Key Points
Biallelic mutations in CSF3R must be considered as a novel genetic subtype in patients with congenital neutropenia.
The p.Arg308Cys mutation in CSF3R leads to altered G-CSF receptor glycosylation and surface expression and abrogated downstream signaling.
Abstract
Severe congenital neutropenia (SCN) is characterized by low numbers of peripheral neutrophil granulocytes and a predisposition to life-threatening bacterial infections. We describe a novel genetic SCN type in 2 unrelated families associated with recessively inherited loss-of-function mutations in CSF3R, encoding the granulocyte colony-stimulating factor (G-CSF) receptor. Family A, with 3 affected children, carried a homozygous missense mutation (NM_000760.3:c.922C>T, NP_000751.1:p.Arg308Cys), which resulted in perturbed N-glycosylation and aberrant localization to the cell surface. Family B, with 1 affected infant, carried compound heterozygous deletions provoking frameshifts and premature stop codons (NM_000760.3:c.948_963del, NP_000751.1:p.Gly316fsTer322 and NM_000760.3:c.1245del, NP_000751.1:p.Gly415fsTer432). Despite peripheral SCN, all patients had morphologic evidence of full myeloid cell maturation in bone marrow. None of the patients responded to treatment with recombinant human G-CSF. Our study highlights the genetic and morphologic SCN variability and provides evidence both for functional importance and redundancy of G-CSF receptor-mediated signaling in human granulopoiesis.
Introduction
Severe congenital neutropenia (SCN) is a heterogeneous disease, characterized by an absolute neutrophil count (ANC) <500 cells/µL and recurrent, life-threatening bacterial infections. Autosomal-dominant, autosomal-recessive, X-linked, and sporadic forms have been described, with mutations in several genes, including ELANE,1 GFI1,2 HAX1,3 G6PC3,4 VPS45,5 and WAS.6 The genetic cause in many SCN patients remains unidentified.7 The mainstay of treatment is recombinant human granulocyte colony-stimulating factor (rhG-CSF), which improves quality of life, increases the ANC in most patients, and decreases their infection frequency.8 Despite this, rhG-CSF–treated patients endure 8% and 21% cumulative incidences of sepsis and leukemic conversion, respectively, after 10 years of treatment.9
G-CSF induces proliferation, differentiation, and survival of myeloid progenitors. It signals through the cell-surface receptor G-CSF receptor (G-CSFR), a member of the type I cytokine receptor superfamily, encoded by CSF3R.10 The extracellular region of G-CSFR contains an immunoglobulin-like domain, a cytokine receptor homology (CRH) domain, and 3 fibronectin domains. The extracellular CRH domain includes highly conserved cysteine residues and the characteristic WSXWS motif of type I cytokine receptors. The cytoplasmic region has no intrinsic kinase activity, but relies on Janus kinase activation for signal transduction through signal transducer and activator of transcription 3 and 5 (STAT3 and STAT5), as well as the Ras–mitogen-activated protein kinase and phosphatidylinositol 3-kinase–Akt pathways.10,11
CSF3R mutations were originally thought to be a cause of SCN.12 However, subsequent studies13,14 showed that CSF3R mutations found in previously reported SCN patients were somatic or de novo in meiosis rather than inherited from the parents of affected individuals. These noninherited, heterozygous CSF3R mutations typically affect the cytoplasmic region of G-CSFR, lead to a truncated variant, and are associated with a hyperproliferative phenotype in mutated cells and the leukemogenic events seen with SCN.15,16
Rarely, somatic or de novo heterozygous CSF3R mutations in the extracellular region of the G-CSFR have been described.17-20 These mutants are characterized by hyporesponsiveness to rhG-CSF and act in a dominant-negative fashion by interfering with proper function of the wild-type (WT) G-CSFR. Additionally, a heterozygous germline CSF3R mutation has been associated with neutrophilia.21
Here, we identify recessively inherited, loss-of-function CSF3R mutations in 4 affected children with SCN from 2 unrelated families, with morphologic evidence of full myeloid cell maturation in bone marrow, thus expanding the spectrum of monogenic disorders associated with SCN.
Materials and methods
Patient information and study approval
Patients were referred to the pediatric centers at Erciyes University (Kayseri), the Dr von Hauner Children’s Hospital at Ludwig Maximilian University (Munich), and the Maternal-Infant Hospital Vall d’Hebron (Barcelona).
Informed consent and assent were obtained according to current ethical and legal guidelines. Ethical review boards from involved institutions (Pediatric Centers at Erciyes University [Kayseri], the Dr von Hauner Children’s Hospital at Ludwig Maximilian University [Munich], and the Maternal-Infant Hospital Vall d’Hebron [Barcelona]) approved this study. This study was conducted in accordance with the Declaration of Helsinki.
Genetic analysis
DNA samples of the parents, I-1 and I-2, of family A were genotyped using the Affymetrix Genome-wide Human SNP array 6.0 (GEO Platform GPL6801) following the manufacturer’s instructions (Affymetrix). Genomic DNA from patient P2 and her parents was used to construct exome libraries using the SureSelect XT Human All Exon V4 + UTRs kit (Agilent Technologies). Barcoded libraries were sequenced using SOLiD 5500 XL next-generation sequencing platform (Life Technologies) to an average coverage depth of 80. Whole-exome sequencing (WES) identified between 110 000 and 120 000 candidate variants for each sample. Further protocol details and primer sequences used to amplify genomic DNA for Sanger sequencing are in the supplemental Methods (see supplemental Data available at the Blood Web site).
Structural analysis of WT G-CSFRWT and mutant G-CSFRArg308Cys proteins
Cell culture
HeLa cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM), supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% penicillin/streptomycin (Pen/Strep), and 2mM L-Glutamine at 37°C and 5% CO2. For starvation, DMEM containing 0.5% heat-inactivated FBS, 1% Pen/Strep, and 2mM L-Glutamine was used. Further information is given in the supplemental Methods.
Plasmid construction and transfection
Transient transfection of cells with G-CSFRWT–enhanced green fluorescent protein (eGFP) and G-CSFRArg308Cys-eGFP was performed using JetPEI transfection reagent (Polyplus-Transfection) according to the manufacturer’s protocol. pMMP-eGFP was used for controls. See the supplemental Methods for details.
Western blot studies
HeLa cell pellets were resuspended in ice-cold 1× cell lysis buffer (Cell Signaling) supplemented with 0.06% protease inhibitor cocktail (Sigma-Aldrich) and 0.005% phenylmethylsulfonyl fluoride (Sigma-Aldrich) and incubated on ice for 60 minutes. Cells were centrifuged at 15 000 rpm for 15 minutes at 4°C. Proteins in the supernatant were electrophoretically separated and blotted. Primary antibodies used included rabbit anti-G-CSFR (H-176) (Santa Cruz Biotechnology), rabbit anti-phosphoSTAT3 (Y705) (Cell Signaling), rabbit anti-phosphoSTAT5 (Y694), and mouse anti-STAT3 (BD). Secondary antibodies included horseradish peroxidase (HRP)-conjugated anti-rabbit (Cell Signaling) and anti-mouse (BD) immunoglobulin G (IgG). Actin was visualized using HRP-conjugated anti-β-actin by Santa Cruz Biotechnology. For glycosylation studies, protein lysate was divided into 3 aliquots for the following conditions: nontreated, EndoH-treated, and PNGase F-treated. All buffers and enzymes were from New England Biolabs (Ipswich). Protein lysate for the samples not subject to treatment and samples intended for EndoH treatment were first incubated with denaturation buffer (5% sodium dodecyl sulfate [SDS], 0.4 M dithiothreitol) for 10 minutes at 95°C. Samples intended for PNGase F treatment were first incubated with 2.5% SDS for 10 minutes at 95°C. Samples were then treated with EndoH and PNGase F according to the manufacturer’s protocol. All protein lysates were boiled at 95°C for 5 minutes with sample buffer prior to SDS–polyacrylamide gel electrophoresis (PAGE) loading.
Fluorescent microscopy for HeLa cells
HeLa cells were seeded onto glass coverslips, washed, and fixed with 3.7% paraformaldehyde for 20 minutes at room temperature. After washing 3 times with phosphate-buffered saline (PBS), cells were permeabilized by 5-minute incubation with 0.5% Nonidet P40 at room temperature. Cells were washed 3 times followed by incubation with 3% bovine serum albumin for 15 minutes at room temperature. Cells were incubated with mouse anti-Calnexin (1:100; BD) for 45 minutes at 37°C and then washed 3 times with PBS. Coverslips were then incubated with goat-anti-mouse Alexa Fluor 594 (1:150; Invitrogen) for 45 minutes at 37°C. Following 3 5-minute PBS washes, nuclei were stained with 4,6 diamidino-2-phenylindole (DAPI) for 1 minute at room temperature. Cells were washed again and mounted using fluorescent mounting medium (Dako).
For confocal imaging, we used a Fluoview FV1000 (Olympus) microscope with XC30 digital camera (Olympus), fitted with a 60× objective. FV10-ASW acquisition software and ImageJ software25 were used for analysis.
Flow cytometry for HeLa cells
Forty-eight hours after transfection, cells were trypsinized, pelleted, and resuspended in ice-cold fluorescence-activated cell sorting (FACS) buffer (2% FBS, 0.1% NaN3 in PBS). Cells were pelleted and resuspended in blocking FACS buffer (10% FBS, 0.1% NaN3 in PBS) for 30 minutes on ice. Cells were washed with FACS buffer and 1 × 106 cells were aliquoted into 5-mL round-bottom Falcons (BD). Cells were stained in 100 μL of FACS buffer with 0.2 μg of phycoerythrin (PE)-conjugated anti-G-CSFR (BD) for 30 minutes on ice in the dark. PE-conjugated IgG1, κ isotype control (BD) was also used. After 3 5-minute washes with FACS buffer, cells were acquired and analyzed using FACSCanto (BD) and FACSDiva software (BD). Data were analyzed with FlowJo software (FlowJo). Further details are given in the supplemental Methods.
G-CSF stimulation
To ensure equal transfection efficiency between each time point, cells were transfected in 10-cm dishes. After 24 hours, cells were trypsinized, counted, and equally reseeded in 35-mm dishes. After attachment, cells were starved for 12 hours. Starvation medium containing G-CSF (100 ng/µL; Dompé Biotec) was then added to cells. After 5, 15, and 30 minutes at 37°C, cells were washed with ice-cold PBS 3 times and harvested by scraping. Cells at time point zero received fresh starvation medium without G-CSF and were immediately washed and harvested. Protein lysate was prepared and analyzed by western blot.
Results
Clinical data
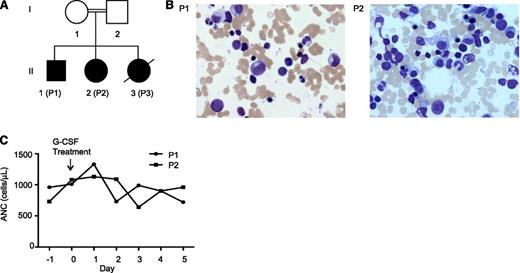
Family A consists of a healthy, consanguineous couple of Turkish descent with 3 SCN-affected children (Figure 1A). The index patient (P2) was diagnosed with SCN and dextrocardia at birth and required multiple hospitalizations for pneumonia, purulent otitis media, and fever of unknown origin. Her ANC to date has ranged between 420 and 2180 cells/µL. P1 was also diagnosed at age 2.5 years with SCN, but not dextrocardia. P3 was diagnosed at birth, but died at 3 months of age due to suspected aspiration pneumonia. Bone marrow examinations of P1 and P2 revealed full maturation of all 3 hematopoietic lineages without evidence of myelodysplasia (Figure 1B). P1 and P2 were treated with rhG-CSF (5 µg/kg per day), but did not show a response (Figure 1C). The father and mother of family A showed no signs of neutropenia, with ANCs of 5780 and 7080 cells/µL, respectively.
Clinical findings of family A. (A) Genealogic pedigree. (B) Giemsa-stained bone marrow smears of P1 and P2. (C) Response of patients P1 and P2 to rhG-CSF treatment.
Clinical findings of family A. (A) Genealogic pedigree. (B) Giemsa-stained bone marrow smears of P1 and P2. (C) Response of patients P1 and P2 to rhG-CSF treatment.
Index patient P4 of family B was a 9-month-old girl born to a healthy, nonconsanguineous couple of Spanish descent (supplemental Figure 1A). P4 was healthy until 2 months of age, when she was hospitalized for a urinary tract infection and treated with parenteral antibiotics. During this admission, P4 was found to be neutropenic (ANC to date: 200-1000 cells/µL). rhG-CSF was started at 5 µg/kg per day and progressively increased to a maximum dose of 40 µg/kg per day, with an ANC after this dose of 200 cells/µL (supplemental Figure 1B). Bone marrow aspirates revealed no maturation arrest and the presence of mature granulocytes. The father and mother of P4 had normal blood counts with ANCs of 3000 and 3900 cells/ µL, respectively. Supplemental Table 1 summarizes the clinical and genetic findings for both families.
Identification of CSF3R mutations
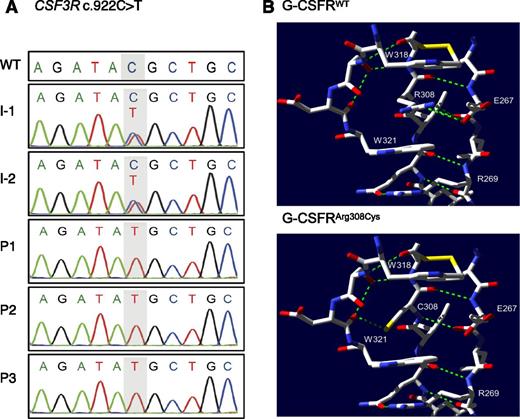
We performed homozygosity mapping and WES as described in the supplemental Methods for both parents and the index patient of family A. Homozygosity mapping identified 4 perfectly segregating runs of homozygosity (ROHs) with size above 1 Mb, which overlapped neither HAX1 nor G6PC3. These 4 regions map to chromosome 1 (33280648-40788961, build 37/hg19), chromosome 8 (19664010-2828292), chromosome 17 (13156053-14915602), and chromosome 18 (100949-2900803). WES detected 9 possible candidate variants, among which 2 variants were inside these ROHs (supplemental Results). We focused on a novel missense variant in CSF3R (NM_000760.3:c.922C>T, NP_000751.1:p.Arg308Cys; supplemental Figure 2B). Sanger sequencing of all family A members confirmed that the p.Arg308Cys CSF3R variant segregated with the disease phenotype in an autosomal-recessive inheritance pattern (Figure 2A). WES did not identify potential mutations in any SCN-causing genes (HAX1, G6PC3, ELANE, GFI1, VPS45, WAS) (supplemental Results, supplemental Table 2). Additionally, WES of P2 identified a homozygous variant in SPAG1 (NM_003114.4:c.1733del, NP_003105.2:p.578GlufsTer601), a causative gene for autosomal-recessive ciliary dyskinesia associated with dextrocardia, which was found in P2 (supplemental Table 3). Ciliary dyskinesia is associated with recurrent sinopulmonary infections.26 It is therefore possible that P2 suffers additionally from a ciliary disorder, which may contribute to her recurrent infections.
Genetic analysis of family A. (A) Sanger sequencing and intrafamilial segregation pattern of c.922C>T CSF3R mutation. (B) Top panel, Selected residues from the WT structure (PDB record 2D9Q) near Arg308. Arg308 participates in a ladder formation between 2 tryptophan residues W318 and W321. Green dashed lines represent hydrogen bonds. Yellow bars represent sulfur residues. Bottom panel, Putative structure of the complex with the Arg308Cys substitution, showing newly created hydrogen bond. Amino acid coordinates within G-CSFR are labeled with respect to the initial methionine of NP_000751.1.
Genetic analysis of family A. (A) Sanger sequencing and intrafamilial segregation pattern of c.922C>T CSF3R mutation. (B) Top panel, Selected residues from the WT structure (PDB record 2D9Q) near Arg308. Arg308 participates in a ladder formation between 2 tryptophan residues W318 and W321. Green dashed lines represent hydrogen bonds. Yellow bars represent sulfur residues. Bottom panel, Putative structure of the complex with the Arg308Cys substitution, showing newly created hydrogen bond. Amino acid coordinates within G-CSFR are labeled with respect to the initial methionine of NP_000751.1.
Sanger sequencing of ELANE, HAX1, G6PC3, GFI1, and SBDS in P4 did not detect mutations. Because of the patient’s hyporesponsiveness to rhG-CSF, CSF3R was sequenced. P4 showed a compound heterozygous mutation in CSF3R. In the first allele, a 16-bp deletion (NM_000760.3:c.948_963del) in exon 8 was found, whereas in the second allele, a 1-bp deletion (NM_000760.3:c.1245del) in exon 10 was found. Both deletions provoke frameshifts and premature stop codons (NP_000751.1:p.Gly316fsTer322 and NP_000751.1:p.Gly415fsTer432, respectively; supplemental Figure 2C). Sanger sequencing of CSF3R revealed that P4’s mother and father were heterozygous for the mutations in exons 8 and 10, respectively (supplemental Figure 1C).
Structural implications of G-CSFR mutations
The G-CSFR p.Arg308Cys substitution is close to the conserved WSXWS motif at positions 318 to 322. As we recently discovered a similar mutation (p.Arg201Leu) preceding the WSXWS motif (positions 214-218) in the type I cytokine receptor IL21R,24 we searched for similarly located disease-causing mutations in other type I cytokine receptors. Interestingly, we found that IL7R, IL11RA, LEPR, IL12RB1, GHR, and MPL also have disease-causing mutations of an arginine shortly preceding the WSXWS motif that act recessively to cause the phenotype (supplemental Results, supplemental Table 4). In most cases, including G-CSFR, the mutation replaces the positively charged arginine with a neutral amino acid.
Bioinformatic analyses using the SIFT27 and PolyPhen28 algorithms gave scores of 0.02 and 0.998, 1.0 (Polyphen gives 2 scores), respectively, predicting that the p.Arg308Cys variant may be deleterious. Of the 19 possible amino acid replacements at position 308, the change to cysteine was most strongly predicted by SIFT to be deleterious. The SIFT analysis differs from the receptor family evaluation (supplemental Table 4) in that SIFT specifically predicts whether each amino acid substitution would be deleterious or tolerated. Supplemental Table 4 shows that the Arg aligned to G-CSFR R308 can be mutated to any of Leu (IL21R), His (LEPR), and Pro (IL12RB1) and cause a disease, suggesting that replacements other than Arg308Cys at this position would also be deleterious.
Next, we predicted the influence of the p.Arg308Cys mutation on G-CSFR protein structure, using the crystal structure (2D9Q) of the G-CSFR/G-CSF complex.23 In the WT protein, the Arg308 residue participates in a Trp/Arg ladder with Trp318, Trp321, and Trp279 (residue not shown), which involves stabilization of the residues Trp318 and Trp321 by π-cation stacking with Arg308 (Figure 2B, top panel). Without Arg308, it is unlikely that the WT orientation of the W residues in the critical WSXWS segment also forms in the mutant structure.
More insight can be gained from a study of the prolactin receptor PRLR,29 which is significantly similar to G-CSFR in sequence (BLAST E-value 6E-16, supplemental Table 4). Dagil et al29 (Figure 3) found that PRLR in the bound state (PDB structure 3D4830 ) has a very different structure from unbound PRLR (PDB structure 2LFG29 ). For example, Trp191 in PRLR, which corresponds to Trp318 in G-CSFR, has a very different relative position in the 2 PRLR structures. Residue Arg183 in PRLR corresponds to Arg308 in G-CSFR. In the unbound state, the Trp/Arg ladder does not form among the relevant residues. Therefore, we suggest that Arg308 is essential for the ladder to form and for G-CSFR to bind to its ligand G-CSF, as shown in 2D9Q, but Arg308 may be unimportant for G-CSFR to fold into its inactive state. The distinction between active and inactive states would explain how our antibody immunoblot could detect the mutant G-CSFRArg308Cys in the inactive state, even if G-CSFRArg308Cys cannot fold properly in the active state to receive the G-CSF signal.
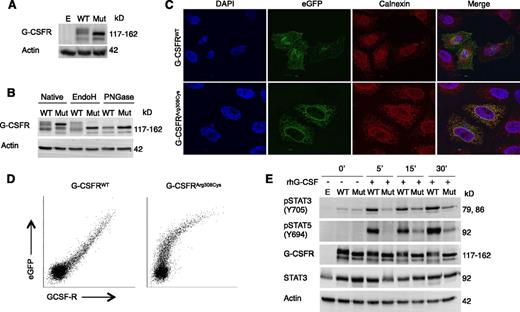
Glycosylation, subcellular localization, and functional analysis of mutant G-CSFRArg308Cys. (A) Western blot of G-CSFR in HeLa cells expressing either WT or mutant G-CSFRArg308Cys (Mut) fused to a C-terminal eGFP. (B) Western blot upon treatment with N-glycosidases EndoH and PNGase F. (C) Confocal microscopy visualizing G-CSFR (eGFP) and ER (Calnexin). Localization of WT-eGFP and mutant G-CSFRArg308Cys-eGFP was imaged with a ×63 objective and 2.1 and ×2.7 zoom, respectively. (D) Flow cytometric analysis of cell surface expression of WT G-CSFR and mutant G-CSFRArg308Cys C-terminal eGFP fusion proteins. Comparison of eGFP and G-CSFR–PE signals reveals a linear relationship in WT, but not in mutant G-CSFRArg308Cys. (E) Western blot of HeLa cells expressing WT-eGFP or Mut-eGFP after incubation with 100 ng/µL rhG-CSF. Empty pMMP vector (E) was used as a control (A,E).
Glycosylation, subcellular localization, and functional analysis of mutant G-CSFRArg308Cys. (A) Western blot of G-CSFR in HeLa cells expressing either WT or mutant G-CSFRArg308Cys (Mut) fused to a C-terminal eGFP. (B) Western blot upon treatment with N-glycosidases EndoH and PNGase F. (C) Confocal microscopy visualizing G-CSFR (eGFP) and ER (Calnexin). Localization of WT-eGFP and mutant G-CSFRArg308Cys-eGFP was imaged with a ×63 objective and 2.1 and ×2.7 zoom, respectively. (D) Flow cytometric analysis of cell surface expression of WT G-CSFR and mutant G-CSFRArg308Cys C-terminal eGFP fusion proteins. Comparison of eGFP and G-CSFR–PE signals reveals a linear relationship in WT, but not in mutant G-CSFRArg308Cys. (E) Western blot of HeLa cells expressing WT-eGFP or Mut-eGFP after incubation with 100 ng/µL rhG-CSF. Empty pMMP vector (E) was used as a control (A,E).
The lower panel of Figure 2B shows the mutant protein with the Arg308Cys substitution. Although no preexisting hydrogen bonds appear to be broken by this substitution, an additional hydrogen bond is created. However, the π-cation stacking that allows the conserved and experimentally verified Trp/Arg ladder to form does not rely on hydrogen bonds, and is abrogated by the substitution.
Aberrant N-glycosylation pattern of mutant G-CSFRArg308Cys
To further characterize the effect the p.Arg308Cys mutation on the G-CSFR protein, HeLa cells were transfected with plasmids encoding a fusion protein of either G-CSFRWT or G-CSFRArg308Cys fused to C-terminal eGFP. Western blot using an anti-G-CSFR antibody showed that the mutated G-CSFRArg308Cys had a decreased molecular weight in comparison with the WT (Figure 3A). This was also seen with eGFP-tagged human WT or mutant G-CSFRArg308Cys–transduced primary myeloid hematopoietic progenitor cells isolated from Csf3r+/− and Csf3r−/− mice; mutant G-CSFRArg308Cys had a clearly reduced molecular weight as compared with WT (supplemental Methods, supplemental Figure 3).
Protein extracts from lysed HeLa cells transfected with eGFP-tagged human WT or mutant G-CSFRArg308Cys were treated with the recombinant glycosidases EndoH (that cleaves mainly high mannose N-glycans) and PNGase F (that removes high mannose, hybrid and complex N-glycans). Subsequent western blotting revealed that G-CSFRWT showed partial insensitivity to EndoH treatment, whereas the mutant G-CSFRArg308Cys was fully sensitive (Figure 3B), suggesting that the N-glycosylation patterns were distinct. In contrast, PNGase F treatment did not reveal differences in cleavage of N-glycans between the WT and mutant proteins.
Mutant G-CSFR is retained in the ER and showed impaired expression on cell surface
In type I cytokine receptors both glycosylation31-33 and the WSXWS motif appear to be critically involved in the intracellular trafficking of receptors to the cell surface.24,34 To investigate the consequences of the p.Arg308Cys mutation in the intracellular trafficking of G-CSFR, HeLa cells transfected with the plasmids encoding the previously mentioned fusion proteins were seeded onto glass coverslips. Confocal microscopy of HeLa cells expressing human G-CSFRWT-eGFP showed a speckled cytosolic pattern and a circumferential distribution at the cell surface (Figure 3C). In contrast, the mutant G-CSFRArg308Cys-eGFP showed a reticular structure with expression around the nucleus, with no proper localization to the plasma membrane. Staining with anti-Calnexin showed colocalization with mutant G-CSFRArg308Cys-eGFP in the endoplasmic reticulum (ER), but not with the G-CSFRWT-eGFP protein.
To confirm this data in hematopoietic cells, we expressed the eGFP-tagged human G-CSFRWT and G-CSFRp.Arg308Cys in murine bone marrow BaF3 cells by plasmid transfection. In BaF3-cells, G-CSFRWT-eGFP showed speckled expression in the cytoplasm, presumably also at the cell surface, and colocalization with Golgi apparatus marker GM130 (supplemental Figure 4). However, the expression pattern of G-CSFRp.Arg308Cys was markedly different from WT and resembled more of the anti-Calnexin staining (supplemental Figure 5). Furthermore, we used murine primary myeloid Csf3r-knockout hematopoietic progenitor cells transduced by retroviral gene transfer. In murine primary hematopoietic progenitor cells, the human G-CSFRWT-eGFP was expressed in speckles all over the cytoplasm, presumably also at cell surface, and colocalized clearly to the Golgi apparatus, whereas the human G-CSFRp.Arg308Cys-eGFP mutant receptor showed clearly distinct localization pattern throughout the entire cytoplasm (supplemental Figure 6).
Furthermore, we stained nonpermeabilized HeLa cells expressing the above-mentioned eGFP-tagged constructs with a PE-conjugated anti-G-CSFR monoclonal antibody and analyzed with flow cytometry. Comparison of the eGFP signal (total G-CSFR expression) and PE signal (surface-bound G-CSFR) revealed a linear relationship for cells expressing the WT receptor, but not for the mutant, suggesting that the mutant G-CSFR lacks proper localization at the cell surface (Figure 3D).
The surface expression of G-CSFR (CD114) was studied in P4 of family B, who harbored both the p.Gly316fsTer322 and p.Gly415fsTer432 mutations. The whole peripheral blood samples of 1 healthy unrelated donor and P4 were stained with G-CSFR (CD114) and CD16 antibodies and detected by flow cytometry. Peripheral blood cells were gated for high CD16 expression and side scatter characteristics, and the expression level of G-CSFR (CD114) was compared between samples. The result showed markedly decreased surface expression of G-CSFR (CD114) in P4 in comparison with a healthy donor (supplemental Figure 7).
Abrogated signaling of mutant G-CSF receptor
We then aimed to characterize the functionality of the mutant G-CSFRArg308Cys. Upon binding of its cognate ligand G-CSF, the G-CSF/G-CSFR complex induces phosphorylation of STAT3 on Tyr705 and STAT5 on Tyr694.10 HeLa cells transiently transfected with the previously mentioned plasmids were serum starved overnight and stimulated with rhG-CSF as described in supplemental Methods. Cell-derived protein lysates were subjected to SDS-PAGE and western blotting to evaluate STAT3 (Tyr705) and STAT5 (Tyr694) phosphorylation. Cells expressing the mutant receptor showed reduced phosphorylation of STAT3 and STAT5, but signal transduction was not completely abrogated (Figure 3E).
Discussion
We herein describe a novel genetic subtype of SCN characterized by recessively inherited loss-of-function CSF3R mutations, full myeloid cell maturation in the bone marrow, and refractoriness to in vivo rhG-CSF treatment. This is in contrast to inherited SCN with mutations in ELANE or HAX1, which show a characteristic myeloid maturation arrest at the promyelocyte-to-myelocyte differentiation step and generally respond to rhG-CSF treatment. Although all patients in this study displayed peripheral neutropenia, full maturation of neutrophil granulocytes in the bone marrow was seen in all 3 patients who underwent a bone marrow examination (P1, P2, P4). G-CSF is a critical regulator of granulopoiesis, as shown in several human and murine studies.35-38 Consistent with our patients, both Csf3r– and G-CSF–deficient mice showed reduced numbers of neutrophils in peripheral blood.35,37 Moreover, no maturation arrest was seen in the bone marrow of the Csf3r– and G-CSF–deficient mice.32,34 Our results support the previous findings observed in genetically engineered mice and suggest the presence of G-CSFR–independent signaling pathways controlling granulopoiesis.35-37
Interestingly, although both patients from family A and B were refractory to in vivo rhG-CSF treatment, in vitro assays with the G-CSFRArg308Cys mutation from family A showed residual response to rhG-CSF stimulation. This may be explained by a small cell population able to express receptor on the surface (Figure 3D, supplemental Figure 7). Alternatively, the residual function seen in vitro may be due to heterotopic surface expression of the mutant G-CSFRArg308Cys in HeLa cells.
We also highlight the relevance of the residues interfacing with the WSXWS motif of G-CSFR for proper glycosylation and folding, as well as cell membrane expression. Mutations in or preceding the WSXWS motif in other type I cytokine receptors such as IL21R and the erythropoietin receptor (EpoR) are known to affect posttranslational glycosylation,24,34 similar to what was seen with the mutated GCSFRArg308Cys. Additionally, previous works on EpoR demonstrated that the WSXWS motif had a stabilizing role and was thus important for efficient receptor folding and surface expression.34 Similar to our findings in the mutated G-CSFRArg308Cys, we recently showed that the loss-of-function p.Arg201Leu IL21R mutation was associated with aberrant ER retention and disrupted surface expression.24 Although our results suggest that a defect in glycosylation leads to misrouting of the p.Arg308Cys, one cannot exclude other processes that may affect the protein conformation and thus localization within the cell. The substitution of Arg with Cys may alter the disulfide bridges and contribute to the abnormal protein distribution seen. The evidence that the Arg residue aligned to Arg308 can be mutated to any of Leu, His, or Pro in other type I cytokine receptors and cause disease (supplemental Table 4) suggests that the effect in G-CSFR is not Cys-specific.
The Arg201 of the IL21R corresponds to the Arg308 of the G-CSFR (supplemental Table 4), and this amino acid position appears to be evolutionarily well conserved. Without this Arg residue, it is unlikely that the WT orientation of the Trp residues in the critical WSXWS motif also forms in the mutant G-CSFR. We conclude that missense mutations at arginine residues shortly preceding the WSXWS motif in type I cytokine receptors are a recurrent deleterious phenomenon, but the precise mechanism may be specific to each receptor (supplemental Table 4). Given the observed ER localization of mutated G-CSFRArg308Cys, we postulate that the p.Arg308Cys mutation destabilizes the WSXWS motif, disrupts the proper folding of the active protein, and inhibits its transport through the ER and Golgi apparatus to the cell membrane.
Taken together, biallelic mutations in CSF3R must be considered as a novel genetic subtype in congenital neutropenia patients.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families for participating in the study and the medical staff for continuous support. The authors thank Prof Ian Wicks and Dr Emma Stuart (Inflammation Division, Walter and Eliza Hall Institute for Medical Research, Parkville, Australia) and Paulette van Strien (Erasmus Medical Center, Rotterdam, The Netherlands) for their kind help with experiments with Csf3r knockout mice. The authors thank Margarita Eustacchio, Barbara Sciskala, and Raffaele Conca for technical and administrative help and Ehsan Bahrami for assistance with cloning.
This work was supported by the European Research Council (ERC Advanced Grant-268608 Explore), the Bundministerium für Bildung und Forschung (E-RARE), the Care-for-Rare Foundation, the German Center for Infection Research (DZIF), and the Spanish Ministry of Health (FIS PS09/01182). This work was also supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM; ZIA LM000097-13).
Authorship
Contribution: A.T., P.M.J., D.M., D.K., N.K., T.R., J.P., D.P., E.M.G., A.A.S., and C.K. designed and performed experiments and analyzed data for family A; J.I.A., J.L.D.D., C.D.d.H.R., J.S.d.T.C., and J.Y. provided clinical care, designed and performed experiments, and analyzed data for family B; I.P.T. provided bones of Csf3r+/− and Csf3r−/− mice; E.U., M. A.O., T.P., and M.K. provided clinical care and performed experiments for family A; and E.M.G. and A.A.S. did bioinformatics analysis and the literature search for mutations in other type I cytokine receptors.
Conflict-of-interest disclosure: The authors declare no competing financial interests
Correspondence: Christoph Klein, Department of Pediatrics, Dr von Hauner Children’s Hospital, Lindwurmstrasse 4, D-80337 Munich, Germany; e-mail: christoph.klein@med.uni-muenchen.de.
References
Author notes
A.T. and P.M.J. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal