to interpret and apply the collective results of prognostic tests for individual patient counseling and to enable clinical scientists to develop risk-adapted therapies for clinical testing. The manuscript represents a major step forward of integrating the most important prognostic tools of the last 30 years into a single model. The lack of such an accurate prognostic system is currently a major clinical problem in CLL.
We demonstrated in the training data set and further confirmed in the validation data set that the prognostic index developed allows a substantial gain of information compared with the conventional clinical staging systems as well as with the most important single risk factors known (unmutated IGHV MS and 17p deletion). The refined prognostic information provided by the index may have potential future application in identifying patients whose projected survival merits alternative or more aggressive treatment approaches (eg, allogeneic stem cell transplantation), identifying early-stage patients who are candidates for trials evaluating the benefits of early intervention/treatment, and/or establishing risk-stratified treatment approaches with new emerging therapies. Relevant clinical phase 3 trials using this prognostic score for risk stratification of clinically early-stage CLL patients are currently being conducted. These and other consecutive trials based on the prognostic index will probably lead to a refined and individualized treatment algorithm in CLL.
We used a well-characterized and prospectively followed population of untreated CLL patients as a training data set to construct a weighted, multivariable prognostic index that includes clinical, biological, and molecular markers and defines 4 different risk groups with significantly different OS rates. These 4 risk groups were reproduced within each Rai/Binet stage and within the subset of patients with unmutated IGHV status or with del(17p), demonstrating the gain of information over the conventional clinical staging systems. The C-statistic of the model was c = 0.75, exceeding the threshold level of 0.70 and signifying prognostic utility at the individual patient level.40
The utility of the prognostic index was also confirmed in an independent validation cohort. Although slight differences in projected survival rates were observed between training and validation data sets, these differences are likely caused by a shorter observation time and high proportion of censored data (87.4%) in the validation cohort. However the C-statistic of the model in the validation cohort was c = 0.83 and analyses of PFS and TFS—which can be seen as disease-specific outcomes and surrogate markers for OS—robustly confirmed the validity and potential of the prognostic index in both cohorts.
We further identified a “very-high-risk” group among CLL patients with an OS after 5 years of only 13.6% to 18.7%. This very-high-risk group comprises only 4% of CLL patients. Although all patients in this risk group are 17p-deleted, it should be emphasized that not all patients with 17p deletion are in this category. Specifically, patients with deletion 17p can be stratified into 2 risk groups (“high risk” or “very high risk”) with very different OS as illustrated in Figures 4B and 7B (P = .001 and P = .04). This finding once again demonstrates the discriminatory power of our index, even in patients traditionally considered as high risk.
Recurring gene mutations affecting ∼10% to 15% of CLL patients such as NOTCH1 and SF3B1 were recently identified by new-generation sequencing.42 Although controversial in the currently reported literature, these markers may have prognostic value.43-48 Prospective clinical trials evaluating the significance of those markers for OS and the additional information in combination with clinical, biological, and genetic markers in CLL are further needed. Novel prognostic markers will continue to be discovered, and accurate risk stratification needs to be an evolving process. The intent of the model presented here is to determine what existing clinical markers have independent value and to consolidate the prognostic value of these markers into a single risk score. As with the historical staging systems that combined clinical and laboratory data, such a platform facilitates evaluation of newly discovered markers—regardless of whether they offer incremental improvement over current knowledge. Of note, prognostication with more traditional markers according to the classification proposed here seems to separate different risk groups more accurately than risk classification based on genetic characteristics exclusively.46 Although 6 of the 8 factors in the comprehensive prognostic model are widely available, IGHV MS and s-TK are not routine clinical assays at many centers. Therefore we evaluated whether we could eliminate these factors from the index or whether a different model could be developed if IGHV MS and/or s-TK were not included in the initial 23 factors considered. In all cases the prognostic value of the index was reduced or lost altogether (data not shown), indicating that the risk measured by s-TK and IGHV MS are distinct from the other parameters. Nonetheless, IGHV analysis is already a routine assay at many clinical sites in both Europe and the US. Similarly, s-TK is also widely available as a routine clinical assay in some European countries and is in the process of being evaluated in American research laboratories as well.26 It is therefore evident that clinical assays assessing these variables are both feasible and indeed already available. Therefore the manuscript helps to eliminate unnecessary tests that do not provide incremental value. Now that the markers with the greatest independent prognostic value are identified, enhanced emphasis can be placed on making these markers more widely available in routine practice rather than developing clinical assays with lower relative value (eg, ZAP-70/CD3849-51 ).
Although attempts to create prognostic models for CLL have been made previously, none of these models incorporated this broad spectrum of markers or derived a risk score from a large, prospectively followed patient cohort. Recently, a multivariable model for OS was developed,29 but without the most robust prognostic factors in CLL (eg, genomic aberrations, IGHV MS). This model reached a C-statistic of c = 0.61 only in our data set below the value needed for clinical utility or relevance to an individual patient.40 Other models using a full array of genetic characteristics (eg, fluorescence in situ hybridization testing in combination with sequencing of TP53, NOTCH1, SF3B1, and BIRC3) have generated C-statistics of c = 0.642 only.46 Our analysis suggests that harnessing the full array of clinical, serum, and molecular characteristics optimizes the accuracy of OS prediction and that, for the first time, the comprehensive index presented here classifies risk accurately enough to be considered potentially useful for the individual patient (c = 0.75-0.83).40
We are aware of some limitations of our analysis. Although our training and validation data set included ∼300 patients >70 years of age, the median age of 60 years in the training set is rather young compared with the reported median age at diagnosis of 72 years. Elderly patients are generally underrepresented in clinical trials such as those from which the index was derived.52 Although the clinical validation cohort from the Mayo Clinic was also somewhat younger than CLL patients on average, the prognostic index was found to reliably predict outcome of CLL patients from around the world, including newly diagnosed patient cohorts. Nonetheless, we recognize that further validation in an extended data set including a larger sample of older, unfit patients is warranted. Further, the fact that choice of therapy was not an independent prognostic factor for OS in the analysis should be interpreted cautiously because our analysis was not designed to extensively investigate the role of different treatment modalities.
In conclusion, we report the development and validation of a novel prognostic index for CLL patients that identifies clinical, serum, and molecular markers with independent prognostic value and combines them into a single risk score. To our knowledge, the index presented here is the first comprehensive prognostic model to simultaneously incorporate a broad spectrum of prognostic markers into a single prognostic index and to reach the C-statistic threshold (c >0.70) necessary to have utility at the individual patient level. The index appears broadly applicable, dramatically improves the accuracy of prognostication over classical CLL clinical staging systems, and holds the potential for the development of more individualized treatment strategies. Clinical trials translating the information gained through the application of the prognostic index into new refined treatment algorithms for CLL patients are currently conducted.
Presented in part at the 14th International Workshop on CLL (Houston, TX, 2011), the 53rd Annual Meeting of the American Society of Hematology (San Diego, CA, 2011), the 119th congress of the German Society of Internal Medicine (Wiesbaden, Hessen, Germany, 2013), the 2013 Annual Meeting of the American Society of Clinical Oncology (Chicago, IL, 2013), and the 12th International Conference on Malignant Lymphoma (Palazzo dei Congressi, Lugano, Switzerland, 2013).
There is an Inside Blood Commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We especially wish to thank the patients and their treating physicians who participated in the trials.
This manuscript was written on behalf of the German CLL Study Group. Studies CLL1, CLL4, and CLL8 were planned and conducted as investigator-initiated trials by the German CLL Study Group and were supported by research grants from German Cancer Aid, Medac Schering Onkologie, and F. Hoffmann-La Roche. T.D.S. is a clinical scholar of the Leukemia Lymphoma Society.
Authorship
Contribution: N.P., J.B., T.E., K.B., and M.H. conceived and designed the study; S.S., H.D., and G.M. performed central laboratory tests; T.E., T.S., K.G.R., S.S., H.D., U.J., M.J.E., G.H., R.B., A.-M.F., C.-M.W., K.F., N.E.K., and M.H. collected and assembled the data; T.S., B.F.E., M.A.B., T.E., S.S., H.D., U.J., M.J.E., G.H., R.B., A.-M.F., C.-M.W., K.F., N.E.K., and M.H. provided the study materials and/or the patients; N.P., J.B., T.S., T.E., K,B., K.G.R., K.F., N.E.K., and M.H. analyzed and interpreted data; and all authors contributed to the writing of and gave final approval for the manuscript.
Conflict-of-interest disclosure: N.P. received Travel Grants from Roche. T.D.S. received research grants from Genentech, Celgene, Glaxo-Smith-Kline, Cephalon, Hospira, and Polyphenon E International. B.E. is a consultant and/or holds an advisory role for Celgene and Pharmacyclics and has received honoraria and research funding from Roche and Mundipharma. S.S. is a consultant and/or holds an advisory role for Roche and Mundipharma, and received honoraria and research funding from both. H.D. received research grants from Roche. U.J. received honoraria and research funding from Roche. M.H. is a consultant and/or holds an advisory role and received research funding from Roche. J.B., M.A.B., T.E., K.B., G.M., K.G.R., M.J.E., G.H., R.B., A.-M.F., C.-M.W., K.F., and N.E.K. declare no competing financial interests.
Correspondence: Michael Hallek, Department I of Internal Medicine and Center of Integrated Oncology Cologne Bonn, University of Cologne, Kerpener Str. 62, 50937 Köln, Germany; e-mail: michael.hallek@uk-koeln.de.
References
Author notes
N.P. and J.B. contributed equally to this study.