Key Points
In adult ALL, oncogenetic markers and minimal residual disease levels are independent outcome predictors.
Both factors should be used for individual treatment stratification.
Abstract
With intensified pediatric-like therapy and genetic disease dissection, the field of adult acute lymphoblastic leukemia (ALL) has evolved recently. In this new context, we aimed to reassess the value of conventional risk factors with regard to new genetic alterations and early response to therapy, as assessed by immunoglobulin/T-cell receptor minimal residual disease (MRD) levels. The study was performed in 423 younger adults with Philadelphia chromosome–negative ALL in first remission (265 B-cell precursor [BCP] and 158 T-cell ALL), with cumulative incidence of relapse (CIR) as the primary end point. In addition to conventional risk factors, the most frequent currently available genetic alterations were included in the analysis. A higher specific hazard of relapse was independently associated with postinduction MRD level ≥10−4 and unfavorable genetic characteristics (ie, MLL gene rearrangement or focal IKZF1 gene deletion in BCP-ALL and no NOTCH1/FBXW7 mutation and/or N/K-RAS mutation and/or PTEN gene alteration in T-cell ALL). These 2 factors allowed definition of a new risk classification that is strongly associated with higher CIR and shorter relapse-free and overall survival. These results indicate that genetic abnormalities are important predictors of outcome in adult ALL not fully recapitulated by early response to therapy. Patients included in this study were treated in the multicenter GRAALL-2003 and GRAALL-2005 trials. Both trials were registered at http://www.clinicaltrials.gov as #NCT00222027 and #NCT00327678, respectively.
Introduction
During the last decade, the management of adults with acute lymphoblastic leukemia (ALL) has markedly evolved. Up to the age of 40 to 60 years, most groups are now using pediatric-inspired approaches or even unmodified pediatric protocols.1-8 Relative to preceding studies, chemotherapy intensity has significantly increased and minimal residual disease (MRD) levels tend to be used to stratify postremission therapy.9,10 This evolution has yielded significant improvement in patient outcome, as reported in the first study of our Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL).11
This evolving context has made it necessary to reassess conventional risk factors in order to define patients who are at higher risk of relapse in current protocols. Early MRD evaluation needs to be included in this process,12 as should the most relevant novel genetic markers originally described in childhood ALL.13 Markers that are of potential prognostic value differ with ALL lineages. In B-cell precursor (BCP) ALL, deletion of the IKZF1 gene has been reported to be associated with a specific gene expression signature and a worse outcome, with a particularly high incidence in Philadelphia chromosome (Ph)-positive ALL.14-19 In T-cell ALL (T-ALL), we and others have reported that mutations of the NOTCH1 pathway are associated with a better prognosis,20,21 later refined by taking into account additional bad-prognosis N/K-RAS gene mutation or PTEN gene alteration.22
With the aim of deriving a modern risk model, we therefore reassessed the prognostic significance of conventional and new risk factors, including MRD and oncogenetics, in 423 adult patients with Ph-negative ALL treated in the pediatric-inspired GRAALL trials.
Methods
Treatments
Between 2003 and 2011, 955 patients with Ph-negative ALL aged 15 to 59 years were treated in the multicenter GRAALL-2003 and 2005 trials (618 BCP-ALL, 337 T-ALL). GRAALL centers and investigators are listed in the supplemental Appendix available at the Blood Web site. Results of the GRAALL-2003 trial (www.clinicaltrials.gov, #NCT00222027), which enrolled 225 patients, have been already reported.11 The GRAALL-2005 trial (www.clinicaltrials.gov, #NCT00327678) enrolled 730 evaluable patients between 2006 and 2011. Patient outcome was updated in January 2013. Overall, the median follow-up was 4.0 years. Treatment protocols are detailed in a supplemental file. Informed consent was obtained from all patients. Both trials were conducted in accordance with the Declaration of Helsinki and approved by local and multicenter research ethical committees. Of the 955 patients, 860 (548 BCP-ALL and 312 T-ALL) reached complete remission (CR) after the first induction cycle. Of these, 257 relapsed and 287 died, including 198 deaths after relapse. A total of 311 patients received allogeneic stem cell transplantation (SCT) in first CR (204 BCP-ALL and 107 T-ALL). Of these, 58 relapsed and 98 died, including 47 deaths after relapse. The patient flowchart and main characteristics are given in the supplemental Appendix (supplemental Figure 1 and supplemental Table 1).
In these GRAALL trials, the following factors were used to classify patients as standard-risk (no factors) or high-risk patients (at least 1 factor): (1) central nervous system (CNS) involvement at diagnosis, (2) low hypodiploidy/near triploidy on karyotype and/or DNA index analysis, (3) complex karyotype (defined as ≥5 unrelated chromosomal abnormalities),23 (4) early resistance to the 1-week steroid prephase, (5) poor bone marrow (BM) blast clearance after 1 additional week of chemotherapy, and (6) late CR achieved after the planned salvage course. Additionally, the following factors were also used to define high-risk BCP-ALL patients: (1) white blood cell (WBC) count ≥30 × 109/L, (2) MLL gene rearrangement (t[4;11] chromosomal translocation and/or MLL-AF4 gene fusion or other MLL rearrangement), (3) t(1;19) chromosomal translocation and/or E2A-PBX1 gene fusion, and (4) immature CD10-negative immunophenotype. In the GRAALL-2003 trial only, an MRD level ≥10−2 was also considered as a high-risk factor, but only 1 patient fell in this category solely for this reason. These factors are detailed in the supplemental Appendix. Per protocol, allogeneic SCT in first CR was offered to high-risk patients aged 55 years or younger if they had an HLA-identical sibling or 10/10 matched unrelated donor. The use of a 9/10 matched unrelated donor was allowed for patients with MLL-rearranged ALL, low-hypodiploidy/near-triploidy ALL, or late CR.
MRD evaluation and study population
Among the 860 patients who reached CR after the first induction cycle, 423 (49%) benefited from MRD1 evaluation, based on immunoglobulin/T-cell receptor (Ig/TCR) gene rearrangements and centrally assessed on BM samples just after the first induction course. The MRD1 time point was 6 weeks after induction initiation. These 423 patients (260 BCP-ALL and 163 T-ALL patients) represent the study population of this work. Other patients had a missing diagnosis and/or MRD1 samples (n = 307 patients), too-low blast percentage in the diagnosis sample (n = 25 patients), lack of informative Ig/TCR marker (n = 22 patients), or insufficient sensitivity by EuroMRD criteria (n = 83 patients). They were older and had a lower WBC count and slightly longer follow-up than the 423 study patients, but, importantly, no difference was observed between both subsets with respect to cumulative incidence of relapse (CIR) (supplemental Table 1). A total of 355 study patients (84%) also had an MRD2 evaluation, assessed after the first consolidation phase (12 weeks after induction initiation). Methods are detailed in the supplemental Appendix. Briefly, (1) DNA was extracted from diagnostic and follow-up BM samples and its quality was assessed and confirmed by albumin gene assay using standardized real-time quantitative polymerase chain reaction (PCR),24 (2) potential Ig/TCR targets were identified using the standardized multiplex PCR established within the BIOMED-2/EuroClonality network,25 and (3) for each patient, preferably 2 independent Ig/TCR targets with a sensitivity of at least 10−4 and a quantitative range of 10−4 for at least 1 of the 2 targets were selected for MRD level monitoring. All MRD data were assessed according to the guidelines developed within the EuroMRD group.24
New genetic markers evaluation
Among study patients, 216 out of 260 BCP-ALL patients (83%) were studied centrally for IKZF1 gene deletions using breakpoint-specific multiplex PCR, as described previously,26 and multiplex-ligation probe assay. Methods are detailed in the supplemental Appendix. A univariable analysis of the impact of IKZF1 gene deletions was also performed in a larger subset of 324 BCP-ALL patients in first CR with available IKZF1 gene status, even if not monitored for MRD levels. This analysis, which is provided in the supplemental Appendix, explains why only focal IKZF1 gene deletions were evaluated as a potential risk factor in the present study. Among T-ALL patients, 125 out of 163 (77%) could be classified centrally according to the 4-gene oncogenetic classifier we recently reported. We have indeed shown that a favorable genetic profile is defined by the presence of NOTCH1/FBXW7 mutation without N/K-RAS mutation or PTEN alteration, whereas high-risk profiles are defined by the absence of NOTCH1/FBXW7 mutation and/or the presence of N/K-RAS mutation and/or PTEN alteration.22 Methods are detailed in the supplemental Appendix. No difference in patient characteristics and outcome was detected between patients evaluated or not evaluated for these new genetics markers in BCP- and T-ALL separately.
Statistical methods
The primary end point was CIR after censoring patients who received allogeneic SCT in first CR at the time of SCT. CIR was estimated taking into account death in first CR as a competing risk. Cause-specific hazard ratios (HRs) with 95% confidence intervals (95% CIs) were given as measures of association between each variable and CIR. In BCP-ALL patients, variables that entered the prognostic analysis were as follows: WBC ≥30 × 109/L, CNS involvement, CD10-negative immature phenotype, t(4;11) translocation or MLL/AF4 or other MLL gene rearrangement, t(1;19) translocation or E2A-PBX1 rearrangement, low hypodiploidy/near triploidy, complex karyotype, focal IKZF1 gene deletion, and MRD1 level ≥10−4. In T-ALL patients, variables that entered the prognostic analysis were as follows: WBC ≥100 × 109/L, pro-T/mature-T phenotype, CNS involvement, complex karyotype, TLX1 gene overexpression defined as TLX1 over ABL expression ratio >1 as described previously,27 high-risk genetic profile as defined above, and MRD1 level ≥10−4. The analysis was then repeated without censoring patients who received allogeneic SCT in first CR at SCT date. Variables associated with P < .15 in univariable analysis, either when using SCT censoring or not, entered the multivariable analysis performed by the Cox models.28 Proportional-hazards assumptions were checked by testing that the log hazard-ratio functions were constant over time. Absence of unacceptable collinearity was checked by calculating the variance inflation factors for each covariable,29 considering a value of 4 as the maximum acceptable level of variance inflation factors. Secondary end points included relapse-free survival (RFS) and overall survival (OS) from CR. RFS and OS from CR were estimated by the Kaplan-Meier method30 and compared by the log-rank test.31 Statistical analyses were performed using the Stata/IC 12.1 software (StataCorp, College Station, TX). All tests were 2 sided, and P < .05 was considered statistically significant.
Results
Patients
The main characteristics of the 423 study patients are given in Table 1. A total of 260 patients had BCP-ALL, and 163 had T-ALL. Their median age was 31.2 years. Overall, 273 of them (65%) could be classified at high risk according to conventional protocol criteria. A total of 158 patients (107 BCP-ALL and 51 T-ALL) actually received allogeneic SCT in first CR. Among them, 35 relapsed and 54 died, including 25 deaths in first CR. Among the remaining 265 patients, 92 relapsed and 78 died, including 8 deaths in first CR.
Study patient characteristics
| . | All patients (n = 423) . | BCP-ALL patients (n = 260) . | T-ALL patients (n = 163) . |
|---|---|---|---|
| Patient-related characteristics | |||
| Trial, 2003/2005 | 103/320 | 66/194 | 37/126 |
| Median age, years (range) | 31.2 (15.2-59.9) | 34.5 (15.2-59.9) | 29.6 (16.3-57.0) |
| Gender, M/F | 264/159 | 147/113 | 117/46 |
| Disease-related characteristics | |||
| Median WBC, 109/L (range) | 15.4 (0.4-456) | 9.3 (0.4-396) | 32.1 (1.4-456) |
| CNS involvement,* Y/N/unknown | 33/386/4 | 15/242/3 | 18/144/1 |
| Complex karyotype,* Y/N/failure/unknown | 32/322/65/4 | 18/200/40/2 | 14/122/25/2 |
| Low hypodiploidy/near triploidy,* Y/N/unknown | 11/363/49 | 11/219/30 | 0/144/19 |
| WBC ≥30 × 109/L (BCP-ALL),* Y/N | — | 67/193 | — |
| CD10-negative immature ALL,* Y/N/unknown | — | 67/183/10 | — |
| MLL gene rearrangement (t[4;11] or other),* Y/N/unknown | — | 29/224/7 | — |
| t(1;19),* Y/N/unknown | — | 14/230/16 | — |
| IKZF1 gene deletion, Y/N/unknown | — | 54/162/44 | — |
| WBC ≥100 × 109/L (T-ALL), Y/N | — | — | 37/126 |
| Pro-T/mature-T ALL,† Y/N/unknown | — | — | 22/129/12 |
| TLX1 overexpression, Y/N/unknown | — | — | 29/98/36 |
| NOTCH1/FBXW7 gene mutation, Y/N/unknown | — | — | 90/46/27 |
| High-risk NOTCH1/FBXW7/RAS/PTEN genetics, Y/N/unknown | — | — | 60/65/38 |
| Response-related characteristics | |||
| Resistance to steroid prephase,* Y/N/unknown | 102/320/1 | 42/218/0 | 60/102/1 |
| Poor early BM blast clearance,* Y/N/unknown | 169/245/9 | 107/146/7 | 62/99/2 |
| MRD1 level ≥10−4, Y/N (%) | 158/265 (37.4%) | 111/149 (42.7%) | 47/116 (28.8%) |
| . | All patients (n = 423) . | BCP-ALL patients (n = 260) . | T-ALL patients (n = 163) . |
|---|---|---|---|
| Patient-related characteristics | |||
| Trial, 2003/2005 | 103/320 | 66/194 | 37/126 |
| Median age, years (range) | 31.2 (15.2-59.9) | 34.5 (15.2-59.9) | 29.6 (16.3-57.0) |
| Gender, M/F | 264/159 | 147/113 | 117/46 |
| Disease-related characteristics | |||
| Median WBC, 109/L (range) | 15.4 (0.4-456) | 9.3 (0.4-396) | 32.1 (1.4-456) |
| CNS involvement,* Y/N/unknown | 33/386/4 | 15/242/3 | 18/144/1 |
| Complex karyotype,* Y/N/failure/unknown | 32/322/65/4 | 18/200/40/2 | 14/122/25/2 |
| Low hypodiploidy/near triploidy,* Y/N/unknown | 11/363/49 | 11/219/30 | 0/144/19 |
| WBC ≥30 × 109/L (BCP-ALL),* Y/N | — | 67/193 | — |
| CD10-negative immature ALL,* Y/N/unknown | — | 67/183/10 | — |
| MLL gene rearrangement (t[4;11] or other),* Y/N/unknown | — | 29/224/7 | — |
| t(1;19),* Y/N/unknown | — | 14/230/16 | — |
| IKZF1 gene deletion, Y/N/unknown | — | 54/162/44 | — |
| WBC ≥100 × 109/L (T-ALL), Y/N | — | — | 37/126 |
| Pro-T/mature-T ALL,† Y/N/unknown | — | — | 22/129/12 |
| TLX1 overexpression, Y/N/unknown | — | — | 29/98/36 |
| NOTCH1/FBXW7 gene mutation, Y/N/unknown | — | — | 90/46/27 |
| High-risk NOTCH1/FBXW7/RAS/PTEN genetics, Y/N/unknown | — | — | 60/65/38 |
| Response-related characteristics | |||
| Resistance to steroid prephase,* Y/N/unknown | 102/320/1 | 42/218/0 | 60/102/1 |
| Poor early BM blast clearance,* Y/N/unknown | 169/245/9 | 107/146/7 | 62/99/2 |
| MRD1 level ≥10−4, Y/N (%) | 158/265 (37.4%) | 111/149 (42.7%) | 47/116 (28.8%) |
Y/N: yes/no.
Risk factor used in GRAALL trials.
According to the European Group for the Immunological Classification of Leukemias.
Early response evaluation
At the postinduction MRD1 time point, the numbers of patients with no detectable MRD1, detectable MRD1 level <10−4, and MRD1 level ≥10−4 were 196 (46.3%), 69 (16.3%), and 158 (37.4%), respectively. The proportion of patients with an MRD1 level ≥10−4 was 42.7% in BCP-ALL (111/260) and 28.8% in T-ALL (47/163). Figure 1A illustrates CIR according to MRD1 level. At 5 years, CIR was estimated at 22.9% (95% CI, 17-31) vs 30.8% (95% CI, 18-49) in patients with negative MRD1 or MRD1 <10−4 (P = .24), whereas it was 60.4% (95% CI, 48-73) in those with MRD1 ≥10−4. The 10−4 MRD1 cutoff was thus retained for prognostic analysis. With regard to the primary CIR end point, the cause-specific HR was 3.20 (95% CI, 2.11-4.84) for patients with an MRD1 level ≥10−4 (P < .001). It was 3.46 (95% CI, 2.00-6.00; P < .001) in BCP-ALL patients and 2.93 (95% CI, 1.50-5.71; P = .002) in T-ALL patients. Similar results were obtained when transplanted patients were not censored at SCT time (data not shown).
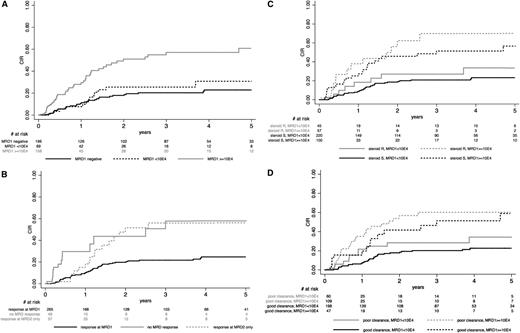
CIR according to early response. CIR, after censoring patients who received allogeneic SCT in first CR at time of SCT, is shown according to (A) postinduction MRD1 level (evaluated at week 6 after initiation of the first induction cycle); (B) postinduction MRD1 and postconsolidation MRD2 (evaluated at week 12 after initiation of the first induction cycle) levels, using a 10−4 MRD cutoff at both time points; (C) resistance or sensitivity to the steroid prephase and MRD1 level, showing that MRD1 response may discriminate high- vs good-risk patients in prephase-resistant (P = .016) as well as sensitive (P < .001) patients (conversely, resistance or sensitivity to the steroid prophase did not significantly define high- vs good-risk patients among those with low or high MRD1 level; P = .30 and .55, respectively); and (D) early BM blast clearance and MRD1 level, showing that MRD1 response may discriminate high- vs good-risk patients both in patients with poor (P = .031) or good (P < .001) early BM blast clearance (conversely, early BM blast clearance did not significantly define high- vs good-risk patients among patients with low or high MRD1 level; P = .06 and .67, respectively).
CIR according to early response. CIR, after censoring patients who received allogeneic SCT in first CR at time of SCT, is shown according to (A) postinduction MRD1 level (evaluated at week 6 after initiation of the first induction cycle); (B) postinduction MRD1 and postconsolidation MRD2 (evaluated at week 12 after initiation of the first induction cycle) levels, using a 10−4 MRD cutoff at both time points; (C) resistance or sensitivity to the steroid prephase and MRD1 level, showing that MRD1 response may discriminate high- vs good-risk patients in prephase-resistant (P = .016) as well as sensitive (P < .001) patients (conversely, resistance or sensitivity to the steroid prophase did not significantly define high- vs good-risk patients among those with low or high MRD1 level; P = .30 and .55, respectively); and (D) early BM blast clearance and MRD1 level, showing that MRD1 response may discriminate high- vs good-risk patients both in patients with poor (P = .031) or good (P < .001) early BM blast clearance (conversely, early BM blast clearance did not significantly define high- vs good-risk patients among patients with low or high MRD1 level; P = .06 and .67, respectively).
Postconsolidation MRD2 level evaluation was available for 355 patients. As expected, MRD2 and MRD1 levels strongly correlated in this cohort. Only 4 out of 249 patients with an MRD1 level <10−4 had an MRD2 level ≥10−4, whereas 81% of the patients with an MRD2 level <10−4 (245/302) had an MRD1 level <10−4 (P < .001). Overall, 265 patients achieved a MRD response <10−4 at MRD1, whereas 57 achieved it at MRD2 only and 49 did not achieve it at either time point. At 5 years, CIR was estimated at 24.7% (95% CI, 19-32) in patients who reached an MRD level <10−4 at MRD1, whereas it was 56.0% (95% CI, 39-75) and 57.8% (95% CI, 37-81) in those who reached this level at MRD2 only or never reached it, respectively (P = .14) (Figure 1B). Similar results were obtained when transplanted patients were not censored at SCT time (not shown).
With regard to the primary CIR end point, resistance to the steroid prephase and poor BM blast clearance were both associated with a higher specific hazard of relapse (cause-specific HR, 1.69 [1.05-2.71] and 2.20 [1.45-3.35]; P = .031 and < 0.001, respectively). For both criteria, HRs were relatively similar in BCP-ALL (1.87 and 2.15, respectively) and T-ALL (1.72 and 2.28, respectively) patients. Similar results were obtained when transplanted patients were not censored at SCT time (not shown). The MRD1 response was nonetheless a better predictor of relapse than earlier morphological response assessment. This is illustrated in Figure 1C-D. For both assessments, MRD1 response allowed to significantly discriminate high-risk vs good-risk patients in the 2 subsets of patients defined by their early morphologic response. Conversely, early morphologic assessment did not significantly define high-risk vs good-risk patients among good or poor MRD1 responders. After adjustment on resistance to the steroid prephase, only the MRD1 response remained significantly predictive of a higher CIR, either in the whole population (P < .001) or in BCP-ALL and T-ALL patients separately (P < .001 and P = .017, respectively). After adjustment on poor BM blast clearance, again only MRD1 response remained significantly predictive of a higher CIR, either in the whole population (P < .001) or in BCP-ALL and T-ALL patients separately (P < .001 and P = .044, respectively). This was also true when not using SCT censoring. For all these reasons, MRD1 response at the 10−4 level was the only response-related factor considered for further prognostic analysis.
Focal IKZF1 gene deletion in BCP-ALL patients
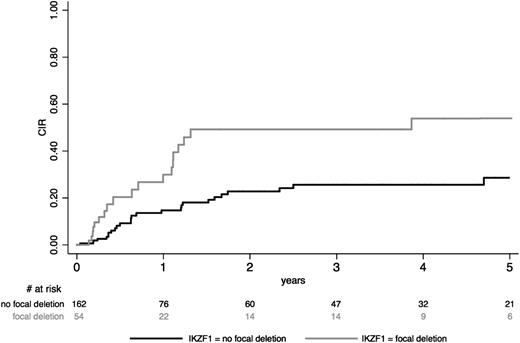
Among the 216 patients studied for IKZF1 gene status, a focal IKZF1 gene deletion was detected in 54 patients (25%) and a complete IKZF1 gene was detected in 15 patients, whereas the 147 remaining patients had no IKZF1 gene deletion. Deletion was monoallelic in the large majority of cases. Among the 54 patients with focal deletion, 28 (52%) had a deletion of exons 4 to 7 and 22 (41%) had a deletion of exons 2 to 7 or 4 to 8, whereas the 4 remaining patients had various other deletions that were undetectable by the multiplex PCR system and only identified by multiplex-ligation probe assay analysis. As explained in the supplemental Appendix, only focal IKZF1 deletions were evaluated as a potential risk factor. Actually, the outcome of patients presenting a complete deletion did not differ from that of patients without any deletion (supplemental Figure 2A). On the other hand, the type of focal deletion did not seem to significantly influence CIR (supplemental Figure 2B). At 5 years, CIR was estimated at 53.9% (95% CI, 38-72) in patients with focal IKZF1 gene deletion vs 28.6% (95% CI, 20-40) in other patients (Figure 2). The corresponding cause-specific HR was 2.65 (95% CI, 1.48-4.73; P = .001). Similar results were obtained when transplanted patients were not censored at SCT time.
CIR according to IKZF1 gene deletion in BCP-ALL study patients. CIR, after censoring patients who received allogeneic SCT in first CR at time of SCT, is shown in BCP-ALL patients according to the presence of focal IKZF1 gene deletion.
CIR according to IKZF1 gene deletion in BCP-ALL study patients. CIR, after censoring patients who received allogeneic SCT in first CR at time of SCT, is shown in BCP-ALL patients according to the presence of focal IKZF1 gene deletion.
NOTCH1/FBXW7/RAS/PTEN gene status in T-ALL patients
Among the 163 T-ALL study patients, 136 were studied for NOTCH1/FBXW7 mutation, and a mutation was found in 90 (66%). A total of 128 of these 136 patients were also tested for N-RAS and K-RAS mutation, which was found in 13 patients (including 4 patients without NOTCH1/FBXW7 mutation). A total of 117 of these 128 patients also were tested for PTEN genomic alteration, which was found in 12 patients (including 7 patients without NOTCH1/FBXW7 mutation). In these 117 patients, 22 (19%) had either N/K-RAS mutation or PTEN alteration, with no patient having both. Overall, 125 patients (77%) could be eventually classified according to our 4-gene (NOTCH1, FBXW7, N/K-RAS, PTEN) classification. Among them, 60 patients (48%) had a high-risk genetic profile, defined by the absence of NOTCH1/FBXW7 mutation and/or N/K-RAS mutation and/or PTEN alteration.22 At 5 years, CIR was 55.7% (39-73) in patients with high-risk genetic profile vs 15.4% (7-32) in other patients. The corresponding cause-specific HR was 5.33 (95% CI, 2.14-13.26; P < .001). Similar results were obtained when transplanted patients were not censored at SCT time.
Prognostic analysis
The prognostic value of conventional risk factors was then examined in this population of patients treated with a pediatric-inspired protocol, first in univariable analysis and then in multivariable analysis, against MRD1 response and the genetic markers mentioned above. Results are summarized in Table 2. Based on univariable analysis results, covariables that entered the multivariable analysis were as follows: (1) in BCP-ALL patients: WBC ≥30 × 109/L, MLL gene rearrangement, IKZF1 gene deletion, and MRD1 level ≥10−4; and (2) in T-ALL patients; WBC ≥100 × 109/L, pro-T/mature-T phenotype, CNS involvement, high-risk genetic profile, and MRD1 level ≥10−4. As shown in Table 2, the following factors were jointly selected as being associated with a worse outcome with regard to the primary CIR end point: (1) MLL gene rearrangement, IKZF1 gene deletion, and MRD1 level ≥10−4 in BCP-ALL patients; and (2) high-risk genetic profile and MRD1 level ≥10−4 in T-ALL patients. The incidence of BCP-ALL patients with an MRD1 level ≥10−4 was higher in the presence of focal IKZF1 gene deletion (65% vs 36%, P = .001), whereas it did not significantly increase in patients with MLL-rearranged ALL (48% vs 42%; P = .56). In T-ALL patients, there was a trend toward a higher incidence of patients with an MRD1 level ≥10−4 in patients with a high-risk genetic profile (37% vs 21.5%, P = .08).
Cause-specific hazards of relapse
| . | Patients, N/tested (SCT patients) . | SCT censoring . | No SCT censoring . | ||
|---|---|---|---|---|---|
| Cause-specific HR (95% CI) . | P value . | Cause-specific HR (95% CI) . | P value . | ||
| Univariable analysis | |||||
| BCP-ALL | |||||
| WBC ≥30 × 109/L* | 67/260 (39/107) | 1.85 (1.01-3.38) | .046 | 1.90 (1.19-3.02) | .007 |
| CNS involvement* | 15/257 (9/107) | 1.41 (0.44-4.56) | .56 | 0.87 (0.32-2.40) | .79 |
| CD10-negative immature ALL* | 67/250 (38/102) | 0.90 (0.45-1.79) | .76 | 0.75 (0.43-1.29) | .29 |
| MLL gene rearrangement (t[4;11] or other)* | 29/253 (18/105) | 2.11 (0.94-4.73) | .07 | 1.50 (0.79-2.85) | .22 |
| t(1;19)* | 14/244 (9/103) | 1.15 (0.28-4.76) | .85 | 1.04 (0.38-2.84) | .95 |
| Low hypodiploidy/near triploidy* | 11/230 (5/100) | 1.43 (0.44-4.60) | .55 | 1.23 (0.45-3.37) | .69 |
| Complex karyotype* | 18/258 (5/107) | 1.04 (0.75-1.44) | .83 | 1.01 (0.76-1.36) | .92 |
| IKZF1 gene deletion | 54/216 (20/88) | 2.65 (1.48-4.73) | .001 | 2.06 (1.25-3.39) | .004 |
| MRD1 level ≥10−4 | 111/260 (59/107) | 3.45 (2.00-6.00) | <.001 | 3.07 (1.92-4.90) | <.001 |
| T-ALL | |||||
| WBC ≥100 × 109/L | 37/163 (13/51) | 1.65 (0.82-3.33) | .16 | 1.77 (0.98-3.21) | .06 |
| CNS involvement* | 18/162 (14/51) | 2.83 (0.82-9.72) | .10 | 1.89 (0.89-4.04) | .10 |
| Pro-T/mature-T ALL | 22/151 (9/47) | 1.34 (0.52-3.47) | .55 | 1.93 (0.95-3.90) | .07 |
| Complex karyotype* | 14/161 (8/51) | 0.86 (0.54-1.35) | .51 | 0.82 (0.54-1.24) | .34 |
| TLX1 overexpression | 29/127 (5/43) | 0.92 (0.41-2.10) | .85 | 0.77 (0.35-1.67) | .51 |
| High-risk NOTCH1/FBXW7/RAS/PTEN genetics | 60/125 (20/39) | 5.33 (2.14-13.26) | <.001 | 4.70 (2.14-10.32) | <.001 |
| MRD1 level ≥10−4 | 47/163 (25/51) | 2.93 (1.50-5.71) | .002 | 2.50 (1.44-4.37) | .001 |
| Multivariable analysis | |||||
| BCP-ALL | |||||
| MRD1 level ≥10−4 | — | 3.21 (1.67-6.18) | <.001 | 2.49 (1.43-4.32) | .001 |
| IKZF1 gene deletion | — | 2.43 (1.29-4.60) | .006 | 1.75 (1.0-3.05) | .05 |
| MLL gene rearrangement (t[4;11] or other)* | — | 3.15 (1.13-8.80) | .028 | 1.73 (0.79-3.77) | .17 |
| WBC ≥30 × 109/L* | — | 1.01 (0.46-2.24) | .98 | 1.37 (0.76-2.47) | .30 |
| T-ALL | |||||
| High-risk NOTCH1/FBXW7/RAS/PTEN genetics | — | 5.59 (1.82-17.19) | .003 | 4.39 (1.75-11.03) | .002 |
| MRD1 level ≥10−4 | — | 2.50 (1.06-5.87) | .036 | 3.13 (1.51-6.50) | .002 |
| WBC ≥100 × 109/L | — | 1.34 (0.54-3.35) | .53 | 1.51 (0.70-3.26) | .29 |
| CNS involvement* | — | 2.49 (0.47-13.3) | .29 | 1.38 (0.51-3.74) | .53 |
| Pro-T/mature-T ALL | — | 1.01 (0.33-3.09) | .98 | 1.22 (0.53-2.80) | .63 |
| . | Patients, N/tested (SCT patients) . | SCT censoring . | No SCT censoring . | ||
|---|---|---|---|---|---|
| Cause-specific HR (95% CI) . | P value . | Cause-specific HR (95% CI) . | P value . | ||
| Univariable analysis | |||||
| BCP-ALL | |||||
| WBC ≥30 × 109/L* | 67/260 (39/107) | 1.85 (1.01-3.38) | .046 | 1.90 (1.19-3.02) | .007 |
| CNS involvement* | 15/257 (9/107) | 1.41 (0.44-4.56) | .56 | 0.87 (0.32-2.40) | .79 |
| CD10-negative immature ALL* | 67/250 (38/102) | 0.90 (0.45-1.79) | .76 | 0.75 (0.43-1.29) | .29 |
| MLL gene rearrangement (t[4;11] or other)* | 29/253 (18/105) | 2.11 (0.94-4.73) | .07 | 1.50 (0.79-2.85) | .22 |
| t(1;19)* | 14/244 (9/103) | 1.15 (0.28-4.76) | .85 | 1.04 (0.38-2.84) | .95 |
| Low hypodiploidy/near triploidy* | 11/230 (5/100) | 1.43 (0.44-4.60) | .55 | 1.23 (0.45-3.37) | .69 |
| Complex karyotype* | 18/258 (5/107) | 1.04 (0.75-1.44) | .83 | 1.01 (0.76-1.36) | .92 |
| IKZF1 gene deletion | 54/216 (20/88) | 2.65 (1.48-4.73) | .001 | 2.06 (1.25-3.39) | .004 |
| MRD1 level ≥10−4 | 111/260 (59/107) | 3.45 (2.00-6.00) | <.001 | 3.07 (1.92-4.90) | <.001 |
| T-ALL | |||||
| WBC ≥100 × 109/L | 37/163 (13/51) | 1.65 (0.82-3.33) | .16 | 1.77 (0.98-3.21) | .06 |
| CNS involvement* | 18/162 (14/51) | 2.83 (0.82-9.72) | .10 | 1.89 (0.89-4.04) | .10 |
| Pro-T/mature-T ALL | 22/151 (9/47) | 1.34 (0.52-3.47) | .55 | 1.93 (0.95-3.90) | .07 |
| Complex karyotype* | 14/161 (8/51) | 0.86 (0.54-1.35) | .51 | 0.82 (0.54-1.24) | .34 |
| TLX1 overexpression | 29/127 (5/43) | 0.92 (0.41-2.10) | .85 | 0.77 (0.35-1.67) | .51 |
| High-risk NOTCH1/FBXW7/RAS/PTEN genetics | 60/125 (20/39) | 5.33 (2.14-13.26) | <.001 | 4.70 (2.14-10.32) | <.001 |
| MRD1 level ≥10−4 | 47/163 (25/51) | 2.93 (1.50-5.71) | .002 | 2.50 (1.44-4.37) | .001 |
| Multivariable analysis | |||||
| BCP-ALL | |||||
| MRD1 level ≥10−4 | — | 3.21 (1.67-6.18) | <.001 | 2.49 (1.43-4.32) | .001 |
| IKZF1 gene deletion | — | 2.43 (1.29-4.60) | .006 | 1.75 (1.0-3.05) | .05 |
| MLL gene rearrangement (t[4;11] or other)* | — | 3.15 (1.13-8.80) | .028 | 1.73 (0.79-3.77) | .17 |
| WBC ≥30 × 109/L* | — | 1.01 (0.46-2.24) | .98 | 1.37 (0.76-2.47) | .30 |
| T-ALL | |||||
| High-risk NOTCH1/FBXW7/RAS/PTEN genetics | — | 5.59 (1.82-17.19) | .003 | 4.39 (1.75-11.03) | .002 |
| MRD1 level ≥10−4 | — | 2.50 (1.06-5.87) | .036 | 3.13 (1.51-6.50) | .002 |
| WBC ≥100 × 109/L | — | 1.34 (0.54-3.35) | .53 | 1.51 (0.70-3.26) | .29 |
| CNS involvement* | — | 2.49 (0.47-13.3) | .29 | 1.38 (0.51-3.74) | .53 |
| Pro-T/mature-T ALL | — | 1.01 (0.33-3.09) | .98 | 1.22 (0.53-2.80) | .63 |
The end point was CIR, after censoring patients who received allogeneic SCT in first CR or not, in BCP-ALL and T-ALL subsets separately.
Univariable and multivariable cause-specific HRs and P values are given;
Risk factors used in GRAALL trials.
New risk classification
Based on these results, high-risk patients could be defined as patients with MRD1 level ≥10−4 and/or unfavorable genetics, defined as (1) t(4;11) translocation or other MLL gene rearrangement and/or IKZF1 gene deletion in BCP-ALL and (2) no NOTCH1/FBXW7 mutation and/or N/K-RAS mutation and/or PTEN alteration in T-ALL patients. With this new definition, the overall percentage of high-risk patients is 59% in both ALL lineages. With regard to the primary CIR end point, the cause-specific HR was 4.38 (95% CI, 2.47-7.76) for high-risk vs standard-risk patients (P < .001). It was 3.89 (95% CI, 1.91-7.90; P < .001) in BCP-ALL patients and 5.31 (95% CI, 2.00-14.07; P = .001) in T-ALL patients. Without censoring at SCT time, the cause-specific HR was 3.78 (95% CI, 2.27-6.30) for high-risk vs standard-risk patients (P < .001). It was 3.17 (95% CI, 1.72-5.83; P < .001) in BCP-ALL patients and 5.33 (95% CI, 2.07-13.7; P = .001) in T-ALL patients.
Figure 3 illustrates CIR without SCT censoring according to the 4 patient subsets that can be defined by genetic characteristics and MRD response (ie, genetics−/MRD−, genetics−/MRD+, genetics+/MRD−, and genetics+/MRD+) in BCP-and T-ALL patients separately. As shown, MRD level seemed to be the predominant predictor in BCP-ALL patients, further refined by genetic features (Figure 3A). In T-ALL patients, conversely, the oncogenetic classification seemed to be the predominant predictor, further refined by MRD response (Figure 3B). In both lineages, patients with high-risk genetic characteristics and poor MRD1 response experienced a worse outcome. Finally, Figure 4 shows the impact of this new risk classification, based on oncogenetics and/or MRD1 level only, on RFS and OS from CR, in BCP-ALL and T-ALL patients separately and without SCT censoring.
CIR according to high-risk genetics and MRD response. CIR is shown for patients also investigated for relevant oncogenetic events, according to the presence of high-risk genetic characteristics (MLL gene rearrangement and/or focal IKZF1 gene deletion in BCP-ALL patients; high-risk NOTCH1/FBXW7/RAS/PTEN profile in T-ALL patients) and MRD1 level (using a 10−4 cutoff). (A) In BCP-ALL patients, MRD1 level discriminates high-risk patients in good- as well as high-risk genetic subgroups (HR, 2.69 [1.28-5.68] and 2.63 [1.23-5.63]; P = .009 and 0.013, respectively), whereas no significant difference in CR was observed in patients with a low MRD1 level, whatever their genetic characteristics (P = .18). (B) In T-ALL patients, a high-risk genetic profile discriminated high-risk patients, whatever the level of MRD1 (HR, 4.27 [1.52-12.01] and 4.04 [1.17-13.97]; P = .006 and 0.027, respectively), whereas no significant difference in CIR was observed in patients with a good-risk genetic profile, whatever their MRD1 level (P = .19).
CIR according to high-risk genetics and MRD response. CIR is shown for patients also investigated for relevant oncogenetic events, according to the presence of high-risk genetic characteristics (MLL gene rearrangement and/or focal IKZF1 gene deletion in BCP-ALL patients; high-risk NOTCH1/FBXW7/RAS/PTEN profile in T-ALL patients) and MRD1 level (using a 10−4 cutoff). (A) In BCP-ALL patients, MRD1 level discriminates high-risk patients in good- as well as high-risk genetic subgroups (HR, 2.69 [1.28-5.68] and 2.63 [1.23-5.63]; P = .009 and 0.013, respectively), whereas no significant difference in CR was observed in patients with a low MRD1 level, whatever their genetic characteristics (P = .18). (B) In T-ALL patients, a high-risk genetic profile discriminated high-risk patients, whatever the level of MRD1 (HR, 4.27 [1.52-12.01] and 4.04 [1.17-13.97]; P = .006 and 0.027, respectively), whereas no significant difference in CIR was observed in patients with a good-risk genetic profile, whatever their MRD1 level (P = .19).
RFS and OS from CR according to the new risk classification. RFS and OS from CR are shown for patients also investigated for relevant oncogenetic events (MLL gene rearrangement and IKZF1 gene deletion in BCP-ALL; NOTCH1/FBXW7/RAS/PTEN anomalies in T-ALL) according to the new risk classification. High-risk patients are defined here as those with high-risk oncogenetics and/or MRD1 response ≥10−4: (A) RFS in BCP-ALL patients (77 vs 44% at 5 years; HR, 2.84 [95% CI, 1.72-4.70]; P ≤ .001); (B) OS from CR in BCP-ALL patients (79 vs 50% at 5 years; HR, 2.78 [95% CI, 1.61-4.80]; P < .001); (C) RFS in T-ALL patients (86 vs 52% at 5 years; HR, 4.20 [95% CI, 1.85-9.51]; P = .001); (D) OS from CR in T-ALL patients (91 vs 62% at 5 years; HR, 4.14 [95% CI, 1.58-10.83]; P = .004).
RFS and OS from CR according to the new risk classification. RFS and OS from CR are shown for patients also investigated for relevant oncogenetic events (MLL gene rearrangement and IKZF1 gene deletion in BCP-ALL; NOTCH1/FBXW7/RAS/PTEN anomalies in T-ALL) according to the new risk classification. High-risk patients are defined here as those with high-risk oncogenetics and/or MRD1 response ≥10−4: (A) RFS in BCP-ALL patients (77 vs 44% at 5 years; HR, 2.84 [95% CI, 1.72-4.70]; P ≤ .001); (B) OS from CR in BCP-ALL patients (79 vs 50% at 5 years; HR, 2.78 [95% CI, 1.61-4.80]; P < .001); (C) RFS in T-ALL patients (86 vs 52% at 5 years; HR, 4.20 [95% CI, 1.85-9.51]; P = .001); (D) OS from CR in T-ALL patients (91 vs 62% at 5 years; HR, 4.14 [95% CI, 1.58-10.83]; P = .004).
Discussion
In the present study, we reassessed the value of conventional and new risk factors, including MRD response and newly described genetic markers, in adult patients with Ph-negative ALL in first CR. We confirm that, as in children, early MRD response is a powerful risk factor that should be used in adult patients treated in modern protocols including prospective treatment stratification on the individual risk. This has been already observed in 6 studies,32-37 published between 2000 and 2013 and including in total more than 1000 patients (102, 196, 116, 212, 161, and 580 patients, respectively). In these studies, MRD levels were evaluated at various early time points using either flow cytometry32,34 or Ig/TCR gene amplification in 4 studies.33,35-37 In all studies but one, MRD was identified as a strong predictor of outcome after adjustment on conventional risk factors in multivariable analysis. However, none of these studies included the more recently identified genetic ALL markers, such as IKZF1 gene deletions in BCP-ALL or NOTCH1 pathway gene mutations in T-ALL, as undertaken here.
Most groups still use conventional risk factors, like WBC count, immunophenotype and standard cytogenetics, in the definition of high-risk vs standard-risk patients.37,38 Although the treatment protocol may have an impact on prognostic factors, the present study provides strong evidence that, at least when using a pediatric-inspired protocol, most conventional risk factors could be safely abandoned in future trials that will rely on prospective MRD monitoring and up-to-date oncogenetic characterization. In fact, the single conventional factor that remained of significant value in our study was the presence of MLL rearrangement, including the recurrent t(4;11) chromosomal translocation.
The present study also confirms the prognosis impact of new ALL genetic subsets in a large multivariate setting including MRD. The number of subsets that have been characterized and evaluated here is still limited, with respect to the long list of genetic events that have been described in this disease.13 It nonetheless includes the most frequent events described to date as influencing patient outcome. Interestingly, even if these genetic anomalies may influence MRD response, they were independently associated with a higher relapse risk in both lineages, meaning that the MRD response does not totally recapitulate the intrinsic risk. One may regret the absence of the so-called BCR-ABL–like ALL subset in the list of markers evaluated here in BCP-ALL patients. This latter subset has been defined recently in pediatric cohorts on the basis of a gene expression signature highly similar to that of Ph-positive ALL associated to a poor outcome.14,39 Nearly half of so-defined BCR-ABL–like cases had IKZF1 deletions and a lower proportion had a high CRLF2 gene expression level. Recently, the Dutch pediatric group has reported that both the BCR-ABL–like signature and IKZF1 gene deletion had independent prognostic value, whereas high CRLF2 gene expression did not.40 The British-American adult Intergroup has also reported an inferior outcome associated with IKZF1 deletions in univariable analysis.41 For the time being, screening for IKZF1 deletions, which can be performed at the individual patient level by routine laboratory assays, appears nonetheless to be the best marker to be used, because prospective multicenter treatment stratification based on gene expression signature does not represent a simple approach.
The statistical independence of high-risk genetics and poor MRD response in predicting relapse underlines the remarkable heterogeneity of ALL, not to mention the fact that even in a given genetic subset, the leukemic clone could be heterogeneous.42-44 One may imagine why MRD may not entirely recapitulate the risk of relapse in this context. The number of BM cells that can be sampled and analyzed limits the detection of residual leukemic cells. Depending on the genetic event and the clonal hierarchy, good apparent treatment response with favorable MRD kinetics may still be associated with the persistence of very low levels of cells endowed with leukemic stem cell capacities that could initiate ALL recurrence in a given patient. Upon examination of the respective roles of genetics and MRD in both lineage subgroups, MRD seems to be the prominent factor in patients with BCP-ALL, whereas the NOTCH1/FBXW7/RAS/PTEN genetic profile seems to be more important in patients with T-ALL (Figure 3).
It thus appears that an accurate genetic characterization of the disease and a prospective evaluation of the BM MRD response may both be required to optimally define individual patient risk. Combination of both factors essentially allows identification of an important fraction of patients with a very good outcome when treated with a pediatric-inspired protocol (5-year OS from CR, approximately 80% and 90% in good-risk BCP-ALL and T-ALL patients, respectively; Figure 4). How they can be used for treatment stratification will depend on the therapeutic option proposed. This should be first elucidated for allogeneic SCT in first CR. By landmark analysis, the German group recently showed that poor MRD responders benefited from SCT in first CR, suggesting that good MRD responders did not.37 Whether this is true within each different ALL genetic subset remains an open issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families for participating in this study and the GRAALL investigators for submitting clinical data and samples; Elodie Boucher, Nathalie Klein, Cyril Melot, Mathieu Sauvezie, and Francis Daniel for their help in data monitoring; Emmanuelle Clappier, Stéphane de Botton, and Philippe Rousselot for reviewing the manuscript; and André Baruchel, Jean Soulier, Jean-Yves Cahn, and Gérard Socié for their longstanding intellectual support.
This work was supported by grants from the Le Programme Hospitalier de Recherche Clinique, French Ministry of Health, and Institut National du Cancer (no. 0200701) (F.H.) (no. AOM 04144 and AOM 08106) (H.D.) in France and from the Swiss State Secretariat for Education, Research and Innovations in Switzerland.
Authorship
Contribution: K.B., V.A., F.H., T.L., X.T., Y.C., N.B., E.D., P.C., A.B., O.R., J.-P.V., M.C.B., M.L., E.M., N.I., and H.D. conceived of and designed the study; V.L. provided administrative support; F.H., T.L., X.T., Y.C., N.B., P.C., A.B., O.R., J.-P.V., N.I., and H.D. provided study materials or patients; K.B., V.A., M.-L.B., J.-M.C., N.G., B.S., E.D., H.C., T.F., V.L., M.C.B., M.L., E.M., N.I., and H.D. collected and assembled data; K.B., S.C., V.A., N.I., and H.D. wrote the manuscript; and all authors were responsible for data analysis and interpretation and the final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Written on behalf of GRAALL, which includes the former France-Belgium Group for Lymphoblastic Acute Leukemia in Adults, the French Western-Eastern Group for Lymphoblastic Acute Leukemia, and the Swiss Group for Clinical Cancer Research. GRAALL participating centers and investigators are listed in the supplemental Appendix.
Correspondence: Hervé Dombret, Department of Hematology, Hôpital Saint-Louis, 1 Ave Claude Vellefaux, 75475 Paris Cedex 10, France; e-mail: herve.dombret@sls.aphp.fr.

![Figure 3. CIR according to high-risk genetics and MRD response. CIR is shown for patients also investigated for relevant oncogenetic events, according to the presence of high-risk genetic characteristics (MLL gene rearrangement and/or focal IKZF1 gene deletion in BCP-ALL patients; high-risk NOTCH1/FBXW7/RAS/PTEN profile in T-ALL patients) and MRD1 level (using a 10−4 cutoff). (A) In BCP-ALL patients, MRD1 level discriminates high-risk patients in good- as well as high-risk genetic subgroups (HR, 2.69 [1.28-5.68] and 2.63 [1.23-5.63]; P = .009 and 0.013, respectively), whereas no significant difference in CR was observed in patients with a low MRD1 level, whatever their genetic characteristics (P = .18). (B) In T-ALL patients, a high-risk genetic profile discriminated high-risk patients, whatever the level of MRD1 (HR, 4.27 [1.52-12.01] and 4.04 [1.17-13.97]; P = .006 and 0.027, respectively), whereas no significant difference in CIR was observed in patients with a good-risk genetic profile, whatever their MRD1 level (P = .19).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/24/10.1182_blood-2014-01-547695/4/m_3739f3.jpeg?Expires=1769091411&Signature=QzJ-siwx3L01d-O57rdGdgdDm4SaURQnrSnQXYPm8AUObFT4EudwU7UF2YN-NYra0Fk4NrZpULM2UwpT50fkgtpnAVoUGGuYtas40Yx9NPNSMohjxuCXHL7zFKPcsz88LnoxdUOQ7BcjtB4b9t79nDHgk3KfRTGKBCSFbqib1mzwzvJ4wvpFHNuJNYbrGJzI4EneAfY0TmInOTHPodpShtt-BkYOTp1ieX8YgjR~YwPLY4XcJXFfs9tcjHMJen53hfWsgtgOLABOjkKMXwVCZSs-XsuPfMSiCdQrdcImnoQoihxUM8-KMLmngiZtrlDLI8vmqddcTlyJMbQHKf~L9A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. RFS and OS from CR according to the new risk classification. RFS and OS from CR are shown for patients also investigated for relevant oncogenetic events (MLL gene rearrangement and IKZF1 gene deletion in BCP-ALL; NOTCH1/FBXW7/RAS/PTEN anomalies in T-ALL) according to the new risk classification. High-risk patients are defined here as those with high-risk oncogenetics and/or MRD1 response ≥10−4: (A) RFS in BCP-ALL patients (77 vs 44% at 5 years; HR, 2.84 [95% CI, 1.72-4.70]; P ≤ .001); (B) OS from CR in BCP-ALL patients (79 vs 50% at 5 years; HR, 2.78 [95% CI, 1.61-4.80]; P < .001); (C) RFS in T-ALL patients (86 vs 52% at 5 years; HR, 4.20 [95% CI, 1.85-9.51]; P = .001); (D) OS from CR in T-ALL patients (91 vs 62% at 5 years; HR, 4.14 [95% CI, 1.58-10.83]; P = .004).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/24/10.1182_blood-2014-01-547695/4/m_3739f4.jpeg?Expires=1769091411&Signature=e55X~2kUA1MTb8XCxU0n4RNsZsOpyJGpDWbS6V0kYg3FS6wAJpfZN36BfGTisChMXWBJxGk5ZKNSOPmITA2AEecTOS3FdsR0HhY6Ck1aB859GIvCvpaTokbA4hvaMQ62wFlpHgVNwmBV9h05W6GyM5URXHfmYYuTtEJIvuToxXMlpM9tBgeDKSKQ6uqwwmLgBxpfayeWjUtT9Ktq38zzZeWXQ6gln9WyGyyHEmrz2Tozq9Bmm8eV5yKIAC9otCMihvFHG65B3JklfM2QPYElbf~P4JsYv~IGTpDAInsLKbLJQech81fZZ-O7oVGj3MO4LUKicxwGsPnPXLrhYWE5WQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal