Key Points
Inherited genetic variation increases risk to developing multiple myeloma through predisposition to MGUS.
Loci identified that increase risk of developing MGUS include 2p23.3, 3p22.1, 3q26.2, 6p21.33, 7p15.3, 17p11.2, and 22q13.1.
Abstract
Monoclonal gammopathy of undetermined significance (MGUS) is present in ∼2% of individuals age >50 years. The increased risk of multiple myeloma (MM) in relatives of individuals with MGUS is consistent with MGUS being a marker of inherited genetic susceptibility to MM. Common single-nucleotide polymorphisms (SNPs) at 2p23.3 (rs6746082), 3p22.1 (rs1052501), 3q26.2 (rs10936599), 6p21.33 (rs2285803), 7p15.3 (rs4487645), 17p11.2 (rs4273077), and 22q13.1 (rs877529) have recently been shown to influence MM risk. To examine the impact of these 7 SNPs on MGUS, we analyzed two case-control series totaling 492 cases and 7306 controls. Each SNP independently influenced MGUS risk with statistically significant associations (P < .02) for rs1052501, rs2285803, rs4487645, and rs4273077. SNP associations were independent, with risk increasing with a larger number of risk alleles carried (per allele odds ratio, 1.18; P < 10−7). Collectively these data are consistent with a polygenic model of disease susceptibility to MGUS.
Continuing Medical Education online
This activity has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education through the joint sponsorship of Medscape, LLC and the American Society of Hematology.
Medscape, LLC is accredited by the ACCME to provide continuing medical education for physicians.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.0 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/blood; and (4) view/print certificate. For CME questions, see page 2593.
Disclosures
The authors, Associate Editor A. Keith Stewart, and CME questions author Laurie Barclay, freelance writer and reviewer, Medscape, LLC, declare no competing financial interests.
Learning objectives
Upon completion of this activity, participants will be able to:
Discuss the effect of inherited genetic variation on the risk for development of multiple myeloma (MM), based on a report of 2 case-control studies and meta-analysis.
Describe loci identified that increase the risk for development of MGUS.
Distinguish how the observed changes in single-nucleotide polymorphisms in these 2 case-control studies may play a role in the development of MM.
Release date: April 17, 2014; Expiration date: April 17, 2015
Introduction
Monoclonal gammopathy of undetermined significance (MGUS) is a benign premalignant clonal condition through which all cases of multiple myeloma (MM) may pass1 where there is no evidence of end organ damage. MGUS is relatively common and is detectable in the plasma of ∼2% of individuals older than age 50 years, and with each advancing year, the risk of MGUS progressing to MM is ∼1%.2
No lifestyle or environmental exposure factors have been identified that are consistently linked to either an increased risk of MM or MGUS or the transition between the two. An increased risk of MM in the relatives of individuals with MGUS is consistent with MGUS having a heritable basis.3 Moreover, recent genome-wide association studies (GWASs) of MM have identified single-nucleotide polymorphisms (SNPs) at 2p23.3 (rs6746082), 3p22.1 (rs1052501), 3q26.2 (rs10936599), 6p21.33 (rs2285803), 7p15.3 (rs4487645), 17p11.2 (rs4273077), and 22q13.1 (rs877529) that confer a modest but significant increase in MM risk.4,5 To explore whether these 7 SNPs influence MM risk through predisposition to MGUS, we analyzed 2 case-control series totaling 492 cases of MGUS and 7306 controls.
Study design
The study was undertaken with relevant ethical review board approval in accordance with the tenets of the Declaration of Helsinki.
The 252 MGUS cases from the United Kingdom (UK) were ascertained by the Haematological Malignancy Diagnostic Service Laboratory in Leeds, the Royal Marsden Hospital, and University College London Hospital in London (119 males; mean age, 73 years; standard deviation [SD] ± 11). MGUS status was evaluated by a clinical blood test between 2005 and 2012, and cases were excluded if an individual progressed to MM prior to the association analysis in 2013. Genotype frequencies were compared with data from the UK Wellcome Trust Case Control Consortium 26 study of 2698 individuals from the 1958 British Birth Cohort7 and 2501 individuals from the UK Blood Service collections typed by using Illumina Human-1.2M-Duo Custom_v1 arrays.
The 240 German MGUS cases were ascertained through the University Clinic in Heidelberg and the University Clinic in Ulm (115 males; mean age, 62 years; SD ± 11). Of 240 German MGUS patients, 2 were diagnosed with MM within 3 years after sampling, and 46 patients were seen only at time of sampling. Genotypes were compared with data from the Heinz-Nixdorf Recall study8 of 2107 individuals typed by using Illumina Human Omni1-Quad Beadchips (1050 males; mean age, 68 years; SD ± 10).
In both series, the diagnosis of MGUS was based on internationally accepted criteria: monoclonal protein concentration <30 g/L, <10% monoclonal plasma cells in the bone marrow (BM), normal plasma calcium and kidney function, no bone destruction, and no anemia. We excluded immunoglobulin M (IgM) MGUS. For all UK cases, DNA was prepared from venous blood collected in EDTA tubes; for 23% of German cases, DNA was prepared from venous blood collected in EDTA tubes; and for the remainder, the source was the CD138– fraction of BM cells, with <5% contamination by tumor cells. Genotyping of cases in both series was conducted by using Kompetitive Allele Specific Polymerase chain reaction chemistry (KASP, LGC); primer sequences are available on request.
Results and discussion
With the exception of rs10936599 in cases, the frequency of SNP genotypes in both cases and controls satisfied the criterion for Hardy-Weinberg equilibrium. The odds ratio (OR) of MGUS associated with each SNP was calculated by unconditional logistic regression. To increase power, we performed a meta-analysis under a fixed-effects model.9,10
An association between genotype and MGUS risk was seen for each of 7 SNPs, which was statistically significant for rs1052501, rs2285803, rs4487645, and rs4273077 with only the rs4273077 association showing between-study heterogeneity (Table 1). The higher ORs for MM shown by most SNPs in the parent MM GWAS may in part be a consequence of a winner’s curse. On the basis of the lower estimate of the effect size of SNPs for MM risk, we had high power to demonstrate a relationship for rs4487645 but <60% power to show associations for rs6746082 or rs10936599; hence, the failure to demonstrate statistically significant associations for all 7 SNPs may reflect study power.
Association between each of the seven SNPs and risk of MGUS
| Chr . | SNP . | Genotype . | MGUS risk . | Myeloma risk . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UK . | German . | Combined . | . | . | |||||||||||||
| Case . | Control . | OR . | 95% CI . | Case . | Control . | OR . | 95% CI . | OR . | 95% CI . | P . | Phet . | I2 . | OR . | 95% CI . | |||
| 2p23.3 | rs6746082 | ||||||||||||||||
| CC | 15 | 238 | 1.0 (ref) | 9 | 108 | 1.0 (ref) | |||||||||||
| AC | 74 | 1758 | 0.67 | 0.38-1.18 | 83 | 725 | 1.37 | 0.67-2.81 | |||||||||
| AA | 152 | 3202 | 0.75 | 0.44-1.30 | 148 | 1274 | 1.39 | 0.69-2.81 | |||||||||
| Per allele | 0.99 | 0.80-1.24 | 1.08 | 0.86-1.36 | 1.04 | 0.92-1.16 | .63 | .67 | 0 | 1.30 | 1.20-1.41 | ||||||
| RAF | 0.78 | 0.79 | 0.79 | 0.78 | |||||||||||||
| 3p22.1 | rs1052501 | ||||||||||||||||
| AA | 161 | 3668 | 1.0 (ref) | 152 | 1482 | 1.0 (ref) | |||||||||||
| GA | 80 | 1391 | 1.31 | 1.00-1.73 | 83 | 576 | 1.40 | 1.06-1.87 | |||||||||
| GG | 11 | 137 | 1.83 | 0.97-3.45 | 5 | 49 | 0.99 | 0.39-2.53 | |||||||||
| Per allele | 1.33 | 1.06-1.66 | 1.26 | 0.99-1.61 | 1.30 | 1.15-1.46 | .002 | .73 | 0 | 1.32 | 1.22-1.43 | ||||||
| RAF | 0.20 | 0.16 | 0.19 | 0.16 | |||||||||||||
| 3q26.2 | rs10936599 | ||||||||||||||||
| AA | 5 | 325 | 1.0 (ref) | 13 | 142 | 1.0 (ref) | |||||||||||
| AG | 111 | 1914 | 3.76 | 1.53-9.31 | 88 | 778 | 1.24 | 0.67-2.27 | |||||||||
| GG | 133 | 2960 | 2.92 | 1.19-7.18 | 139 | 1187 | 1.28 | 0.71-2.32 | |||||||||
| Per allele | 1.02 | 0.83-1.26 | 1.08 | 0.87-1.35 | 1.05 | 0.94-1.17 | .54 | .71 | 0 | 1.35 | 1.25-1.47 | ||||||
| RAF | 0.76 | 0.75 | 0.76 | 0.75 | |||||||||||||
| 6p21.33 | rs2285803 | ||||||||||||||||
| GG | 117 | 2699 | 1.0 (ref) | 99 | 1047 | 1.0 (ref) | |||||||||||
| AG | 102 | 2055 | 1.15 | 0.88-1.50 | 113 | 833 | 1.43 | 1.08-1.91 | |||||||||
| AA | 24 | 444 | 1.25 | 0.79-1.96 | 28 | 226 | 1.31 | 0.84-2.04 | |||||||||
| Per allele | 1.13 | 0.93-1.37 | 1.24 | 1.01-1.51 | 1.18 | 1.07-1.31 | .019 | .50 | 0 | 1.30 | 1.19-1.43 | ||||||
| RAF | 0.31 | 0.28 | 0.35 | 0.31 | |||||||||||||
| 7p15.3 | rs4487645 | ||||||||||||||||
| AA | 26 | 646 | 1.0 (ref) | 24 | 226 | 1.0 (ref) | |||||||||||
| AC | 93 | 2333 | 0.99 | 0.64-1.54 | 104 | 887 | 1.10 | 0.69-1.76 | |||||||||
| CC | 132 | 2216 | 1.48 | 0.96-2.27 | 117 | 992 | 1.11 | 0.70-1.76 | |||||||||
| Per allele | 1.32 | 1.08-1.60 | 1.11 | 0.90-1.36 | 1.21 | 1.10-1.34 | .007 | .24 | 28 | 1.24 | 1.16-1.32 | ||||||
| RAF | 0.71 | 0.65 | 0.70 | 0.68 | |||||||||||||
| 17p11.2 | rs4273077 | ||||||||||||||||
| AA | 197 | 4221 | 1.0 (ref) | 163 | 1690 | 1.0 (ref) | |||||||||||
| AG | 47 | 926 | 1.09 | 0.78-1.51 | 64 | 390 | 1.70 | 1.25-2.32 | |||||||||
| GG | 8 | 48 | 3.57 | 1.67-7.65 | 11 | 27 | 4.22 | 2.06-8.67 | |||||||||
| Per allele | 1.31 | 1.00-1.72 | 1.87 | 1.45-2.41 | 1.58 | 1.38-1.80 | 1.64 × 10−6 | .03 | 80 | 1.28 | 1.18-1.39 | ||||||
| RAF | 0.13 | 0.10 | 0.18 | 0.11 | |||||||||||||
| 22q13.1 | rs877529 | ||||||||||||||||
| GG | 73 | 1633 | 1.0 (ref) | 72 | 692 | 1.0 (ref) | |||||||||||
| AG | 134 | 2560 | 1.17 | 0.88-1.57 | 117 | 1026 | 1.09 | 0.80-1.49 | |||||||||
| AA | 43 | 1000 | 0.96 | 0.65-1.41 | 51 | 389 | 1.26 | 0.86-1.84 | |||||||||
| Per allele | 1.00 | 0.84-1.20 | 1.12 | 0.93-1.36 | 1.06 | 0.97-1.16 | .40 | .41 | 0 | 1.22 | 1.14-1.31 | ||||||
| RAF | 0.44 | 0.44 | 0.46 | 0.43 | |||||||||||||
| Chr . | SNP . | Genotype . | MGUS risk . | Myeloma risk . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UK . | German . | Combined . | . | . | |||||||||||||
| Case . | Control . | OR . | 95% CI . | Case . | Control . | OR . | 95% CI . | OR . | 95% CI . | P . | Phet . | I2 . | OR . | 95% CI . | |||
| 2p23.3 | rs6746082 | ||||||||||||||||
| CC | 15 | 238 | 1.0 (ref) | 9 | 108 | 1.0 (ref) | |||||||||||
| AC | 74 | 1758 | 0.67 | 0.38-1.18 | 83 | 725 | 1.37 | 0.67-2.81 | |||||||||
| AA | 152 | 3202 | 0.75 | 0.44-1.30 | 148 | 1274 | 1.39 | 0.69-2.81 | |||||||||
| Per allele | 0.99 | 0.80-1.24 | 1.08 | 0.86-1.36 | 1.04 | 0.92-1.16 | .63 | .67 | 0 | 1.30 | 1.20-1.41 | ||||||
| RAF | 0.78 | 0.79 | 0.79 | 0.78 | |||||||||||||
| 3p22.1 | rs1052501 | ||||||||||||||||
| AA | 161 | 3668 | 1.0 (ref) | 152 | 1482 | 1.0 (ref) | |||||||||||
| GA | 80 | 1391 | 1.31 | 1.00-1.73 | 83 | 576 | 1.40 | 1.06-1.87 | |||||||||
| GG | 11 | 137 | 1.83 | 0.97-3.45 | 5 | 49 | 0.99 | 0.39-2.53 | |||||||||
| Per allele | 1.33 | 1.06-1.66 | 1.26 | 0.99-1.61 | 1.30 | 1.15-1.46 | .002 | .73 | 0 | 1.32 | 1.22-1.43 | ||||||
| RAF | 0.20 | 0.16 | 0.19 | 0.16 | |||||||||||||
| 3q26.2 | rs10936599 | ||||||||||||||||
| AA | 5 | 325 | 1.0 (ref) | 13 | 142 | 1.0 (ref) | |||||||||||
| AG | 111 | 1914 | 3.76 | 1.53-9.31 | 88 | 778 | 1.24 | 0.67-2.27 | |||||||||
| GG | 133 | 2960 | 2.92 | 1.19-7.18 | 139 | 1187 | 1.28 | 0.71-2.32 | |||||||||
| Per allele | 1.02 | 0.83-1.26 | 1.08 | 0.87-1.35 | 1.05 | 0.94-1.17 | .54 | .71 | 0 | 1.35 | 1.25-1.47 | ||||||
| RAF | 0.76 | 0.75 | 0.76 | 0.75 | |||||||||||||
| 6p21.33 | rs2285803 | ||||||||||||||||
| GG | 117 | 2699 | 1.0 (ref) | 99 | 1047 | 1.0 (ref) | |||||||||||
| AG | 102 | 2055 | 1.15 | 0.88-1.50 | 113 | 833 | 1.43 | 1.08-1.91 | |||||||||
| AA | 24 | 444 | 1.25 | 0.79-1.96 | 28 | 226 | 1.31 | 0.84-2.04 | |||||||||
| Per allele | 1.13 | 0.93-1.37 | 1.24 | 1.01-1.51 | 1.18 | 1.07-1.31 | .019 | .50 | 0 | 1.30 | 1.19-1.43 | ||||||
| RAF | 0.31 | 0.28 | 0.35 | 0.31 | |||||||||||||
| 7p15.3 | rs4487645 | ||||||||||||||||
| AA | 26 | 646 | 1.0 (ref) | 24 | 226 | 1.0 (ref) | |||||||||||
| AC | 93 | 2333 | 0.99 | 0.64-1.54 | 104 | 887 | 1.10 | 0.69-1.76 | |||||||||
| CC | 132 | 2216 | 1.48 | 0.96-2.27 | 117 | 992 | 1.11 | 0.70-1.76 | |||||||||
| Per allele | 1.32 | 1.08-1.60 | 1.11 | 0.90-1.36 | 1.21 | 1.10-1.34 | .007 | .24 | 28 | 1.24 | 1.16-1.32 | ||||||
| RAF | 0.71 | 0.65 | 0.70 | 0.68 | |||||||||||||
| 17p11.2 | rs4273077 | ||||||||||||||||
| AA | 197 | 4221 | 1.0 (ref) | 163 | 1690 | 1.0 (ref) | |||||||||||
| AG | 47 | 926 | 1.09 | 0.78-1.51 | 64 | 390 | 1.70 | 1.25-2.32 | |||||||||
| GG | 8 | 48 | 3.57 | 1.67-7.65 | 11 | 27 | 4.22 | 2.06-8.67 | |||||||||
| Per allele | 1.31 | 1.00-1.72 | 1.87 | 1.45-2.41 | 1.58 | 1.38-1.80 | 1.64 × 10−6 | .03 | 80 | 1.28 | 1.18-1.39 | ||||||
| RAF | 0.13 | 0.10 | 0.18 | 0.11 | |||||||||||||
| 22q13.1 | rs877529 | ||||||||||||||||
| GG | 73 | 1633 | 1.0 (ref) | 72 | 692 | 1.0 (ref) | |||||||||||
| AG | 134 | 2560 | 1.17 | 0.88-1.57 | 117 | 1026 | 1.09 | 0.80-1.49 | |||||||||
| AA | 43 | 1000 | 0.96 | 0.65-1.41 | 51 | 389 | 1.26 | 0.86-1.84 | |||||||||
| Per allele | 1.00 | 0.84-1.20 | 1.12 | 0.93-1.36 | 1.06 | 0.97-1.16 | .40 | .41 | 0 | 1.22 | 1.14-1.31 | ||||||
| RAF | 0.44 | 0.44 | 0.46 | 0.43 | |||||||||||||
Relationship between each SNP and risk of multiple myeloma (MM) is shown by using data on 4692 MM cases from the previously published GWAS.
Chr, chromosome; I2, variation attributable to heterogeneity; Phet, heterogeneity P value; RAF, risk allele frequency; ref, reference; UK, United Kingdom.
To the best of our knowledge, only one previous study based on an analysis of rs6746082, rs1052501, and rs4487645 genotypes in 388 cases and 378 controls has examined the association between MM risk variants and MGUS.11 Importantly, in that study, 14% of the MGUS cases had an IgM paraprotein, and study power was <55%. Despite this, an association between rs1052501 and risk was seen (P = .04). Pooling data from this published study with the two series reported here provided enhanced support for the associations with rs6746082, rs1052501, and rs4487645 with risk of MGUS; ORs in the combined analysis were 1.09 (95% confidence interval [CI], 0.95 to 1.24; P = .23; P for heterogeneity [Phet] = .23), 1.30 (95% CI, 1.14 to 1.50; P = 1.72 × 10−4; Phet = .94), and 1.18 (95% CI, 1.05 to 1.34; P = .006; Phet = .39), respectively.
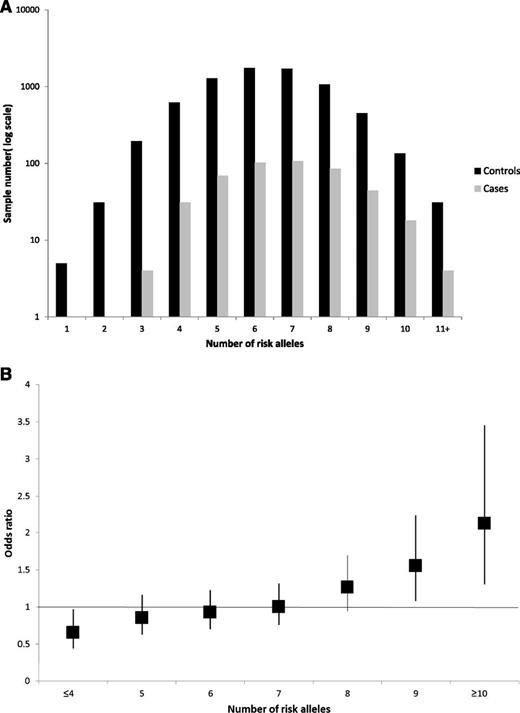
We investigated the combined effect of pairs of loci by logistic regression modeling with evidence for interactive effects between SNPs being assessed by a likelihood ratio test. This analysis provided no evidence for interactive effects between any of the loci (ie, P > .05 after adjustment for multiple testing; supplemental Table 1, available on the Blood Web site), consistent with each locus having an independent role in MGUS risk. By using data from both series, the risk of MGUS was seen to increase with an increasing numbers of risk alleles for the 7 loci carried, counting two for a homozygote and one for a heterozygote, assuming equal weights (OR per allele, 1.18; 95% CI, 1.11 to 1.25; Ptrend < 10−7). The proportion of cases and controls grouped according to the number of risk alleles carried is detailed in Figure 1A, which shows a shift toward a higher number of risk alleles in the cases. For those with 10 or more risk alleles, there is a greater than twofold increase in risk compared with those with the median number of risk alleles (Figure 1B). These ORs may be underestimates because the model assumes equal weighting across SNPs. Our data provide only estimates of the effect on susceptibility attributable to the variations since some of the controls in both series may have MGUS, albeit small numbers, which would negatively bias ORs.
(A) Proportion of cases and controls grouped according to the number of risk alleles carried, (B) Plot of the increasing ORs for MGUS with increasing number of risk alleles. (A) Distribution of risk alleles in controls (black) and MGUS cases (gray) for the seven loci rs6746082, rs1052501, rs10936599, rs2285803, rs4487645, rs4273077, and rs877529. (B) The ORs are relative to the median number of 7 risk alleles; vertical bars correspond to 95% confidence intervals. The distribution of risk alleles follows a normal distribution in both case and controls, with a shift toward a higher number of risk alleles in cases. Horizontal line denotes the null value (OR, 1.0).
(A) Proportion of cases and controls grouped according to the number of risk alleles carried, (B) Plot of the increasing ORs for MGUS with increasing number of risk alleles. (A) Distribution of risk alleles in controls (black) and MGUS cases (gray) for the seven loci rs6746082, rs1052501, rs10936599, rs2285803, rs4487645, rs4273077, and rs877529. (B) The ORs are relative to the median number of 7 risk alleles; vertical bars correspond to 95% confidence intervals. The distribution of risk alleles follows a normal distribution in both case and controls, with a shift toward a higher number of risk alleles in cases. Horizontal line denotes the null value (OR, 1.0).
Since we are not able to evaluate the association with progression directly, we examined whether there was a significant difference in the strength of the association between MGUS and MM. To perform this analysis, we made use of previously published data from our MM GWAS. Because subgroups were not mutually exclusive and common control series were used, we performed an analysis by using the R statistical package ASSET (association analysis based on subsets).12 In this analysis, there was no statistically significant difference (ie, P > .05; supplemental Figure 1) between the effect seen in MGUS and MM risk consistent with variation at these loci influencing the risk of MM through the development of clonal plasma cells typical of MGUS. However, we acknowledge that our power to detect a difference between MGUS and MM is limited.
The only SNP resulting in an amino acid change was rs1052501, responsible for A542T in ULK4, a key regulator of mammalian target of rapamycin (mTOR)–mediated autophagy. Many of the other SNPs are intronic to genes which have strong biologic plausibility for a role in MM development, notably rs4273077, which maps to the gene encoding TNFRSF13B which is important for the regulation of normal B-cell homeostasis and a determinant of circulating IgG levels.13 rs877529 is intronic to the gene CBX7, which acts through Ink4a/Arf cooperating with Myc to promote aggressive B-cell lymphomagenesis.14 Other risk SNPs examined here are not necessarily strong candidates for being directly causal but are likely to be correlated with functional variants.
In conclusion, these findings provide further insights into inherited susceptibility to MGUS and its relationship to MM.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to all study participants and investigators.
This work was supported in the United Kingdom (UK) by Myeloma UK, Leukaemia Lymphoma Research and Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund), the National Institute for Health Research Biomedical Research Centre at the Royal Marsden National Health Service Hospitals Trust, by the Heinz-Nixdorf Foundation (Heinz Nixdorf Recall [HNR] study; genotyping of HNR patients supported by the German Centre for Neurodegenerative Disorders in Bonn). UK control genotypes were generated by the Wellcome Trust Sanger Institute. Supported in Germany by Dietmar-Hopp-Stiftung in Walldorf, the German Ministry of Education and Science (Gliomics 01ZX1309B), German Cancer Aid, the University Hospital Heidelberg, the German Ministry of Education and Science, and the German Research Council (Projects SI 236/8-1, SI236/9-1, and ER 155/6-1).
Authorship
Contribution: C.D., D.B.B., and N.B.D. conducted research; A.C.R., B.A.W., and D.C.J. collected clinical data; A.C.R., F.E.D., G.J.M., and K.Y. collected samples; P.B. and J.V. performed genotyping; R.S.H. and F.J.H. performed statistical analysis; N.W., E.D., and A.F. performed genotyping; H.G. and D.H. collected clinical data; D.H. and N.W. managed Heidelberg MGUS samples; C.L. ascertained and managed Ulm MGUS samples; L.E. and K.-H.J. performed ascertainment and management of Heinz Nixdorf Recall study samples; P.H., T.W.M., and M.M.N. performed and coordinated GWASs of German controls; and R.S.H., G.J.M., K.H., and H.G. conceived the study and drafted the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard S. Houlston, 15 Cotswold Rd, Sutton-on-Surrey SM2 5NG, United Kingdom; e-mail: richard.houlston@icr.ac.uk.
References
Author notes
N.W. and D.C.J. contributed equally to this work.
H.G., G.J.M., K.H., and R.S.H. contributed equally to this work as principal investigators.